ABSTRACT
HIV-1 latency is characterized by reversible silencing of viral transcription driven by the long terminal repeat (LTR) promoter of HIV-1. Cellular and viral factors regulating LTR activity contribute to HIV-1 latency, and certain repressive cellular factors modulate viral transcription silencing. Nef-associated factor 1 (Naf1) is a host nucleocytoplasmic shuttling protein that regulates multiple cellular signaling pathways and HIV-1 production. We recently reported that nuclear Naf1 promoted nuclear export of unspliced HIV-1 gag mRNA, leading to increased Gag production. Here we demonstrate new functions of Naf1 in regulating HIV-1 persistence. We found that Naf1 contributes to the maintenance of HIV-1 latency by inhibiting LTR-driven HIV-1 gene transcription in a nuclear factor kappa B-dependent manner. Interestingly, Naf1 knockdown significantly enhanced viral reactivation in both latently HIV-1-infected Jurkat T cells and primary central memory CD4+ T cells. Furthermore, Naf1 knockdown in resting CD4+ T cells from HIV-1-infected individuals treated with antiretroviral therapy significantly increased viral reactivation upon T-cell activation, suggesting an important role of Naf1 in modulating HIV-1 latency in vivo. Our findings provide new insights for a better understanding of HIV-1 latency and suggest that inhibition of Naf1 activity to activate latently HIV-1-infected cells may be a potential therapeutic strategy.
IMPORTANCE HIV-1 latency is characterized mainly by a reversible silencing of LTR promoter-driven transcription of an integrated provirus. Cellular and viral proteins regulating LTR activity contribute to the modulation of HIV-1 latency. In this study, we found that the host protein Naf1 inhibited HIV-1 LTR-driven transcription of HIV genes and contributed to the maintenance of HIV-1 latency. Our findings provide new insights into the effects of host modulation on HIV-1 latency, which may lead to a potential therapeutic strategy for HIV persistence by targeting the Naf1 protein.
KEYWORDS: HIV-1 latency, HIV-1 Nef-associated factor 1, NF-κB, transcription
INTRODUCTION
Persistence of HIV-1 infection due to viral latency is the major barrier to the eradication of AIDS (1–5). Although combination antiretroviral therapy effectively reduces HIV-1 replication and results in a significant viral load reduction, to a level below the limit of detection for most infected individuals, a reservoir of infected cells still persists (1, 2). Transcriptional inefficiency of HIV-1 gene expression is an important mechanism of viral latency (2, 6, 7). HIV-1 gene transcription and viral reactivation from latency require the activity of nuclear factor kappa B (NF-κB), whose modulation by cellular factors contributes to the establishment and maintenance of HIV-1 latency (7–14).
Nef-associated factor 1 (Naf1), also known as ABIN-1 (A20-binding inhibitor of NF-κB activation 1) and TNFAIP3 (tumor necrosis factor alpha [TNF-α]-induced protein 3)-interacting protein 1 (15), was discovered by yeast two-hybrid screening and pulldown assays using HIV-1 Nef as bait (16). Naf1 has two isoforms, Naf1-α and Naf1-β, and Naf1-α is the canonical isoform (16). Naf1 is expressed ubiquitously in human tissues, with high levels of expression in peripheral blood lymphocytes (16, 17). Naf1 can increase cell surface CD4 expression (16), inhibit NF-κB activation (15, 18, 19), and modulate HIV-1 infection and viral production (17, 20). Naf1 also interacts with the HIV-1 matrix protein, and the overexpression of Naf1 inhibits HIV-1 infection (17). In association with A20, Naf1 facilitates A20-mediated deubiquitination of NF-κB essential modifier/IκB kinase γ and NF-κB inhibition in response to TNF-α stimulation (21). However, the potential role of Naf1 in regulating HIV-1 reactivation from latency has not been reported.
Here we report that Naf1 maintains HIV-1 latency by suppressing viral gene expression in an NF-κB-dependent manner. Naf1 knockdown in primary CD4+ T cells significantly increased HIV-1 replication and enhanced viral reactivation in primary CD4+ central memory T cells (TCM) latently infected with HIV-1. Furthermore, Naf1 knockdown in resting CD4+ T cells from HIV-1-infected individuals on antiretroviral therapy significantly increased viral reactivation upon CD4+ T-cell activation. Overall, our findings suggest an important role for Naf1 in modulating HIV-1 latency in vivo.
RESULTS
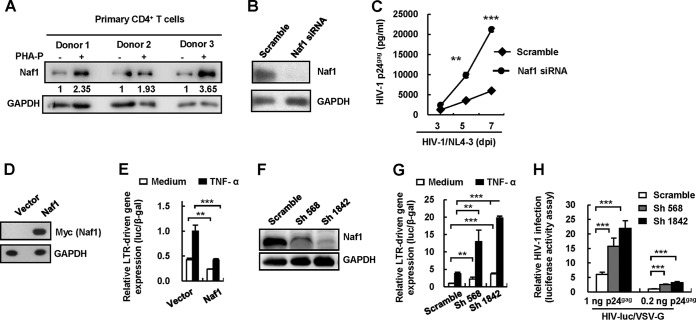
Naf1 suppresses HIV-1 long terminal repeat (LTR)-driven gene expression and inhibits HIV-1 replication.
Naf1 has been shown to be a nucleocytoplasmic shuttling protein (17, 20). We recently reported that nuclear Naf1 promoted nuclear export of unspliced HIV-1 gag mRNA, leading to increased Gag production (in that study, a Gag-expressing plasmid was used) (20). To further determine the overall effect of endogenous Naf1 on HIV-1 replication in this study, the replication-competent HIV-1NL4-3 strain was used to infect primary CD4+ T cells (Fig. 1A to C). We first detected endogenous Naf1 expression in resting CD4+ T cells and found that T-cell activation enhanced Naf1 expression (Fig. 1A). We used a specific small interfering RNA (siRNA) to achieve significant knockdown of endogenous Naf1 in activated primary CD4+ T cells (Fig. 1B), and we observed that Naf1 knockdown increased HIV-1NL4-3 replication in primary CD4+ T cells (Fig. 1C). These results suggest that endogenous Naf1 suppresses HIV-1 replication in primary CD4+ T cells.
FIG 1.
Naf1 suppresses HIV-1 LTR-driven gene expression and viral replication. (A) Endogenous expression of Naf1 in primary CD4+ T cells as detected by immunoblotting. The densities of bands were analyzed with the plug-ins of ImageJ software, and the values relative to that for GAPDH were calculated. (B and C) Naf1 knockdown increases HIV-1 replication. Phytohemagglutinin P (PHA-P)-activated primary CD4+ T cells were transfected with Naf1-specific siRNA or an off-target control, and then cells were infected with replication-competent HIV-1NL4-3. The levels of HIV-1 p24gag in the supernatants were quantified by ELISA. Results are representative of three independent repeats. dpi, days postinfection. (D and E) Naf1 overexpression inhibits HIV-1 LTR-driven gene expression. The myc-tagged plasmid pCMV-Tag 3B/Naf1 or vector and an HIV-1NL4-3 LTR promoter-driven luciferase reporter plasmid were cotransfected into HEK293T cells, and a β-Gal-expressing vector was used to normalize transfection efficiency. At 24 h posttransfection, cells were treated with or without TNF-α for an additional 24 h, and then cells were harvested and reporter gene expression assessed. Results are representative of five independent repeats. (F and G) Naf1 knockdown significantly increases TNF-α-induced LTR-driven gene expression. The endogenous Naf1 in HEK293T cells was knocked down by use of Naf1-specific shRNA. Cells were transfected with an HIV-1NL4-3 LTR promoter-driven luciferase reporter plasmid, and reporter gene expression was detected as described above. (H) Naf1 knockdown promotes HIV-1 infection. The endogenous Naf1 in HEK293T cells was knocked down by use of Naf1-specific shRNA, cells (1 × 105) were infected with pseudotyped HIV-luc/VSV-G for 24 h (using amounts of virus equivalent to 0.2 or 1 ng p24gag), and viral infections were quantified by detection of luciferase activity. Results in panels G and H are representative of four independent experiments. Data are presented as means ± standard deviations (SD). **, P < 0.01; ***, P < 0.001 (unpaired t test).
The HIV-1 LTR promoter plays an essential role in driving viral transcription and productive infection (22–24). To determine the mechanism of Naf1 inhibition of HIV-1 replication, we investigated whether Naf1 could inhibit LTR activity. We performed a cotransfection assay in HEK293T cells by using a luciferase reporter driven by the full-length LTR promoter from HIV-1NL4-3. We treated the transfected cells with or without TNF-α and then examined the effect of Naf1 on LTR-driven transcription. Treatment with TNF-α can enhance LTR activity (25). We observed that the overexpression of Naf1 (Fig. 1D) significantly inhibited LTR-driven basal gene expression (2.0-fold; P < 0.01) and that TNF-α stimulated gene expression (2.5-fold; P < 0.001) (Fig. 1E). To examine whether endogenous cellular Naf1 could inhibit LTR-driven transcription, we knocked down endogenous Naf1 expression in HEK293T cells by using specific short hairpin RNAs (shRNAs) (Fig. 1F), and we found that Naf1 knockdown increased LTR-driven basal gene expression (2.3- to 3.8-fold; P < 0.01), as well as TNF-α-induced LTR-driven gene expression (3.4- to 5.3-fold; P < 0.001), compared to that in control cells (Fig. 1G). Furthermore, when these Naf1 knockdown cells were infected with pseudotyped HIV-luc/VSV-G for 24 h (using amounts of virus equivalent to 0.2 or 1 ng p24gag), significantly increased HIV-1 infection (2.5- to 3.6-fold; P < 0.001) was also observed upon Naf1 knockdown (Fig. 1H). Together, these data suggest that Naf1 suppresses HIV-1 LTR-driven gene expression and inhibits HIV-1 replication.
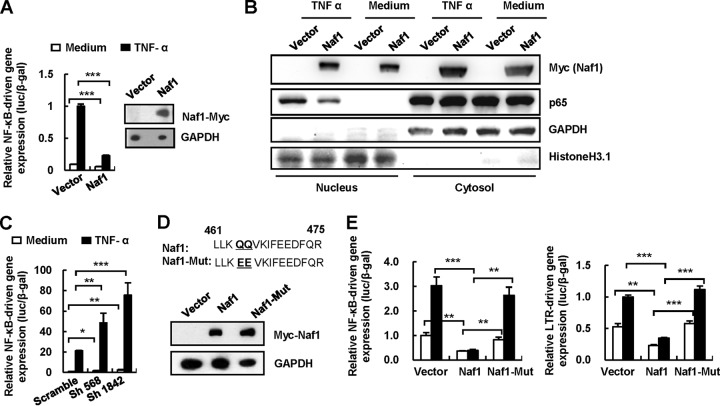
Naf1 suppresses HIV-1 LTR-driven gene expression by inhibiting NF-κB activation.
The HIV-1 LTR promoter often contains two adjacent NF-κB binding sites that are crucial for initiating viral transcription (26). NF-κB activity is required for efficient cellular and HIV-1 gene transcription, and Naf1 can inhibit NF-κB activation (15, 18, 19, 27). We hypothesized that Naf1 suppresses NF-κB-dependent HIV-1 LTR-driven gene expression. To investigate this, we first confirmed the Naf1-mediated inhibition of NF-κB activity. We coexpressed myc-tagged full-length Naf1 and an NF-κB reporter in HEK293T cells (Fig. 2A) and normalized the transfection efficiency by using a β-galactosidase (β-Gal)-expressing vector. We then treated the cells with TNF-α to activate NF-κB. In the absence of TNF-α stimulation, transient expression of Naf1 reduced NF-κB reporter gene expression 2-fold, while Naf1 overexpression suppressed TNF-α-induced NF-κB activation 5-fold (P < 0.001) (Fig. 2A). We further demonstrated that TNF-α stimulation increased the nuclear import of the NF-κB p65 subunit in cells, while Naf1 overexpression counteracted the effect (Fig. 2B). When the endogenous cellular Naf1 in HEK293T cells was knocked down using specific shRNAs, the basal level of NF-κB activation increased (1.6- to 2.4-fold; P < 0.05) compared to that in control cells (Fig. 2C). More significant effects on NF-κB activation were obtained in the TNF-α-treated cells (2.3- to 3.6-fold; P < 0.001). These results confirm that Naf1 controls the basal level of NF-κB activity and, further, inhibits TNF-α-stimulated NF-κB activation.
FIG 2.
Naf1 suppresses NF-κB-dependent HIV-1 LTR-driven gene expression. (A) Naf1 overexpression inhibits NF-κB activation. The myc-tagged plasmid pCMV-Tag 3B/Naf1 or vector and an NF-κB reporter plasmid were cotransfected into HEK293T cells, and a β-Gal-expressing vector was used to normalize the transfection efficiency. At 24 h posttransfection, cells were treated with or without TNF-α for an additional 24 h, and then cells were harvested and reporter gene expression assessed. (B) Naf1 blocks NF-κB p65 nuclear import. HEK293T cells were transfected with myc-tagged pCMV-Tag 3B/Naf1 or vector for 24 h, and then cells were stimulated with or without TNF-α (20 ng/ml) for 30 min. The cytoplasmic and nuclear protein extracts were then isolated, and NF-κB p65 was detected by immunoblotting. The histone H3.1 and GAPDH proteins were used to validate the nuclear and cytoplasmic extracts, respectively, and one blot representative of three repeats is shown. (C) Naf1 knockdown significantly increases TNF-α-induced NF-κB activation. The endogenous Naf1 in HEK293T cells was knocked down by use of Naf1-specific shRNA. Cells were transfected with an NF-κB reporter plasmid, and reporter gene expression was detected as described above. (D and E) Inhibition of NF-κB activation and HIV-1 LTR-promoted gene expression by a Naf1 mutant. A Naf1 mutant was constructed and its expression confirmed by immunoblotting (D), and the inhibition of NF-κB activation and HIV-1 LTR-promoted gene expression by mutant (Mut) and wild-type Naf1 was detected as described above (E). Data are presented as means ± SD. Results in panels A, C, and E are representative of at least four independent experiments. *, P < 0.05; **, P < 0.01; ***, P < 0.001 (unpaired t test).
To investigate whether Naf1-suppressed HIV-1 LTR transcription is NF-κB dependent, we constructed a Naf1 mutant. It has been shown that the short ABINs homology domain 2 (AHD2) of Naf1 is highly conserved and is necessary for the inhibition of NF-κB activation (28). We generated a Naf1 mutant that is unable to inhibit NF-κB (28) by changing two conserved glutamines (QQ) in AHD2, at residues 464 and 465, to glutamic acids (EE), and we performed a cotransfection assay using HEK293T cells (Fig. 2D). Unlike wild-type (WT) Naf1, the Naf1 mutant neither inhibited the activation of NF-κB nor suppressed HIV-1 LTR-driven gene expression (Fig. 2E). These data suggest that Naf1-mediated suppression of HIV-1 LTR gene transcription is dependent on the inhibition of NF-κB activation.
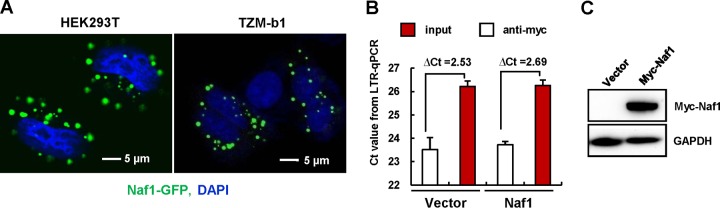
Naf1 has been reported to be a shuttling protein that moves between the cytoplasm and the nucleus (17, 20). The cytoplasmic and nuclear localization of Naf1 was confirmed by confocal microscopy (Fig. 3A). To investigate whether Naf1 represses the HIV-1 LTR through binding to its promoter sequence, a cross-linked chromatin immunoprecipitation (ChIP) assay was performed on HeLa cell-derived TZM-bl cells, which contain an integrated HIV-1 LTR sequence for driving reporter gene expression (29). No obvious enrichment of the LTR fragments was found in the immunoprecipitation experiment (Fig. 3B and C), indicating no direct binding between Naf1 and the HIV-1 LTR sequence. Overall, these data suggest that Naf1 suppresses NF-κB-dependent HIV-1 LTR-driven gene expression.
FIG 3.
Detection of Naf1 binding of the HIV-1 LTR promoter by ChIP assay. (A) HEK293T or TZM-bl cells were transfected with the pEGFP-Naf1 expression plasmid for 48 h, and cells were observed by confocal microscopy. Nuclei were stained with DAPI (blue). (B and C) TZM-bl cells (5 × 106) were transfected with pCMV-Tag3B/Naf1 or vector for 48 h, a cross-linked ChIP assay was performed to detect the binding of Naf1 to the HIV-1 LTR promoter (B), and Naf1 expression was detected by immunoblotting (C). Results are representative of three independent repeats.
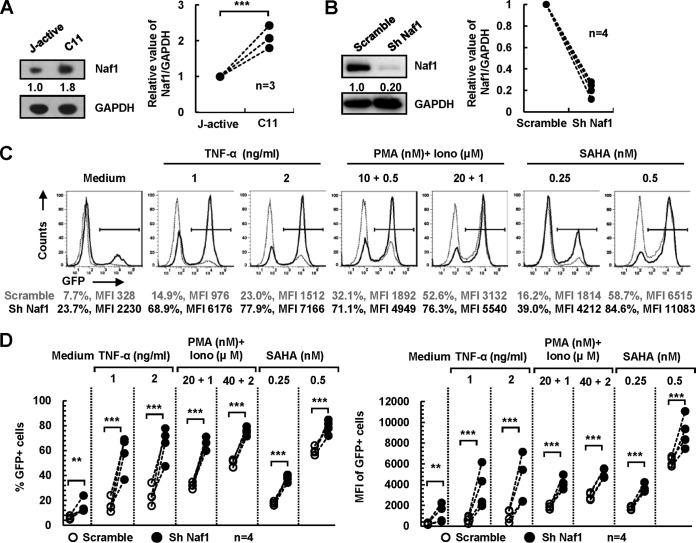
Naf1 knockdown increases HIV-1 reactivation from an integrated HIV-1 proviral DNA.
HIV-1 latency is characterized mainly by a reversible silencing of LTR-driven transcription of an integrated provirus (2, 6, 7). The inhibitory effect of Naf1 on HIV-1 LTR-driven gene expression suggests a potential role for Naf1 in HIV-1 latency. Thus, we sought to investigate the effect of endogenous Naf1 on the expression of an integrated HIV-1 proviral DNA in CD4+ T cells by using a latently HIV-1-infected Jurkat T-cell clone (C11) which harbors an HIV-1 proviral DNA encoding green fluorescent protein (GFP) (30). Upon TNF-α stimulation, C11 cells express GFP, which serves as an indication of reactivation of HIV-1 from latency (30, 31). We observed that C11 cells expressed a higher level of Naf1 protein than that in an actively HIV-1-infected Jurkat T-cell clone (Fig. 4A). To evaluate the effect of Naf1 on HIV-1 reactivation from latency, we knocked down endogenous Naf1 in C11 cells by using specific shRNAs (Fig. 4B), and we assessed HIV-1 reactivation by measuring GFP expression upon stimulation with or without different latency-reversing agents. In the absence of stimulation, Naf1 knockdown increased HIV-1 reactivation 3-fold (P < 0.05) (Fig. 4C and D), while in the presence of TNF-α stimulation, Naf1 knockdown significantly enhanced HIV-1 reactivation (3.4- to 4.6-fold) compared to that in scramble shRNA cells (P < 0.05) (Fig. 4C and D). Furthermore, Naf1 knockdown also significantly increased HIV-1 reactivation upon stimulation with phorbol-12-myristate-13-acetate (PMA) and ionomycin or SAHA (Fig. 4C and D). These results suggest that endogenous Naf1-mediated suppression of HIV-1 gene expression from proviral DNA contributes to viral latency in CD4+ T cells.
FIG 4.
Naf1 knockdown increases HIV-1 reactivation from an integrated HIV-1 proviral DNA. (A) Naf1 expression in latently (C11) or actively (J-active) HIV-1-infected Jurkat T cells. The densities of bands were analyzed with the plug-ins of ImageJ software, the values relative to the GAPDH level were calculated, and the results from 3 independent repeats were summarized and analyzed (right panel). (B to D) Naf1 knockdown promotes HIV-1 reactivation. (B) C11 cells were infected with lentiviruses containing Naf1 shRNA or a scramble control for 72 h, and at 24 h postinfection, puromycin was added to the medium for selection. The calculated data from Western blots showing Naf1 knockdown for 4 independent repeats were summarized and analyzed (right panel). (C and D) Cells were stimulated with or without TNF-α, PMA/ionomycin, or suberoylanilide hydroxyamic acid (SAHA) for an additional 24 h, HIV-1 reactivation was measured by detecting GFP expression, and the percentage of GFP+ cells and the mean fluorescence intensity (MFI) were calculated. Data from 4 independent repeats were summarized and analyzed.
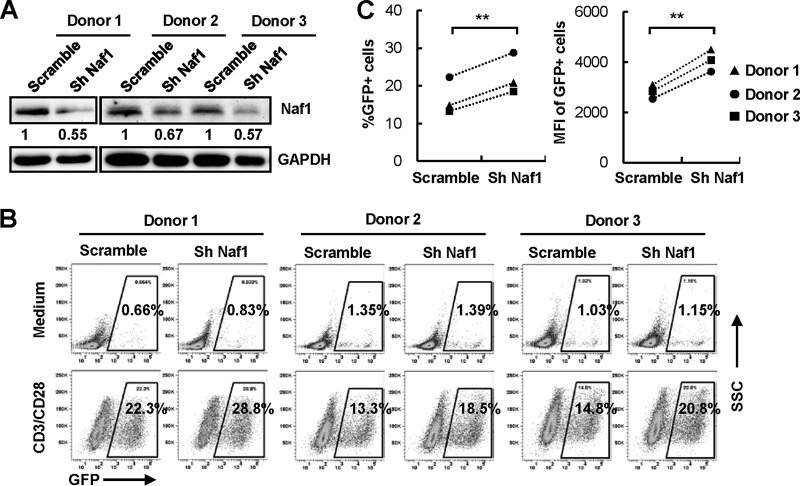
Naf1 knockdown promotes viral reactivation from latently HIV-1-infected CD4+ TCM.
Previous analyses using patient cells revealed that the HIV-1 reservoir resides mainly in resting CD4+ memory T cells (32–34), and central memory T cells (TCM) are thought to be one of the most important reservoirs (35, 36). To examine the role of Naf1 in regulating HIV-1 latency in CD4+ TCM, we used an established method to generate a large number of latently HIV-1-infected human primary CD4+ T cells (37), which are an important tool for the investigation of many aspects of HIV-1 latency, viral reactivation mechanisms, and the signaling pathways involved (38). We knocked down Naf1 expression in latently HIV-1-infected TCM derived from three donors by using a specific shRNA (Fig. 5A) and then activated latent GFP-reporter HIV-1 in these cells by use of anti-CD3/CD28 antibodies. Compared to control cells, we found that Naf1 knockdown increased HIV-1 reactivation 29 to 40% upon T-cell activation (Fig. 5B and C), although no obvious enhancements were observed for unstimulated cells (medium control). These results suggest that Naf1 negatively regulates HIV-1 gene transcription to maintain viral latency in latently HIV-1-infected CD4+ central memory T cells.
FIG 5.
Naf1 knockdown promotes viral reactivation from latently HIV-1-infected TCM. Latently HIV-eGFP/VSV-G-infected TCM were infected with lentiviruses containing Naf1-specific shRNA or a scramble control for 72 h (A) and then were stimulated with anti-CD3/CD28 antibodies for an additional 72 h, viral reactivation was measured by detecting GFP expression (B), and the percentage of GFP+ cells and the MFI were calculated (C). For panels B and C, data from 4 independent repeats were summarized and analyzed. **, P < 0.01 (paired t test). For panel A, the densities of bands were analyzed with the plug-ins of ImageJ software, and the values relative to that for GAPDH were calculated.
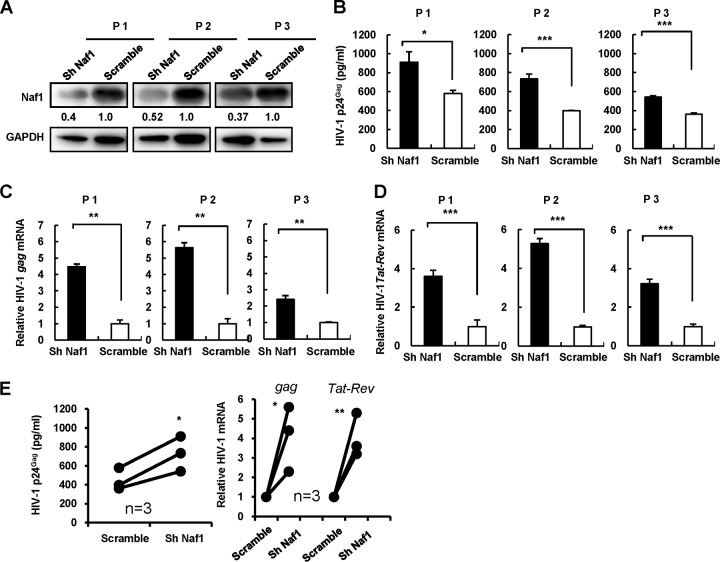
Naf1 knockdown increases viral reactivation in primary resting CD4+ T cells from HIV-1-infected subjects.
To confirm the role of Naf1 in maintaining HIV-1 latency in vivo, we examined the effect of Naf1 knockdown on HIV-1 activation in resting CD4+ T cells from HIV-1-infected individuals. Resting CD4+ T cells are the major source of HIV-1 latency, and their activation leads to HIV-1 reactivation from viral latency (39). These patients underwent combination antiretroviral therapy for more than 3 years and had undetectable plasma viral loads in a standard real-time PCR (RT-PCR)-based assay (Roche). Silencing of endogenous Naf1 expression in resting CD4+ T cells from three HIV-1-infected individuals by use of a specific shRNA resulted in 48 to 63% decreases of Naf1 expression (Fig. 6A) and significantly increased HIV-1 reactivation upon T-cell activation by phytohemagglutinin-P (PHA-P), as monitored by detecting HIV-1 particles released into the supernatants (Fig. 6B) or quantifying the level of HIV-1 gag or tat-rev mRNA (Fig. 6C and D). These results further suggest that Naf1 contributed to HIV-1 latency in resting CD4+ T cells from infected individuals treated with antiretroviral drugs.
FIG 6.
Naf1 knockdown increases HIV-1 reactivation in primary resting CD4+ T cells from patients. Resting CD4+ T cells were isolated from antiretroviral-treated patients and coinfected with lentiviruses containing Naf1-specific shRNA or a scramble control and lentiviruses containing Vpx for 3 days. The patient numbers of three patients (P1 to P3) are noted. (A) Naf1 expression levels were detected by immunoblotting, and GAPDH was used as a loading control. The densities of bands were analyzed with the plug-ins of ImageJ software, and the values relative to that for GAPDH were calculated. HIV-1 reactivation in CD4+ T cells upon PHA-P stimulation was detected by either quantifying p24gag levels in the supernatants by ELISA (B) or quantifying cell-associated HIV-1 gag and tat-rev mRNAs, respectively (C and D). (E) The p24gag levels and the expression levels of cell-associated gag and tat-rev mRNAs from 3 donors' cells were summarized and analyzed. *, P < 0.05; **, P < 0.01; ***, P < 0.001 (unpaired t test).
DISCUSSION
In this study, we demonstrated that the host factor Naf1 maintains HIV-1 latency by suppressing viral gene expression in an NF-κB-dependent manner. HIV-1 latency is characterized by a reversible silencing of viral LTR promoter-driven transcription of an integrated provirus (2, 6, 7). The LTR promoters of the majority of HIV-1 isolates contain two adjacent NF-κB binding sites that are crucial for initiating viral transcription (8, 11). Although other transcription factors also bind to the HIV-1 LTR promoter and regulate viral gene expression, NF-κB, as a master regulator of viral and gene transcription, is one of the most critical cellular factors in regulating HIV-1 latency (7–14). Lower levels of nuclear NF-κB in resting CD4+ T cells than in activated cells contribute to the establishment and maintenance of HIV-1 latency (14, 40). Activation of NF-κB signaling has been proposed as an attractive target for the development of agents to reverse HIV-1 latency (41); however, NF-κB signaling modulates multiple cellular events, and the nonspecific activation of bystander T cells by NF-κB signal-activating agents should be evaluated cautiously (42). Naf1 is a native repressor of NF-κB signaling in normal cells by blocking the nuclear import of the p65 subunit of NF-κB. Thus, studying Naf1-mediated regulation of NF-κB signaling in CD4+ central memory T cells can help us to understand host modulation of HIV-1 latency.
Besides the negative regulation of NF-κB activation, Naf1 has multiple regulatory cellular functions in receptor-mediated signaling pathways (43). Naf1 can also be incorporated into HIV-1 virions and interacts with the matrix protein of HIV-1 (17). The HIV-1 matrix protein is involved in viral assembly by directing Gag and Gag-Pol polyproteins to the plasma membrane during viral production (44). These data may suggest that Naf1 possesses multifaceted and likely cell-type-dependent roles in regulating HIV-1 infection. Naf1 has been shown to be a nucleocytoplasmic shuttling protein (17, 20). Recently, we found that nuclear Naf1 could promote nuclear export of unspliced HIV-1 gag mRNA, leading to increased Gag production for viral assembly and release (20). In this study, we found a new role for Naf1 in regulating HIV-1 infection, as we showed that Naf1 in the cytosol inhibited the activity of the integrated HIV-1 LTR by suppressing NF-κB signaling.
Naf1 is encoded by the TNIP1 gene, and single-nucleotide polymorphisms (SNPs) of TNIP that lead to a loss of control of NF-κB activation have been reported to be associated with multiple inflammation-related autoimmune diseases (43, 45–49). Naf1 is ubiquitously expressed in human tissues and cell types, particularly in peripheral blood lymphocytes (17). For HIV-1-infected individuals, whether the SNPs of TNIP have a potential relationship with viral load and/or disease progression is worth further study.
In summary, our in vitro study revealed a novel mechanism of NF-κB-dependent Naf1 function in maintaining HIV-1 latency in primary CD4+ T cells. We further confirmed this observation by using resting CD4+ T cells isolated from chronically HIV-1-infected patients who underwent effective antiretroviral therapy. Our findings provide new insights into HIV-1 pathogenesis that suggest potential therapeutic strategies to inhibit Naf1 activity in latently HIV-1-infected cells.
MATERIALS AND METHODS
Ethics statement.
HIV-1-infected outpatients who had undergone combination antiretroviral therapy for more than 3 years and had undetectable HIV-1 viral loads in plasma were recruited at the First Affiliated Hospital of Henan University of Traditional Chinese Medicine (HUTCM), China. Resting CD4+ T cells were isolated from peripheral blood mononuclear cells (PBMCs) by previously described methods (4, 50). The Medical Ethics Review Committee of the First Affiliated Hospital of HUTCM approved the study, and signed informed consent was obtained from each research participant.
Antibodies.
A monoclonal antibody (MAb) specific for Naf1 was described previously (51). An anti-c-myc MAb (clone 9E10) and an anti-Flag MAb (clone M2) were purchased from Sigma-Aldrich. An anti-glyceraldehyde-3-phosphate dehydrogenase (anti-GAPDH) MAb (clone 3B3) and an anti-histone H3.1 polyclonal antibody (pAb) were purchased from Abmart (Shanghai, China). An anti-p65 pAb was purchased from Cell Signaling.
Plasmids.
The myc-tagged Naf1 expression plasmid pCMV-Tag3B-Naf1 was described previously (51). Naf1 was cloned into the lentiviral vector pCDH-CMV (System Bioscience) to generate Flag-tagged Naf1-expressing stable cell lines and into a pEGFP-N1 vector which expressed a Naf1-GFP fusion protein. The Naf1 site-specific mutant (Naf1-Mut) expression plasmid pCMV-Tag 3B-Naf1-Mut was generated with a Mutant Best kit (TaKaRa Biotechnology). pGL3-LTR-luc was constructed by PCR amplification of the corresponding DNA fragment from pNL4-3 and subsequently cloned into the pGL3-basic vector (Promega, Madison, WI). The 3κB-luc NF-κB reporter plasmid has been described previously (52) and was kindly provided by Chen Wang, Shanghai Institute of Biochemistry and Cell Biology, Chinese Academy of Sciences, Shanghai, China.
Cell culture.
Human PBMCs were obtained from the Blood Center of Shanghai, China. CD4+ T cells were purified from PBMCs by use of anti-CD4-specific antibody-coated microbeads (Miltenyi Biotec) as described previously (53). CD4+ T cells were activated with 5 μg/ml PHA-P (Sigma-Aldrich) and cultured in the presence of 20 IU/ml recombinant interleukin-2 (IL-2) (R&D Systems) for 48 h. A latently HIV-1-infected Jurkat T-cell clone (C11) that harbors an HIV-1 proviral DNA encoding GFP was described previously (30). TNF-α was purchased from Invitrogen. PMA and ionomycin were purchased from Sigma. The C11 cell clone was cultured in RPMI 1640 medium containing 10% fetal bovine serum (FBS) (HyClone) with 100 U/ml penicillin and 100 μg/ml streptomycin. The human embryonic kidney cell line HEK293T was maintained in Dulbecco's modified Eagle's medium (DMEM) (HyClone) containing 10% FBS with 100 U/ml penicillin and 100 μg/ml streptomycin, and this cell line was a kind gift from Vineet KewalRamani (National Cancer Institute, Frederick, MD) and has been described previously (54).
HIV-1 stocks.
Single-cycle infectious HIV-Luc/VSV-G or HIV-eGFP/VSV-G was generated by calcium phosphate-mediated cotransfection of HEK293T cells with the luciferase reporter–HIV-1 proviral vector pLAI-Δ-env-Luc or pHIV-eGFP, respectively, and an expression plasmid for the vesicular stomatitis virus G (VSV-G) protein. Replication-competent HIV-1 strain NL4-3 was generated by transfection of HEK293T cells with the proviral construct pNL4-3. Harvested supernatants of transfected cells that contained viral particles were filtered and titrated by use of a p24gag capture enzyme-linked immunosorbent assay (ELISA). HIV-1 p24gag-specific monoclonal antibodies were a kind gift from Yong-Tang Zheng (Kunming Institute of Zoology, Chinese Academy of Sciences, China).
Assays of reporter gene expression.
HEK293T cells were seeded in a 24-well cell culture plate. Transfection was performed using Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions, and the plasmid 3κB-luc or pGL3-LTR-luc (50 ng) was used. At 6 h posttransfection, the medium, which contained a mixture of plasmids and transfection reagent, was replaced with fresh DMEM supplemented with 10% FBS. When indicated, cells were treated with TNF-α (50 ng/ml) at 24 h posttransfection and further cultured for an additional 24 h. Cells were harvested for luciferase assay with a luciferase assay system (Promega). The pCMV-β-Galactosidase control vector (5 ng) (BD Clontech) was cotransfected for normalization of transfection efficiency, and β-galactosidase activity was determined using the Beta-Glo assay system (Promega).
siRNA- and shRNA-mediated gene silencing.
The sequences of siRNA duplexes were as follows: for Naf1 siRNA, 5′-GCU UUU GGA AGA GUC CCA GTT-3′ (sense) and 5′-CUG GGA CUC UUC CAA AAG CTC-3′ (antisense); and for scramble siRNA, 5′-UUC UCC GAA CGU GUC ACG UTT-3′ (sense) and 5′-ACG UGA CAC GUU CGG AGA ATT-3′ (antisense). siRNAs (4 nM) were transfected into CD4+ T cells by use of an Amaxa human T cell Nucleofector kit (Lonza) with program T-023. Naf1 shRNA was subcloned into the pLKO.1-puro shRNA expression vector. The targeted sequences of shRNAs were as follows: Naf1-sh 1842, 5′-AAT CAG AGC TCC CAA GTG ATG-3′; Naf1-sh 568, 5′-AAG CCA TCA CAG ATG GGA GAA-3′; and scramble, 5′-TTC TCC GAA CGT GTC ACG TAT-3′. Calcium phosphate-mediated transfection of HEK293T cells was used to generate shRNA lentiviruses as previously described (55). To generate a cell line for stable Naf1 knockdown, lentiviruses containing Naf1 shRNA were used to infect HEK293T cells. At 24 h postinfection, puromycin (1 μg/ml) was added to the medium for selection. For Naf1 knockdown in the latently HIV-1-infected Jurkat T-cell clone, lentiviruses containing Naf1 shRNA were used for infection, and puromycin was added to the medium for selection at 24 h postinfection.
Immunoblotting.
Cells were lysed for 1 h at 4°C in ice-cold lysis buffer (50 mM HEPES, pH 7.4, 150 mM NaCl, 0.5 mM EGTA, 1% protease inhibitor cocktail [Sigma], 1 mM sodium orthovanadate, 1 mM NaF, 1% [vol/vol] Triton X-100, and 10% [vol/vol] glycerol). After centrifugation for 10 min at 12,000 × g, the supernatant was boiled in reducing SDS sample loading buffer and analyzed by SDS-PAGE. For immunoblotting, the indicated specific primary antibodies were used, followed by horseradish peroxidase-conjugated goat anti-mouse IgG or goat anti-rabbit IgG (Sigma) as the secondary antibody. The gray density of the gel was analyzed by using the plug-ins of ImageJ software.
Confocal microscopy.
Cells were seeded onto poly-l-lysine-coated microscope slides (Polyscience) and transfected with the Naf1-GFP-expressing plasmid pEGFP-N1-Naf1 for 24 h. Nuclei were stained with 4′,6-diamidino-2-phenylindole (DAPI). Slides were mounted with fluorescence mounting medium (Dako) and observed using a laser scanning confocal microscope (Leica SP5).
ChIP assay.
Chromatin immunoprecipitation (ChIP) experiments were performed according to the protocol provided with an EZ-ChIP chromatin immunoprecipitation kit (Millipore) (31). HeLa cell-derived TZM-bl indicator cells, which contain an integrated HIV-1 LTR promoter, were transfected with pCMV-Tag3B or pCMV-Tag3B-Naf1 for 48 h by use of Lipofectamine 2000. Cells were cross-linked in 1% formaldehyde for 10 min at room temperature and quenched with 0.125 M glycine for 5 min. After lysis, chromatin was sheared by use of a sonicator for a total of 12 min (10 s on and 10 s off) on ice to obtain DNA fragments of 200 to 1,000 bp. Ten percent of the total sheared chromatin DNA was used as input DNA. Other sheared chromatin was incubated with an antibody against c-myc or mouse IgG (Millipore) as a negative control. The immunoprecipitated DNA was analyzed by real-time PCR (ABI Prism 7900 real-time PCR system) for 30 cycles with Taq master mix (Invitrogen), using primers specific for the 5′ LTR of HIV-1 (forward, 5′-TAG CAG TGG CGC CCG AAC AGG-3′; and reverse, 5′-GCC TCT TGC CGT GCG CGC TTC-3′).
Assays of HIV-1 reactivation from latently infected cells.
We used the primary TCM model established by Vicente Planelles's lab (37, 38), with slight modifications. Briefly, naive CD4+ T cells were isolated by magnetically activated cell sorting (MACS) microbead negative sorting using a naive T-cell isolation kit (Miltenyi Biotec). Naive CD4+ T cells were stimulated with anti-CD3/CD28 antibody-coated microbeads for 3 days. Microbeads were then removed, and cells were cultured for an additional 4 days. Cells were infected with VSV-G-pseudotyped defective HIV (DHIV)-GFP (dEnv) and cultured for an additional 7 days to establish latency. Latently infected cells were infected with lentiviruses containing Naf1 shRNA or the scramble control for 3 days and were stimulated with or without anti-CD3/CD28 antibody-coated microbeads for an additional 3 days for detection of latency reactivation by flow cytometry or were harvested for immunoblotting.
Assays of viral activation in resting CD4+ T cells from HIV-1-infected individuals.
Purified resting CD4+ T cells (1 × 106/sample) isolated from HIV-1-infected individuals who had undergone combination antiretroviral therapy were coinfected with lentiviruses containing Naf1 shRNA or the scramble control (50 ng p24gag) and lentiviruses containing Vpx (multiplicity of infection [MOI] = 0.5) for 6 h (56), and after washing, the cells were cultured for another 3 days. Cells were collected for immunoblotting or stimulated with PHA-P (5 μg/ml) for 13 days. HIV-1 activation was detected by measuring the released HIV-1 particles in the supernatants by p24gag capture ELISA or by RT-PCR for quantification of the HIV-1 transcription products. For RT-PCR, total cellular RNA was extracted by use of a QIAamp RNeasy minikit (Qiagen), and the HIV-1 gag or tat-rev mRNA was quantified and normalized to the GAPDH mRNA level. The primer sequences were as follows: for gag, forward primer 5′-GTG TGG AAA ATC TCT AGC AGT GG-3′ and reverse primer 5′-CGC TCT CGC ACC CAT CTC-3′; for tat-rev, forward primer 5′-ATG GCA GGA AGA AGC GGA G-3′ and reverse primer 5′-ATT CCT TCG GGC CTG TCG-3′; and for GAPDH, forward primer 5′-ATC CCA TCA CCA TCT TCC AGG-3′ and reverse primer 5′-CCT TCT CCA TGG TGG TGA AGA C-3′.
Statistical analysis.
Statistical analysis was performed using the paired or unpaired Student t test with SigmaStat 2.0 (Systat Software, San Jose, CA).
ACKNOWLEDGMENTS
This work was supported by grants to J.-H.W. from the National Grant Program on Key Infectious Disease (grant 2014ZX10001003), the National Natural Science Foundation of China (grant 81572001), and the NSFC-NIH (grant 81561128009) and by an NIH grant (AI120209) to L.W. from the NIH/NIAID. L.W. is also supported in part by NIH grants (CA181997, AI104483, and AI127667) and by the Public Health Preparedness for Infectious Diseases Program of The Ohio State University.
The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
We thank Vicente Planelles for sharing HIV-1 proviral DNA constructs, Vineet KewalRamani, Chen Wang, and Yong-Tang Zheng for the kind gifts of cells or reagents, and Xia Jin (Institut Pasteur of Shanghai, Chinese Academy of Sciences, China) for editorial assistance and suggestions.
REFERENCES
- 1.Trono D, Van Lint C, Rouzioux C, Verdin E, Barre-Sinoussi F, Chun TW, Chomont N. 2010. HIV persistence and the prospect of long-term drug-free remissions for HIV-infected individuals. Science 329:174–180. doi: 10.1126/science.1191047. [DOI] [PubMed] [Google Scholar]
- 2.Margolis DM. 2010. Mechanisms of HIV latency: an emerging picture of complexity. Curr HIV/AIDS Rep 7:37–43. doi: 10.1007/s11904-009-0033-9. [DOI] [PubMed] [Google Scholar]
- 3.Coiras M, Lopez-Huertas MR, Perez-Olmeda M, Alcami J. 2009. Understanding HIV-1 latency provides clues for the eradication of long-term reservoirs. Nat Rev Microbiol 7:798–812. doi: 10.1038/nrmicro2223. [DOI] [PubMed] [Google Scholar]
- 4.Han Y, Wind-Rotolo M, Yang HC, Siliciano JD, Siliciano RF. 2007. Experimental approaches to the study of HIV-1 latency. Nat Rev Microbiol 5:95–106. doi: 10.1038/nrmicro1580. [DOI] [PubMed] [Google Scholar]
- 5.Lassen K, Han Y, Zhou Y, Siliciano J, Siliciano RF. 2004. The multifactorial nature of HIV-1 latency. Trends Mol Med 10:525–531. doi: 10.1016/j.molmed.2004.09.006. [DOI] [PubMed] [Google Scholar]
- 6.Shen L, Siliciano RF. 2008. Viral reservoirs, residual viremia, and the potential of highly active antiretroviral therapy to eradicate HIV infection. J Allergy Clin Immunol 122:22–28. doi: 10.1016/j.jaci.2008.05.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mbonye U, Karn J. 2014. Transcriptional control of HIV latency: cellular signaling pathways, epigenetics, happenstance and the hope for a cure. Virology 454–455:328–339. doi: 10.1016/j.virol.2014.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Alcami J, Lain de Lera T, Folgueira L, Pedraza MA, Jacque JM, Bachelerie F, Noriega AR, Hay RT, Harrich D, Gaynor RB. 1995. Absolute dependence on kappa B responsive elements for initiation and Tat-mediated amplification of HIV transcription in blood CD4 T lymphocytes. EMBO J 14:1552–1560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dahabieh MS, Ooms M, Brumme C, Taylor J, Harrigan PR, Simon V, Sadowski I. 2014. Direct non-productive HIV-1 infection in a T-cell line is driven by cellular activation state and NFkappaB. Retrovirology 11:17. doi: 10.1186/1742-4690-11-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fernandez G, Zaikos TD, Khan SZ, Jacobi AM, Behlke MA, Zeichner SL. 2013. Targeting IkappaB proteins for HIV latency activation: the role of individual IkappaB and NF-kappaB proteins. J Virol 87:3966–3978. doi: 10.1128/JVI.03251-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Williams SA, Chen LF, Kwon H, Ruiz-Jarabo CM, Verdin E, Greene WC. 2006. NF-kappaB p50 promotes HIV latency through HDAC recruitment and repression of transcriptional initiation. EMBO J 25:139–149. doi: 10.1038/sj.emboj.7600900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Coiras M, Lopez-Huertas MR, Rullas J, Mittelbrunn M, Alcami J. 2007. Basal shuttle of NF-kappaB/I kappaB alpha in resting T lymphocytes regulates HIV-1 LTR dependent expression. Retrovirology 4:56. doi: 10.1186/1742-4690-4-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McNamara LA, Ganesh JA, Collins KL. 2012. Latent HIV-1 infection occurs in multiple subsets of hematopoietic progenitor cells and is reversed by NF-kappaB activation. J Virol 86:9337–9350. doi: 10.1128/JVI.00895-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cary DC, Fujinaga K, Peterlin BM. 2016. Molecular mechanisms of HIV latency. J Clin Invest 126:448–454. doi: 10.1172/JCI80565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cohen S, Ciechanover A, Kravtsova-Ivantsiv Y, Lapid D, Lahav-Baratz S. 2009. ABIN-1 negatively regulates NF-kappaB by inhibiting processing of the p105 precursor. Biochem Biophys Res Commun 389:205–210. doi: 10.1016/j.bbrc.2009.08.074. [DOI] [PubMed] [Google Scholar]
- 16.Fukushi M, Dixon J, Kimura T, Tsurutani N, Dixon MJ, Yamamoto N. 1999. Identification and cloning of a novel cellular protein Naf1, Nef-associated factor 1, that increases cell surface CD4 expression. FEBS Lett 442:83–88. doi: 10.1016/S0014-5793(98)01631-7. [DOI] [PubMed] [Google Scholar]
- 17.Gupta K, Ott D, Hope TJ, Siliciano RF, Boeke JD. 2000. A human nuclear shuttling protein that interacts with human immunodeficiency virus type 1 matrix is packaged into virions. J Virol 74:11811–11824. doi: 10.1128/JVI.74.24.11811-11824.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Verstrepen L, Carpentier I, Verhelst K, Beyaert R. 2009. ABINs: A20 binding inhibitors of NF-kappa B and apoptosis signaling. Biochem Pharmacol 78:105–114. doi: 10.1016/j.bcp.2009.02.009. [DOI] [PubMed] [Google Scholar]
- 19.Heyninck K, De Valck D, Vanden Berghe W, Van Criekinge W, Contreras R, Fiers W, Haegeman G, Beyaert R. 1999. The zinc finger protein A20 inhibits TNF-induced NF-kappaB-dependent gene expression by interfering with an RIP- or TRAF2-mediated transactivation signal and directly binds to a novel NF-kappaB-inhibiting protein ABIN. J Cell Biol 145:1471–1482. doi: 10.1083/jcb.145.7.1471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ren XX, Wang HB, Li C, Jiang JF, Xiong SD, Jin X, Wu L, Wang JH. 2016. HIV-1 Nef-associated factor 1 enhances viral production by interacting with CRM1 to promote nuclear export of unspliced HIV-1 gag mRNA. J Biol Chem 291:4580–4588. doi: 10.1074/jbc.M115.706135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mauro C, Pacifico F, Lavorgna A, Mellone S, Iannetti A, Acquaviva R, Formisano S, Vito P, Leonardi A. 2006. ABIN-1 binds to NEMO/IKKgamma and co-operates with A20 in inhibiting NF-kappaB. J Biol Chem 281:18482–18488. doi: 10.1074/jbc.M601502200. [DOI] [PubMed] [Google Scholar]
- 22.Schiralli Lester GM, Henderson AJ. 2012. Mechanisms of HIV transcriptional regulation and their contribution to latency. Mol Biol Int 2012:614120. doi: 10.1155/2012/614120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jeeninga RE, Hoogenkamp M, Armand-Ugon M, de Baar M, Verhoef K, Berkhout B. 2000. Functional differences between the long terminal repeat transcriptional promoters of human immunodeficiency virus type 1 subtypes A through G. J Virol 74:3740–3751. doi: 10.1128/JVI.74.8.3740-3751.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tripathy MK, Abbas W, Herbein G. 2011. Epigenetic regulation of HIV-1 transcription. Epigenomics 3:487–502. doi: 10.2217/epi.11.61. [DOI] [PubMed] [Google Scholar]
- 25.Spina CA, Anderson J, Archin NM, Bosque A, Chan J, Famiglietti M, Greene WC, Kashuba A, Lewin SR, Margolis DM, Mau M, Ruelas D, Saleh S, Shirakawa K, Siliciano RF, Singhania A, Soto PC, Terry VH, Verdin E, Woelk C, Wooden S, Xing S, Planelles V. 2013. An in-depth comparison of latent HIV-1 reactivation in multiple cell model systems and resting CD4+ T cells from aviremic patients. PLoS Pathog 9:e1003834. doi: 10.1371/journal.ppat.1003834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Manson McManamy ME, Hakre S, Verdin EM, Margolis DM. 2014. Therapy for latent HIV-1 infection: the role of histone deacetylase inhibitors. Antivir Chem Chemother 23:145–149. doi: 10.3851/IMP2551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nabel G, Baltimore D. 1987. An inducible transcription factor activates expression of human immunodeficiency virus in T cells. Nature 326:711–713. doi: 10.1038/326711a0. [DOI] [PubMed] [Google Scholar]
- 28.Heyninck K, Kreike MM, Beyaert R. 2003. Structure-function analysis of the A20-binding inhibitor of NF-kappa B activation, ABIN-1. FEBS Lett 536:135–140. doi: 10.1016/S0014-5793(03)00041-3. [DOI] [PubMed] [Google Scholar]
- 29.Wei X, Decker JM, Liu H, Zhang Z, Arani RB, Kilby JM, Saag MS, Wu X, Shaw GM, Kappes JC. 2002. Emergence of resistant human immunodeficiency virus type 1 in patients receiving fusion inhibitor (T-20) monotherapy. Antimicrob Agents Chemother 46:1896–1905. doi: 10.1128/AAC.46.6.1896-1905.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Qu X, Wang P, Ding D, Li L, Wang H, Ma L, Zhou X, Liu S, Lin S, Wang X, Zhang G, Liu S, Liu L, Wang J, Zhang F, Lu D, Zhu H. 2013. Zinc-finger-nucleases mediate specific and efficient excision of HIV-1 proviral DNA from infected and latently infected human T cells. Nucleic Acids Res 41:7771–7782. doi: 10.1093/nar/gkt571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ding D, Qu X, Li L, Zhou X, Liu S, Lin S, Wang P, Kong C, Wang X, Liu L, Zhu H. 2013. Involvement of histone methyltransferase GLP in HIV-1 latency through catalysis of H3K9 dimethylation. Virology 440:182–189. doi: 10.1016/j.virol.2013.02.022. [DOI] [PubMed] [Google Scholar]
- 32.Chun TW, Carruth L, Finzi D, Shen X, DiGiuseppe JA, Taylor H, Hermankova M, Chadwick K, Margolick J, Quinn TC, Kuo YH, Brookmeyer R, Zeiger MA, Barditch-Crovo P, Siliciano RF. 1997. Quantification of latent tissue reservoirs and total body viral load in HIV-1 infection. Nature 387:183–188. doi: 10.1038/387183a0. [DOI] [PubMed] [Google Scholar]
- 33.Wong JK, Hezareh M, Gunthard HF, Havlir DV, Ignacio CC, Spina CA, Richman DD. 1997. Recovery of replication-competent HIV despite prolonged suppression of plasma viremia. Science 278:1291–1295. doi: 10.1126/science.278.5341.1291. [DOI] [PubMed] [Google Scholar]
- 34.Finzi D, Hermankova M, Pierson T, Carruth LM, Buck C, Chaisson RE, Quinn TC, Chadwick K, Margolick J, Brookmeyer R, Gallant J, Markowitz M, Ho DD, Richman DD, Siliciano RF. 1997. Identification of a reservoir for HIV-1 in patients on highly active antiretroviral therapy. Science 278:1295–1300. doi: 10.1126/science.278.5341.1295. [DOI] [PubMed] [Google Scholar]
- 35.Chomont N, El-Far M, Ancuta P, Trautmann L, Procopio FA, Yassine-Diab B, Boucher G, Boulassel MR, Ghattas G, Brenchley JM, Schacker TW, Hill BJ, Douek DC, Routy JP, Haddad EK, Sekaly RP. 2009. HIV reservoir size and persistence are driven by T cell survival and homeostatic proliferation. Nat Med 15:893–900. doi: 10.1038/nm.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Brenchley JM, Hill BJ, Ambrozak DR, Price DA, Guenaga FJ, Casazza JP, Kuruppu J, Yazdani J, Migueles SA, Connors M, Roederer M, Douek DC, Koup RA. 2004. T-cell subsets that harbor human immunodeficiency virus (HIV) in vivo: implications for HIV pathogenesis. J Virol 78:1160–1168. doi: 10.1128/JVI.78.3.1160-1168.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bosque A, Planelles V. 2011. Studies of HIV-1 latency in an ex vivo model that uses primary central memory T cells. Methods 53:54–61. doi: 10.1016/j.ymeth.2010.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bosque A, Planelles V. 2009. Induction of HIV-1 latency and reactivation in primary memory CD4+ T cells. Blood 113:58–65. doi: 10.1182/blood-2008-07-168393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Coleman CM, Wu L. 2009. HIV interactions with monocytes and dendritic cells: viral latency and reservoirs. Retrovirology 6:51. doi: 10.1186/1742-4690-6-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hiscott J, Kwon H, Genin P. 2001. Hostile takeovers: viral appropriation of the NF-kappaB pathway. J Clin Invest 107:143–151. doi: 10.1172/JCI11918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jiang G, Dandekar S. 2015. Targeting NF-kappaB signaling with protein kinase C agonists as an emerging strategy for combating HIV latency. AIDS Res Hum Retroviruses 31:4–12. doi: 10.1089/aid.2014.0199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhu X, Liu S, Wang P, Qu X, Wang X, Zeng H, Chen H, Zhu H. 2014. Oxaliplatin antagonizes HIV-1 latency by activating NF-kappaB without causing global T cell activation. Biochem Biophys Res Commun 450:202–207. doi: 10.1016/j.bbrc.2014.05.088. [DOI] [PubMed] [Google Scholar]
- 43.Ramirez VP, Gurevich I, Aneskievich BJ. 2012. Emerging roles for TNIP1 in regulating post-receptor signaling. Cytokine Growth Factor Rev 23:109–118. doi: 10.1016/j.cytogfr.2012.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bell NM, Lever AM. 2013. HIV Gag polyprotein: processing and early viral particle assembly. Trends Microbiol 21:136–144. doi: 10.1016/j.tim.2012.11.006. [DOI] [PubMed] [Google Scholar]
- 45.Kawasaki A, Ito S, Furukawa H, Hayashi T, Goto D, Matsumoto I, Kusaoi M, Ohashi J, Graham RR, Matsuta K, Behrens TW, Tohma S, Takasaki Y, Hashimoto H, Sumida T, Tsuchiya N. 2010. Association of TNFAIP3 interacting protein 1, TNIP1 with systemic lupus erythematosus in a Japanese population: a case-control association study. Arthritis Res Ther 12:R174. doi: 10.1186/ar3134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bowes J, Orozco G, Flynn E, Ho P, Brier R, Marzo-Ortega H, Coates L, McManus R, Ryan AW, Kane D, Korendowych E, McHugh N, FitzGerald O, Packham J, Morgan AW, Bruce IN, Barton A. 2011. Confirmation of TNIP1 and IL23A as susceptibility loci for psoriatic arthritis. Ann Rheum Dis 70:1641–1644. doi: 10.1136/ard.2011.150102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Allanore Y, Saad M, Dieude P, Avouac J, Distler JH, Amouyel P, Matucci-Cerinic M, Riemekasten G, Airo P, Melchers I, Hachulla E, Cusi D, Wichmann HE, Wipff J, Lambert JC, Hunzelmann N, Tiev K, Caramaschi P, Diot E, Kowal-Bielecka O, Valentini G, Mouthon L, Czirjak L, Damjanov N, Salvi E, Conti C, Muller M, Muller-Ladner U, Riccieri V, Ruiz B, Cracowski JL, Letenneur L, Dupuy AM, Meyer O, Kahan A, Munnich A, Boileau C, Martinez M. 2011. Genome-wide scan identifies TNIP1, PSORS1C1, and RHOB as novel risk loci for systemic sclerosis. PLoS Genet 7:e1002091. doi: 10.1371/journal.pgen.1002091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ellinghaus E, Stuart PE, Ellinghaus D, Nair RP, Debrus S, Raelson JV, Belouchi M, Tejasvi T, Li Y, Tsoi LC, Onken AT, Esko T, Metspalu A, Rahman P, Gladman DD, Bowcock AM, Helms C, Krueger GG, Koks S, Kingo K, Gieger C, Wichmann HE, Mrowietz U, Weidinger S, Schreiber S, Abecasis GR, Elder JT, Weichenthal M, Franke A. 2012. Genome-wide meta-analysis of psoriatic arthritis identifies susceptibility locus at REL. J Invest Dermatol 132:1133–1140. doi: 10.1038/jid.2011.415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Strange A, Capon F, Spencer CC, Knight J, Weale ME, Allen MH, Barton A, Band G, Bellenguez C, Bergboer JG, Blackwell JM, Bramon E, Bumpstead SJ, Casas JP, Cork MJ, Corvin A, Deloukas P, Dilthey A, Duncanson A, Edkins S, Estivill X, Fitzgerald O, Freeman C, Giardina E, Gray E, Hofer A, Huffmeier U, Hunt SE, Irvine AD, Jankowski J, Kirby B, Langford C, Lascorz J, Leman J, Leslie S, Mallbris L, Markus HS, Mathew CG, McLean WH, McManus R, Mossner R, Moutsianas L, Naluai AT, Nestle FO, Novelli G, Onoufriadis A, Palmer CN, Perricone C, Pirinen M, Plomin R, Potter SC, Pujol RM, Rautanen A, Riveira-Munoz E, Ryan AW, Salmhofer W, Samuelsson L, Sawcer SJ, Schalkwijk J, Smith CH, Stahle M, Su Z, Tazi-Ahnini R, Traupe H, Viswanathan AC, Warren RB, Weger W, Wolk K, Wood N, Worthington J, Young HS, Zeeuwen PL, Hayday A, Burden AD, Griffiths CE, Kere J, Reis A, McVean G, Evans DM, Brown MA, Barker JN, Peltonen L, Donnelly P, Trembath RC. 2010. A genome-wide association study identifies new psoriasis susceptibility loci and an interaction between HLA-C and ERAP1. Nat Genet 42:985–990. doi: 10.1038/ng.694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kim M, Hosmane NN, Bullen CK, Capoferri A, Yang HC, Siliciano JD, Siliciano RF. 2014. A primary CD4(+) T cell model of HIV-1 latency established after activation through the T cell receptor and subsequent return to quiescence. Nat Protoc 9:2755–2770. doi: 10.1038/nprot.2014.188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang HB, Wang JT, Zhang L, Geng ZH, Xu WL, Xu T, Huo Y, Zhu X, Plow EF, Chen M, Geng JG. 2007. P-selectin primes leukocyte integrin activation during inflammation. Nat Immunol 8:882–892. doi: 10.1038/ni1491. [DOI] [PubMed] [Google Scholar]
- 52.Diao L, Zhang B, Fan J, Gao X, Sun S, Yang K, Xin D, Jin N, Geng Y, Wang C. 2005. Herpes virus proteins ICP0 and BICP0 can activate NF-kappaB by catalyzing IkappaBalpha ubiquitination. Cell Signal 17:217–229. doi: 10.1016/j.cellsig.2004.07.003. [DOI] [PubMed] [Google Scholar]
- 53.Wang JH, Janas AM, Olson WJ, Wu L. 2007. Functionally distinct transmission of human immunodeficiency virus type 1 mediated by immature and mature dendritic cells. J Virol 81:8933–8943. doi: 10.1128/JVI.00878-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wu L, Martin TD, Vazeux R, Unutmaz D, KewalRamani VN. 2002. Functional evaluation of DC-SIGN monoclonal antibodies reveals DC-SIGN interactions with ICAM-3 do not promote human immunodeficiency virus type 1 transmission. J Virol 76:5905–5914. doi: 10.1128/JVI.76.12.5905-5914.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Dong C, Kwas C, Wu L. 2009. Transcriptional restriction of human immunodeficiency virus type 1 gene expression in undifferentiated primary monocytes. J Virol 83:3518–3527. doi: 10.1128/JVI.02665-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Berger G, Durand S, Goujon C, Nguyen XN, Cordeil S, Darlix JL, Cimarelli A. 2011. A simple, versatile and efficient method to genetically modify human monocyte-derived dendritic cells with HIV-1-derived lentiviral vectors. Nat Protoc 6:806–816. doi: 10.1038/nprot.2011.327. [DOI] [PubMed] [Google Scholar]