ABSTRACT
Formation of the cytoplasmic viral assembly compartment (cVAC) is an important step for efficient human cytomegalovirus (HCMV) assembly. To do this, the virus must alter and repurpose the normal cellular balance of membrane and protein flux, a process that is not well understood. Although a recent screen identified three viral proteins essential for cVAC formation, less is known about the contribution of cellular factors. We show that HCMV infection increases the protein level of a cellular trafficking factor, syntaxin 5 (STX5), a member of the syntaxin family of SNARE proteins. STX5 is recruited to the cVAC in infected cells and is required for the efficient production of infectious virions. We find that STX5 is important for normal cVAC morphology and the proper localization of viral proteins. A previously identified inhibitor of trafficking, Retro94, causes the mislocalization of STX5, an altered cVAC morphology, and dispersal of viral proteins. The presence of Retro94 results in severely impaired production of infectious virions, with a decrease as great as 5 logs. We show that this inhibition is conserved among different strains of HCMV and the various cell types that support infection, as well as for murine CMV. Thus, our data identify a key cellular trafficking factor important for supporting HCMV infection.
IMPORTANCE Human cytomegalovirus (HCMV) infection causes severe disease and mortality in immunocompromised individuals, including organ transplant and AIDS patients. In addition, infection of a developing fetus may result in lifelong complications such as deafness and learning disabilities. Understanding in detail the processes involved in HCMV replication is important for developing novel treatments. One of these essential processes, assembly of infectious virions, takes places in the cytoplasmic viral assembly compartment. We identify a cellular protein, syntaxin 5, important for generating this compartment, and show that it is required for the efficient production of infectious virions. We also show that a small molecule that disrupts this protein also significantly reduces the amount of infectious virions that are generated. Thus, by pinpointing a cellular protein that is important in the replication cycle of HCMV, we identified a novel target that can be pursued for therapeutic intervention.
KEYWORDS: Golgi apparatus, HCMV, SNARE, assembly, assembly compartment, cytomegalovirus, protein trafficking
INTRODUCTION
Human cytomegalovirus (HCMV), also referred to as human herpesvirus 5, is a ubiquitous virus that causes severe disease and mortality in the immunocompromised. Additionally, infection of a developing fetus can result in lifetime complications that include learning disabilities and deafness. One hallmark of HCMV infection is the profound reorganization of cellular organelles to form a cytoplasmic viral assembly compartment (cVAC). The cVAC consists of loosely associated organelle-specific vesicles organized in concentric layers (1, 2). Viral proteins important for tegument acquisition and envelopment localize to the cVAC. It has been proposed that as nucleocapsids exit the nucleus, they traverse the layers of the cVAC where tegument acquisition and envelopment occur (1). Conditions that perturb cVAC morphology result in reduced production of infections virions (3–6). Thus, although not essential for virus assembly and maturation, the cVAC is required for efficient production of infectious HCMV virions.
Despite identification of a number of viral and cellular proteins associated with the cVAC, little is known about the mechanisms governing the gross reorganization of cellular organelles during cVAC formation. A recent small interfering RNA (siRNA) screen of candidate viral proteins identified three HCMV proteins important for cVAC biogenesis: UL48, UL103, and UL94 (7). In endothelial cells, the absence of either UL135 or UL136 prevents the reorganization of cellular membranes into the cVAC, implicating a role for these two proteins as well (3). These viral proteins likely act in concert with cellular proteins to direct the events that lead to the formation and maintenance of the cVAC. The contribution of cellular factors to cVAC biogenesis is incompletely defined, and only a few cellular proteins have been identified to date, including Rab11, Bicaudal D1, FIP4, BiP, and dynein (8–11). Very little is known about the mechanistic contribution of these factors to the cVAC: only that disrupting their function disrupts the canonical morphology of the compartment. However, these factors play important roles in the trafficking of cellular proteins and membranes, suggesting that the virus may usurp these factors to form the cVAC. Investigating a role for additional cellular trafficking proteins will be essential for elucidating the mechanism of cVAC formation.
Generation of the cVAC also requires the expression of viral microRNAs (miRNAs) (4). Interestingly, these miRNAs target multiple factors of the cellular secretory pathway, including both Rab and SNARE (soluble N-ethylmaleimide-sensitive factor [NSF] attachment protein receptor) proteins, Rab11A and Rab5C, SNAP23 (synaptosomal-associated protein 23), and VAMP3 (vesicle-associated membrane protein 3). SNARE proteins mediate membrane fusion and are important at most intracellular trafficking steps. Thus, it is conceivable that HCMV regulates the cellular SNARE repertoire to bias membrane flow toward generation of the cVAC. In addition to the SNARE proteins identified in the miRNA study, other SNAREs are modulated during HCMV infection. Two SNARE proteins associated with exocytosis, syntaxin 3 (STX3) and SNAP23, are required for production and/or release of infectious particles (12, 13); however, the requirement of SNAP23 for virus release needs to be reconciled with its reported downregulation by HCMV miRNAs (4). The essential roles in trafficking of the SNARE proteins make them targets for modulation by HCMV as the virus optimizes conditions for producing infectious virions.
Here, we investigated a role for a subset of key SNARE proteins involved in transport to, from, and within the Golgi apparatus in forming the HCMV cVAC. We found that STX5 is upregulated during infection concomitant with cVAC formation and identified an essential role for STX5 in the production of infectious virions. The reduced production of virus correlated with a morphologically altered cVAC devoid of viral proteins. Furthermore, we determined that a compound that displaces STX5 from its cVAC localization impaired HCMV replication. These findings identify STX5 as a critical SNARE for cVAC formation.
RESULTS
STX5 is upregulated during HCMV infection and localizes to the cVAC.
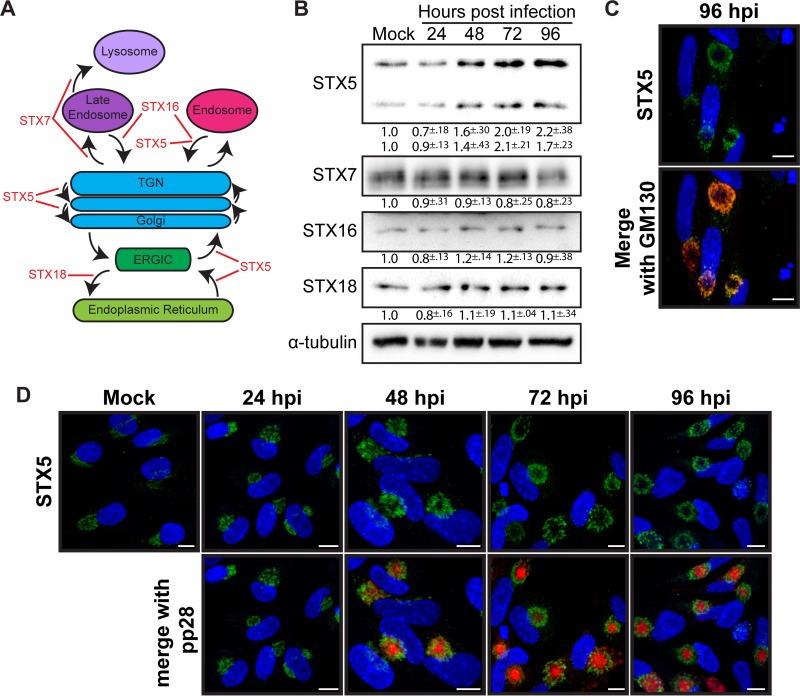
A striking morphological characteristic of the cVAC is reorganization of the Golgi apparatus into a juxtanuclear ring. We hypothesized that cellular proteins involved in regulating membrane flow to and from the Golgi apparatus, including the membrane fusion-promoting SNARE proteins, are important for cVAC formation and that HCMV may regulate these proteins during infection. SNARE proteins are structurally classified into Qa, Qb, Qc, and R SNAREs (14), and we initially focused on the Qa SNAREs of the syntaxin family associated with Golgi apparatus-related trafficking (Fig. 1A). Protein levels of STX7, which regulates trafficking to the late endosome and lysosome, of STX16, which is involved in retrograde transport from endosomes to the trans-Golgi network (TGN), and of STX18, involved in anterograde trafficking from the endoplasmic reticulum (ER) to the ER-Golgi intermediate complex (ERGIC), were stable throughout the course of infection (Fig. 1B). In contrast, both the long and short isoforms of STX5 protein were increased during infection, reaching peak levels late in infection (Fig. 1B). Since the increase in STX5 corresponds temporally with the presence of the cVAC, we chose to focus our studies on STX5. We first investigated whether the increase in protein corresponded with an increase in transcript levels. We found no significant change in the transcript levels of any of the above-mentioned SNAREs, including STX5, suggesting that the observed changes in protein levels occur at a posttranscriptional level (data not shown).
FIG 1.
HCMV regulation of Golgi apparatus SNAREs during virion production. (A) Schematic showing Qa SNAREs involved in Golgi apparatus-related trafficking. (B) Western blot analysis of Qa SNAREs from lysates harvested from uninfected cells (Mock) or cells infected with HCMV (AD169) (MOI of 3) for 24, 48, 72, and 96 hpi. Values below depict fold change from mock levels, calculated from three independent experiments. (C) Immunofluorescence analysis showing STX5 (green) and GM130 (red) in HCMV-infected cells at 96 hpi. (D) Immunofluorescence analysis showing STX5 (green) localization in uninfected (Mock) or HCMV-infected cells at 24, 48, 72, and 96 hpi relative to pp28 tagged with mCherry (red). Nuclei shown in panels C and D are labeled with DAPI (blue). Scale bar, 10 μm.
We next investigated the localization of STX5 during infection. In uninfected cells, STX5 is localized to the Golgi apparatus where it functions in retrograde, anterograde, and intra-Golgi compartment trafficking. During infection, STX5 retained this Golgi apparatus association and was present in the Golgi ring of the cVAC, colocalizing with the Golgi compartment marker GM130 (Fig. 1C). Temporally, by 24 h postinfection (hpi), normal Golgi apparatus morphology is already altered, and STX5 is accumulated at a juxtanuclear position, presumably marking the site of the future assembly compartment. At 48 hpi STX5 remains in this juxtanuclear location, with nascent rings beginning to form concurrent to pp28 accumulation inside the ring. At 72 and 96 hpi, STX5 is localized to the Golgi rings surrounding pp28 that are associated with the cVAC (Fig. 1D). Thus, STX5 localization during infection is consistent with a potential role in the formation and maintenance of the cVAC.
STX5 is functionally important for HCMV infection.
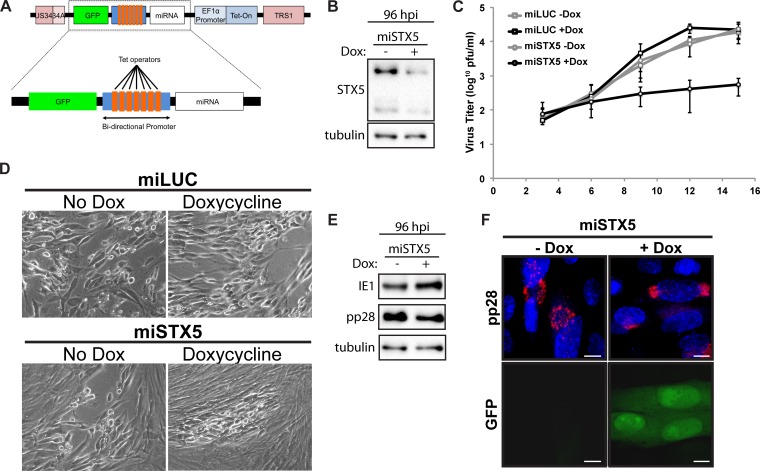
Since STX5 protein is increased late in infection and localizes to a ring of the cVAC, we hypothesized that it is functionally important for infection. To test the hypothesis that HCMV requires STX5 for infection, we designed an miRNA sequence targeting STX5 and engineered it into the viral genome between the UL34a and TRS1 loci. Since our hypothesis predicts that expression of this construct would be detrimental to the production of infectious virions, we sought to utilize an inducible promoter to drive the expression of our miRNA. Since previous attempts at utilizing inducible promoters in the viral genome have reported a large degree of leaky expression, we utilized a bidirectional promoter to express both green fluorescent protein (GFP) and our miRNA cassette, providing a way to visually monitor expression of our inducible construct (Fig. 2A). Comparison of STX5 protein levels from cells infected with an miRNA targeting STX5 (miSTX5) at 96 hpi revealed that STX5 protein was greatly reduced in the presence of doxycycline compared to the level in the no-doxycycline control, validating our knockdown strategy (Fig. 2B).
FIG 2.
STX5 is required for efficient infection. (A) Schematic showing design of the inducible miRNA-expressing locus inserted between UL34a and TRS1 of the AD169 strain. (B) Western blot analysis of STX5 protein at 96 hpi with miSTX5 (MOI of 3) in the presence or absence of doxycycline (Dox). (C) Growth curve analysis of viral titers from miLUC or miSTX5 infections treated with or without doxycycline (10 μg/ml) at 3, 6, 9, 12, and 15 dpi (MOI of 0.05). Data shown are composites of three independent replicates. (D) Bright-field images of CPE associated with infections from the experiment shown in panel C at 9 dpi. (E) Western blot analysis of IE1 and pp28 protein at 96 hpi with miSTX5 (MOI of 3) in the presence or absence of doxycycline. (F) Immunofluorescence analysis of pp28 and expression of GFP at 96 hpi in miSTX5-infected cells (MOI of 3) either untreated or treated with doxycycline (10 μg/ml). Nuclei were labeled with DAPI (blue). Scale bar, 10 μm.
Using our miSTX5 virus and a control virus expressing a microRNA against luciferase (miLUC), we infected cells at a low multiplicity of infection (MOI; 0.05 PFU/ml). Doxycycline was added to a subset of both miSTX5 and miLUC samples at 2 hpi, and the medium was spiked with fresh doxycycline every 3 days throughout infection. Cells and supernatant were harvested for growth curve analysis at 3, 6, 9, 12, and 15 days postinfection (dpi). The miLUC virus, with or without doxycycline, and the miSTX5 virus without doxycycline progressed similarly throughout infection, and the titers of all three viruses were comparable at 15 dpi. In contrast, viral titers from the miSTX5 doxycycline-treated samples were decreased by approximately 2 logs compared to the levels in untreated miSTX5 samples (Fig. 2C). The lower viral titer corresponded to decreased viral spread and less cytopathic effect (CPE) in the monolayer (Fig. 2D). Thus, STX5 is functionally important for HCMV infection.
Since STX5 is a cellular trafficking protein associated with Golgi apparatus morphology and is upregulated during the late stages of infection, corresponding with the loss of the canonical Golgi apparatus structure and the appearance of the Golgi ring, we reasoned that STX5 might be important for this Golgi apparatus rearrangement. Western blot analysis showed no major difference in levels of the immediate early protein IE1 or the late protein pp28 (Fig. 2E). This suggests that the block in the production of infectious virions is at a late stage in infection, consistent with our hypothesis. We next determined the localization of pp28 during infection. We detected pp28 in the cVAC of the untreated miSTX5 virus at 96 hpi (Fig. 2E). However, pp28 localization was dispersed during infection with the miSTX5 virus when doxycycline was added to induce STX5 knockdown, represented by the coinduction of GFP (Fig. 2E). This observation is consistent with a role for STX5 in the proper formation of the viral assembly compartment.
STX5 modulation by Retro94.
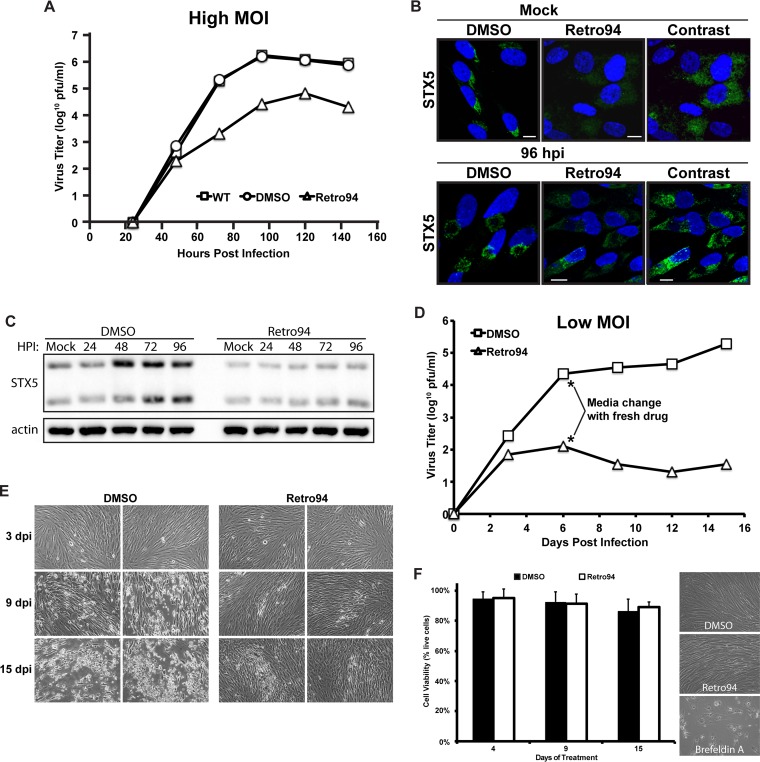
To verify our previous observations, we sought an alternative method to study a role for STX5 during infection. We took advantage of a compound, Retro94, that promotes the mislocalization of STX5. The parent compound, Retro2, was one of two compounds, identified in a recent screen, that blocked the trafficking and subsequent toxicity of bacterial Shiga-like toxins (15). An analysis of trafficking factors revealed that STX5 was displaced from the Golgi apparatus after treatment with Retro2. Subsequent optimization of Retro2 led to a more potent version of the inhibitor, named Retro94 (16). Since STX5 is upregulated by HCMV during infection and is functionally important, we hypothesized that Retro94 would inhibit HCMV replication. Addition of Retro94 at 2 h after infection at a high MOI (3 PFU/cell) reduced production of infectious virions by greater than 2 logs (Fig. 3A). We next investigated the localization of STX5 after treatment to verify that Retro94 could still alter its localization in infected cells. As previously reported (16), STX5 lost its Golgi compartment localization after treatment with Retro94 in uninfected cells and was dispersed throughout the cell (Fig. 3B). In infected cells, STX5 was also dispersed throughout the cell and no longer localized to the cVAC ring (Fig. 3B). We also detected a decrease in STX5 staining intensity in the Retro94-treated samples. Western blot analysis confirmed that STX5 levels were reduced following Retro94 treatment (Fig. 3C). Thus, Retro94, due to its modulation of both STX5 localization and protein level, can be used as a tool for studying the role of STX5 during infection.
FIG 3.
Retro94 blocks the efficient production of infectious HCMV virions. (A) Growth curve analysis of HCMV (AD169; MOI of 3) in untreated (wild type, WT), vehicle-treated (DMSO), or Retro94-treated cells. Samples were harvested at 24, 48, 72, 96, 120, and 144 hpi. Data shown are from one of three independent replicates. (B) Immunofluorescence detection of STX5 in uninfected (Mock) and HCMV-infected cells (96 hpi) in the presence of DMSO or Retro94. The far right panel features enhanced contrast of Retro94 samples for easier visualization of STX5 displacement. Nuclei are labeled with DAPI (blue). Scale bar, 10 μm. (C) Western blot analysis of STX5 protein in uninfected (Mock) and HCMV-infected cells at 24, 48, 72, and 96 hpi. Samples were treated with DMSO or Retro94 (10 μM) from 2 hpi. The mock sample was treated with Retro94 for 96 h. (D) Growth curve analysis of HCMV (MOI of 0.05) in DMSO- or Retro94-treated cells. Samples were harvested at 3, 6, 9, 12, and 15 dpi. Asterisks indicate the exchange of medium with fresh Retro94 at day 6. Data shown are from one of four independent replicates. (E) Bright-field images of CPE associated with infections from the experiment shown in panel C. (F) The viability of cells treated with DMSO or Retro94 for 4, 9, and 15 days. Fresh Retro94 was added at day 6. Bright-field images of cell monolayers at 4 days posttreatment are shown to the right of the graph. Brefeldin A was included as a positive control for cell death.
We next performed growth curve analyses on low-MOI infections (0.05 PFU/cell) of cells treated with Retro94 or the vehicle control (dimethyl sulfoxide [DMSO]). Since the stability of Retro94 in medium remains to be determined, we exchanged medium at 6 days postinfection (dpi) and added fresh drug. While the control-treated infection continued to progress after the medium change, the Retro94-treated samples never recovered. At 15 dpi, there was a 5-log reduction in the amount of infectious virus produced (Fig. 3D). Visualization of the monolayer at 3 dpi showed little difference between the Retro94- and control-treated samples, with small foci of infection detectable in both treatments (Fig. 3E). However, by 9 dpi the control-treated infection had spread, with areas of visible clearing present. In contrast, the spread of the infection was limited in Retro94-treated cells. By 15 dpi CPE was widespread in the control-treated monolayer, while much of the Retro94-treated monolayer remained intact, with only a few distinct foci of infection that were slightly larger than those present at 9 dpi (Fig. 3E). Thus, both knockdown of STX5 by miRNA and modulation of STX5 by Retro94 led to reduced production of infectious virions.
Taken together, these results suggest a direct role for STX5 in HCMV replication. However, the extended treatment of cells with drug for the duration of infection may result in unhealthy cells that are unable to support infection. We found that treatment of cells with Retro94 for 4, 9, or 15 days had no effect on cell viability or cell morphology (Fig. 3F). This is in contrast to another trafficking inhibitor that is known to be toxic after extended treatment, brefeldin A, in which very few cells in the monolayer remained after 96 h of treatment. Thus, the effect of Retro94 on HCMV replication is unlikely to be a result of toxicity to the cells.
Retro94 alters formation of the cytoplasmic viral assembly compartment.
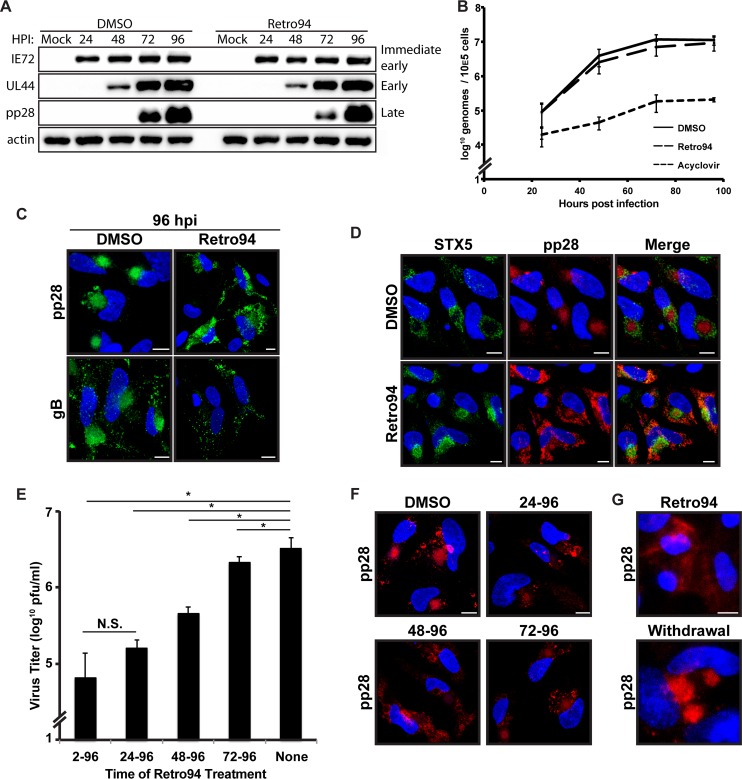
Results with our miSTX5 virus indicated that STX5 may be important for properly organizing the cVAC. This would be a unique role for STX5 during virus infections as Retro94 has previously been used to block the entry of several viruses, including polyomavirus and papillomavirus (17), indicating a role for STX5 in the entry of these viruses. Since HCMV does not share the entry pathway of these viruses, we anticipated that Retro94 would not affect HCMV entry. Rather, we hypothesized that the Retro94-mediated modulation of STX5 would affect a later stage in replication, such as the organizing of the cVAC, consistent with our miSTX5 results. To further investigate this, we performed a steady-state analysis of viral proteins. Western blot analysis of lysates from control- and Retro94-treated samples showed that steady-state levels of immediate early and late proteins were not affected, confirming that infection was established and had progressed to production of late proteins (Fig. 4A). The presence of late proteins also indirectly indicated that DNA replication had occurred. To verify this, we analyzed levels of viral DNA in infected cells treated with DMSO, Retro94, or acyclovir. Levels of viral DNA were similar in DMSO- and Retro94-treated cells (Fig. 4B). This was in contrast to the markedly reduced levels in acyclovir-treated cells. These results support a post-DNA replication, late block to infection.
FIG 4.
Retro94 alters morphology of the HCMV assembly compartment. (A) Western blot analysis of uninfected cells (Mock) or HCMV-infected cells (AD169; MOI of 3) treated with DMSO or Retro94 (10 μM) at 24, 48, 72, and 96 hpi. (B) DNA replication analysis of cells infected at an MOI of 3 and harvested at 24, 48, 72, and 96 hpi. Samples were treated with DMSO, Retro94 (10 μM), or acyclovir (100 μg/ml). Data are composites of three independent replicates. (C) Localization of viral proteins in HCMV-infected cells at 96 hpi in the presence of DMSO or Retro 94 (10 μM). pp28 was detected by tagging with mCherry, and gB was detected by immunofluorescence. (D) Immunofluorescence detection of STX5 (green) in HCMV-infected cells (MOI of 3) at 96 hpi treated with DMSO or Retro94. pp28-mCherry is shown in red for reference. (E) Virus titers at 96 hpi of HCMV-infected cells treated with Retro94 at 2, 24, 48, or 72 hpi. *, P < 0.05; N.S., not significant. (F) Localization of pp28-mCherry in samples corresponding to those shown in panel E. (G) Immunofluorescence detection of pp28 in HCMV-infected cells subjected to either treatment with Retro94 throughout infection or treatment with Retro94 and subsequent withdrawal of Retro94 at 9 dpi followed by recovery for an additional 9 days. Nuclei shown in panels C, D, F, and G are labeled with DAPI (blue). Scale bar, 10 μm.
Since STX5 is a cellular trafficking factor that is localized to the Golgi ring of the cVAC and appears to affect a late stage in infection, we investigated whether disruption of STX5 with Retro94 alters cVAC morphology. We investigated the localization of three viral proteins known to localize to the juxtanuclear cVAC, i.e., pp28, pp150, and glycoprotein B. In the presence of Retro94, all three viral proteins were dispersed throughout the cytoplasm and were no longer present in the cVAC (Fig. 4C and data not shown). Since both STX5 and the viral proteins were dispersed throughout the cytoplasm in the presence of Retro94, we investigated whether STX5 and pp28 colocalized. There was not a high degree of colocalization, and for the most part they were dispersed to distinct regions of the cell (Fig. 4D).
To confirm that STX5 is involved in cVAC formation and maintenance, we performed a temporal analysis on the effects of adding Retro94 at various times postinfection. In the previous experiments, Retro94 was either added simultaneously with virus or after the adsorption phase at 2 hpi. We hypothesized that the greatest effect on replication would be when Retro94 was added prior to cVAC formation and that the efficiency of the block to infectious virion formation would decrease later in infection after the cVAC is already formed. In contrast, if Retro94 was blocking entry, we would expect Retro94 to be inefficient at blocking production of infectious virions when added late in infection. Consistent with our hypothesis, the greatest inhibition was observed when Retro94 was added early in infection, at 2 or 24 hpi, time points that precede the formation of the cVAC (Fig. 4E). The efficacy of Retro94 was reduced when it was added at 48 hpi, when components of the cVAC are beginning to take shape in the juxtanuclear position. Addition of Retro94 at 72 hpi, when the assembly compartment is fully formed and virion production is well under way, resulted in a modest, but reproducible and significant 2-fold reduction in infectious virion production. These results support a late role for STX5 in HCMV infection, in contrast to its role in entry that was observed for other viruses.
We next sought to correlate this inhibition of infectious virion production with the morphology of the assembly compartment. Assembly compartments were not present in cells treated with Retro94 at 24 or 48 hpi (Fig. 4F). In contrast, when Retro94 was added at 72 hpi, pp28 was found clustered in spheres, reminiscent of an assembly compartment. However, these spheres were lighter in intensity, with some pp28 dispersed from these regions (Fig. 4F). Thus, the observed decrease in virus titers correlated with the status of the assembly compartment.
To provide additional evidence that STX5 is involved in directly modulating cVAC morphology and that these observations were not a result of general cell toxicity, we monitored the status of the cVAC in cells that were treated with Retro94 and then allowed to recover after Retro94 withdrawal. In these cells, pp28 accumulated in a juxtanuclear location resembling the canonical HCMV cVAC (Fig. 4G). Thus, even after 9 days of Retro94 treatment, the cells were healthy and viable enough to form an assembly compartment. This result, in combination with the earlier toxicity results, provides solid evidence that the results with Retro94 represent a direct effect on the virus and not a result of general cellular toxicity.
Juxtanuclear membrane accumulation occurs in Retro94-treated cells.
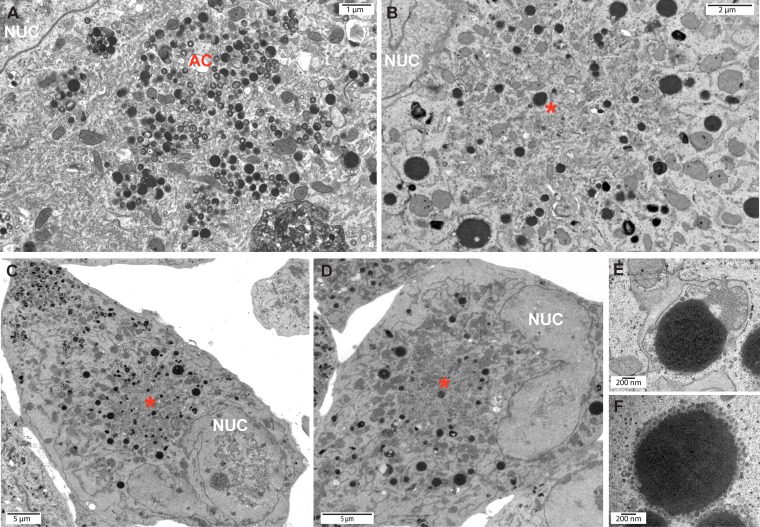
To further examine the effect of STX5 on cVAC formation and viral assembly, we performed electron microscopy (EM) on infected cells treated with Retro94 or vehicle. In vehicle-treated cells, a juxtanuclear region of intense viral activity was observed. There were numerous capsids at various stages of tegumentation and envelopment present in this region, as well as dense bodies, indicative of a fully formed and functioning assembly compartment (Fig. 5A). In Retro94-treated cells, an area with comparable viral activity was not observed, despite the presence of juxtanuclear membrane that resembled the membrane accumulation normally associated with the cVAC (Fig. 5B to D). Thus, the absence of STX5 disrupts the cytoplasmic viral activity associated with a normal infection.
FIG 5.
Retro94 results in reduced cytoplasmic viral activity. Electron micrographs of HCMV-infected cells (AD169; MOI of 3) at 96 hpi treated with DMSO (A) or Retro94 (B to F) at 2 hpi. Asterisks in panels B to D indicate areas of juxtanuclear accumulation of membranes. (E and F) High-magnification images of large, dense, membrane-enwrapped structures found at the periphery of areas of juxtanuclear membrane accumulation. AC, assembly compartment. Nuc, nucleus.
The juxtanuclear accumulation of extra membrane, particularly evident at lower magnification (Fig. 5C and D), implicates this area as a potential cVAC, despite the absence of capsids and enveloped virions. Although nucleocapsids were present in the nucleus in both DMSO- and Retro94-treated cells, very few capsids were observed in the cytoplasm after Retro94 treatment. When present, they were found throughout the cytoplasm and did not preferentially associate with the region of juxtanuclear membranes. Instead, large, dense structures greater than 1 μm long were found at the periphery of these membranous areas (Fig. 5E and F). One hypothesis is that these structures are degradative organelles responsible for clearing cytoplasmic nucleocapsids that are unable to mature in the absence of STX5. Thus, under conditions when STX5 function is perturbed, membrane is delivered to a juxtanuclear location, but these membranes are not associated with the viral activity normally observed at this location. The lack of viral activity at these membranous sites correlates with the observed decrease in infectious virion production in the presence of Retro94.
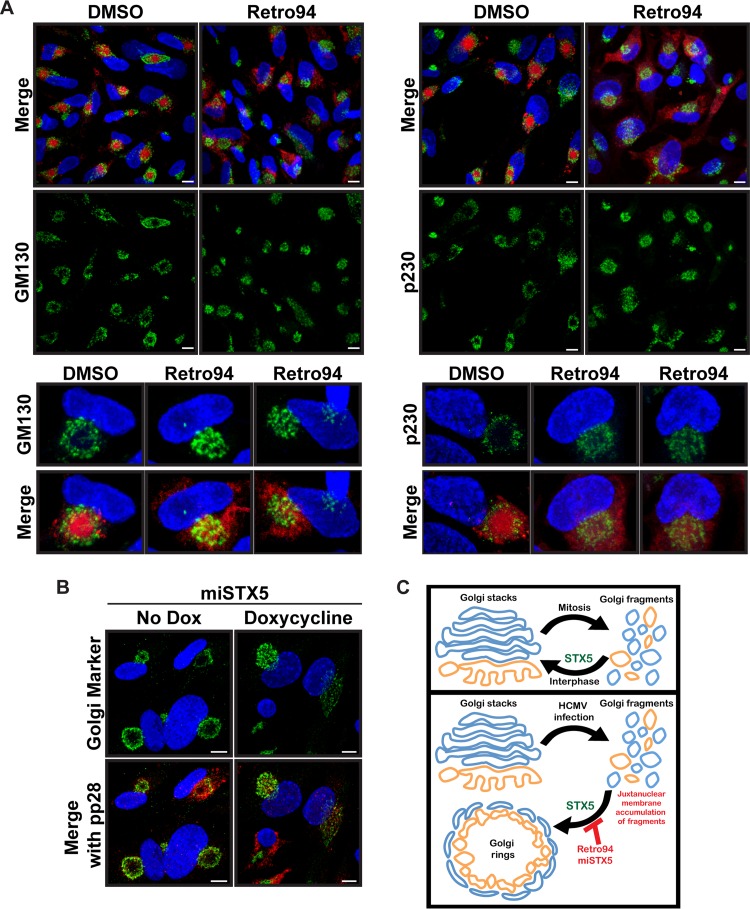
We hypothesized that the juxtanuclear membranes we observed were analogous to the cellular membranes that accumulate to form the layers of the cVAC. To test this hypothesis, we investigated the location of various cellular cVAC markers. We found that markers of the Golgi apparatus (GM130), TGN (p230), and endosomes (EEA1) all retained their juxtanuclear localizations (Fig. 6A and data not shown). However, the Golgi and TGN rings characteristic of normal cVAC morphology were collapsed. We next asked whether a similar phenotype was observed with the miSTX5 virus. As expected, in the absence of doxycycline the Golgi apparatus formed the ring typically associated with the cVAC. However, the Golgi rings did not form in the presence of doxycycline, instead accumulating in a collapsed juxtanuclear position similar to what was observed with Retro94 treatment (Fig. 6B). Thus, in the absence of STX5 the canonical Golgi apparatus morphology is disrupted, and membranes accumulate in a juxtanuclear location, but the Golgi rings associated with proper cVAC morphology are not present. Thus, both the proper organization of the membrane layers associated with the cVAC and the correct localization of viral proteins are disrupted in the absence of functional STX5.
FIG 6.
Juxtanuclear accumulation of Golgi fragments in the absence of STX5. (A) Immunofluorescence of Golgi complex (GM130) and TGN (p230) markers (green) in HCMV-infected cells (AD169; MOI of 3, 96 hpi) treated with DMSO or Retro94. Lower insets show single cells enlarged from the images above. A merged image with pp28-mCherry (red) is shown for reference. (B) Immunofluorescence of Golgi apparatus markers p115 (green) and pp28 (red) in cells infected with miSTX5 in the presence or absence of doxycycline at 96 hpi. (C) Proposed model for the role of STX5 during HCMV infection. Nuclei shown in panels A and B are labeled with DAPI (blue). Scale bar, 10 μm.
Retro94 is a potent inhibitor of cytomegalovirus production.
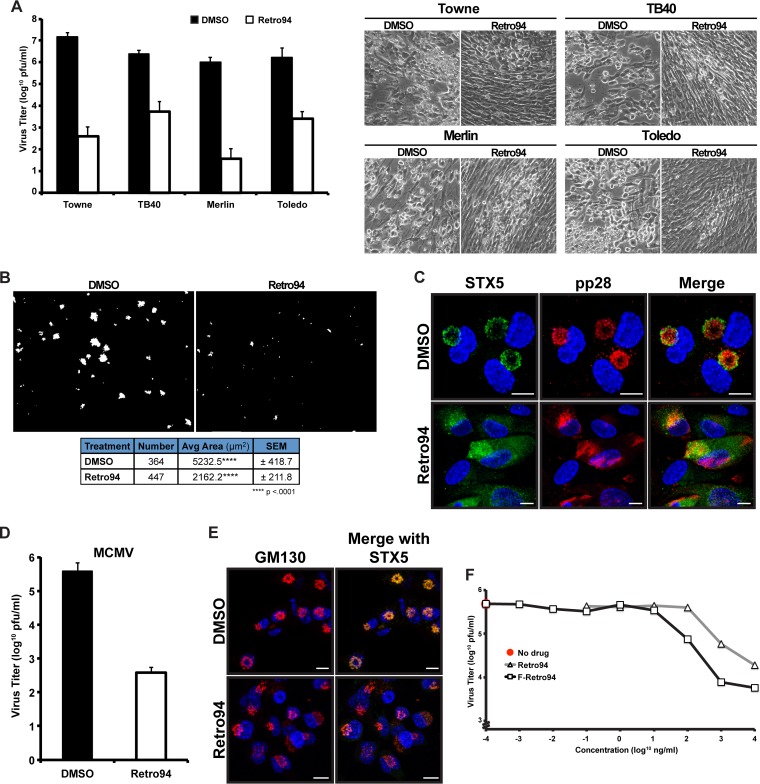
Our data show that Retro94 inhibits production of infectious virions in the context of the laboratory strain AD169. We wondered if this inhibition was conserved among other HCMV strains. To test this, we infected fibroblasts with the Towne, TB40, Merlin, and Toledo strains of HCMV and treated cells with Retro94 or DMSO at 2 hpi. Growth curve analysis on cells and cell supernatants showed that viral titers of all four strains were reduced by several logs in the presence of Retro94 (Fig. 7A). This corresponded to reduced CPE in Retro94-treated samples (Fig. 7A). Thus, Retro94 inhibits the replication of multiple strains of HCMV.
FIG 7.
The Retro94-dependent inhibition of HCMV is not strain, cell type, or species specific. (A) Viral titers at 15 dpi of fibroblasts infected with various strains of HCMV and treated with DMSO or Retro94 at 2 hpi. The medium was changed, and fresh Retro94 was added at 6 dpi. Bright-field images representing CPE of infections are shown on the right. (B) HCMV-infected ARPE-19 cells treated with DMSO or Retro94 (10 μM). Infected cells were detected by monitoring mCherry (red) expressed by TB40 virus. Scale bar, 1 mm. The table below the images shows the number of foci detected, average area, and standard error of the mean (SEM). (C) Immunofluorescence analysis of STX5 (green) and pp28 (red) in HCMV-infected ARPE-19 cells at 96 hpi in the presence of DMSO or Retro94 (10 μM). (D) Viral titers at 6 dpi of MCMV-infected fibroblasts treated with DMSO or Retro94. (E) Immunofluorescence analysis of STX5 (green) and GM130 (red) in MCMV-infected NIH 3T3 cells at 72 hpi in the presence of DMSO or Retro94 (10 μM). (F) Growth curve analysis of HCMV (MOI of 3) in the DMSO control (No drug) or in cells treated with Retro94 and F-Retro94 at increasing concentrations. Samples were harvested at 96 hpi. Representative data shown are from one of four (Retro94) or one of three (F-Retro94) independent replicates. Values in A and D are composites of three independent experiments.
We next wanted to investigate if Retro94 inhibited infection of an alternative cell type, particularly since the effect of Retro94 on HCMV replication acts through altering localization of the cellular protein STX5. To test this, we infected epithelial ARPE-19 cells with a viral strain of TB40 that expresses mCherry to enable us to monitor the spread of infection. Three weeks after infection the area representing virus infection, as indicated by the presence of mCherry, in both the DMSO- and Retro94-treated cells was calculated. The area of the DMSO-treated cells was more than twice the area of the Retro94-treated cells (Fig. 7B). Thus, Retro94 inhibits the spread of HCMV in both fibroblasts and epithelial cells. Similar to treatment in fibroblast cells, treatment with Retro94 dispersed both STX5 and pp28 throughout the cytoplasm of epithelial cells (Fig. 7C), indicating that Retro94 is affecting replication in ARPE-19 cells in a manner analogous to that in fibroblasts.
Since Retro94 has the ability to block various strains of HCMV in multiple cell types, we next asked if this block was species specific. We found that Retro94 inhibited production of infectious MCMV, decreasing the titer by nearly 3 logs (Fig. 7D). We next wanted to show that Retro94 was acting in a similar manner to prevent production of murine cytomegalovirus (MCMV) infectious virions. Staining with GM130 and STX5 showed that MCMV does reorganize the Golgi apparatus into a ring, similar to HCMV, although morphologically these rings appeared more compact. Treatment with Retro94 dispersed STX5 and collapsed the Golgi rings, similar to what was observed for HCMV (Fig. 7E). Taken together, these data demonstrate that Retro94 is a general inhibitor of cytomegaloviruses.
Due to the potent inhibition of several logs by Retro94 on the production of HCMV infectious virions, we sought to define the concentration requirements of this inhibition. Tenfold dilutions of Retro94 were added at 2 hpi to cells infected at an MOI of 3, and growth curve analyses were performed at 96 hpi. The dilution at which a significant inhibition in viral titers was first present was 1 μM (Fig. 7F). The 50% inhibitory concentration (IC50) was calculated to be in the nanomolar range, ∼130 nM. Recent studies on Retro94 have identified derivatives that exhibit more potency, including addition of a fluorine (18). We found that this derivative, F-Retro94, was even more effective in reducing HCMV titers, decreasing the IC50 by about 1 log to ∼15 nM. Thus, our studies have led to the identification of a potent inhibitor of HCMV that is well tolerated by cells.
DISCUSSION
Here, we identify an important role for STX5, a cellular SNARE, in HCMV replication. STX5 protein is upregulated late in infection, concurrent with cVAC formation, and localizes to the Golgi ring associated with this compartment. We found that when STX5 function is disrupted, by either miRNA knockdown or pharmacological intervention, production of HCMV infectious virions is significantly reduced. We show that this block to infection is at a late stage and corresponds with the aberrant formation of the cVAC. Not all SNARE proteins are required for HCMV infection, as evidenced by the complete disappearance of the Qc SNARE GS15 during infection (data not shown). Since protein levels of other SNARE proteins are differentially regulated, this suggests that HCMV generates a distinct SNARE repertoire that optimizes trafficking conditions for efficient replication. This is consistent with a recent study identifying several HCMV miRNAs that target cellular trafficking factors, including members of the SNARE family (4). These miRNAs also affected cVAC morphology. Thus, an important step for HCMV replication is the preparation of cellular trafficking factors to generate the cVAC and to deliver factors required for supporting the efficient assembly and transport of virions.
SNARE complexes require a tight interaction between four SNARE motifs from four, or sometimes three, different SNARE proteins. STX5 is normally localized to the Golgi compartment where it participates in different t-SNARE complexes. The identification of other SNAREs that function in conjunction with STX5 during infection is an important next step to elucidating the STX5-dependent mechanism that mediates viral protein localization and proper cVAC morphology. Future experiments are needed to investigate this in further detail. However, the identification of SNARE partners that interact with STX5 during infection is complicated by complexes that spontaneously form after detergent lysis and that are, in fact, nonfunctional. For example, although STX5 binds to other SNARE proteins, these complexes are not fusogenic when reconstituted into liposomes (19). In fact, this study showed that only 2 out of a potential 147 yeast SNARE combinations containing the STX5 homologue, Sed5, promoted fusion. Caution must be used in identifying the STX5-interacting partners, particularly any potential novel SNARE complexes, relevant to the role of STX5 during infection. These proteomic studies, however, may be useful in identifying important regulatory proteins of STX5, such as tethering factors and Sec1/Munc18-like (SM) proteins. While the role of STX5 in infection is likely to involve its SNARE activity, it has also been reported that the long chain of STX5 affects ER morphology and that this is accomplished independently of the normal SNARE function of STX5 (20). Thus, we cannot rule out a SNARE-independent contribution of STX5 to HCMV infection, which may explain why STX5 is upregulated independently of other SNARE proteins.
One intriguing result is the observation that viral proteins were dispersed throughout the cells but that markers of cellular membranes were still present in a juxtanuclear position. However, Retro94 prevented the reorganization of these membranes into the layers that form the cVAC. This suggests that formation of these layers is dependent upon STX5 directly or upon viral proteins that require STX5 for localization to this region. Thus, an interesting mechanistic question remains about the relationship between viral proteins, cellular membranes, and STX5 in the context of forming the cVAC. Does STX5 deliver the viral proteins that in turn organize the membranes, or does STX5 directly organize the membranes to promote the accumulation of viral proteins? Addressing this and other similar questions will further elucidate the molecular details surrounding the formation of this complex structure that is essential for the efficient production of infectious virions.
While it is unclear at this point whether STX5 is playing a direct or indirect role in organizing Golgi membranes during infection, a direct role for STX5 in properly organizing cVAC membranes would not be unexpected since it has been implicated in maintaining organelle morphology. For example, a single amino acid substitution of a phosphorylated residue on the yeast homologue Sed5 resulted in the normally dispersed yeast Golgi apparatus to form an ordered structure similar to the mammalian Golgi apparatus (21). Interestingly, another study showed that STX5 levels affected Golgi apparatus order in the presence of a drug that fragments the Golgi apparatus (22). STX5 is also linked to rebuilding Golgi apparatus morphology following mitosis. The Golgi apparatus is fragmented during mitosis and is subsequently reorganized into stacks from these fragments. STX5 is the only known factor to participate in both the p97/VCP and NSF-mediated pathways of Golgi apparatus reassembly following mitosis (23). During infection, the Golgi apparatus undergoes similar morphological alterations. First, the canonical Golgi apparatus must be fragmented, and we observe disordering of the Golgi compartment as early as 24 hpi (Fig. 1D). This process is unaffected by Retro94 and thus appears to be STX5 independent. During infection these fragments are not rebuilt into Golgi stacks but, instead, reassemble into the observed ring. The appearance of this ring correlates temporally with the increase in STX5 protein. Thus, STX5 may promote construction of the Golgi ring in a similar manner as it does to reassemble the Golgi stacks after mitosis (Fig. 6C). In the absence of functional STX5, we observed the accumulation of Golgi complex fragments in a juxtanuclear position, unable to form the Golgi rings. Future studies investigating a direct role for STX5 and Golgi apparatus reassembly pathways in Golgi ring formation are warranted.
Identifying an important role for STX5 in infection allowed us to take advantage of a compound that disrupts STX5 to block the efficient production of HCMV virions. The parent compound of this derivative was originally identified for its ability to block the entry of bacterial toxins, which require retrograde transport for entry (15). Viruses such as polyomavirus and papillomaviruses require the same retrograde transport for entry into cells, and consequently these compounds effectively blocked the establishment of these viral infections (17, 24, 25). Efficacy against viruses such as HCMV that do not share this entry pathway has not been previously reported. The low-nanomolar concentration required for 50% inhibition of infectious virion production, coupled with a lack of toxicity to cells, suggests that Retro94 has potential to be investigated as a novel therapeutic approach for HCMV.
MATERIALS AND METHODS
Tissue culture.
Normal human dermal fibroblasts ([NHDFs] 106-05N; Cell Applications, Inc.) and normal human lung fibroblasts, MRC-5 cells (ATCC CCL-171), were maintained in Dulbecco's modified Eagle's medium (DMEM) (Lonza). Human retinal pigmented epithelial cells (ARPE-19), maintained in DMEM/Ham's F-12 medium (1:1 mix) (Lonza), and mouse embryonic fibroblasts (NIH 3T3), maintained in DMEM (Lonza), were obtained from David Spector (20, 23). All media were supplemented with 10% fetal bovine serum, 100 U/ml penicillin, 100 g/ml streptomycin, and 2 mM GlutaMAX (Gibco). Cells were maintained at 37°C with 5% CO2. Trypan blue was used for studies assaying cell viability.
Cloning and bacterial artificial chromosome (BAC) mutagenesis.
pTRE3G-Bi and pEF1a-Tet3G (catalog number 631340; Clontech) were used to create inducible miRNA expression vectors. A DNA oligonucleotide encoding the full-length miRNA construct with BamHI and NotI overhangs was inserted into pTRE3G-Bi MCS1 using BamHI and NotI enzymes (5′-GATCCTGGAGGCTTGCTGAAGGCTGTATGCTCAGGACCCATGGCCTGTTACTAGCACTCACATGGAACAAATGGCCCA). The miRNA construct was created with an internal NcoI sequence to insert specific miRNA sequences into the full-length construct. miRNAs targeting STX5 and luciferase were designed using Block-iT RNAi Designer (ThermoFisher Scientific), and complementary DNA oligonucleotides encoding the gene-specific sequences were annealed and subsequently inserted using BamHI and NcoI enzymes, as follows: miSTX5, 5′-GATCCTGGAGGCTTGCTGAAGGCTGTATGCTGAGACACCATGCAGAACATTGAGTTTTGGCCACTGACTGACTCAATGTTGCATGGTGTCTCAGGACC (sense) and 5′-CATGGGTCCTGAGACACCATGCAACATTGAGTCAGTCAGTGGCCAAAACTCAATGTTCTGCATGGTGTCTCAGCATACAGCCTTCAGCAAGCCTCCAG (antisense); miLUC, 5′-GATCCTGGAGGCTTGCTGAAGGCTGTATGCTGAAATCGCTGATTTGTGTAGTCGTTTTGGCCACTGACTGACGACTACACATCAGCGATTTCAGGACC (sense) and 5′-CATGGGTCCTGAAATCGCTGATGTGTAGTCGTCAGTCAGTGGCCAAAACGACTACACAAATCAGCGATTTCAGCATACAGCCTTCAGCAAGCCTCCAG (antisense). To insert enhanced GFP (EGFP) into pTRE3G-Bi MCS2, EGFP was amplified using the forward primer 5′-GCATGAATTCATGGTGAGCAAGGGCGAG-3′ and the reverse primer 5′-GCATTCTAGATTACTTGTACAGCTCGTC-3′ and inserted using EcoRI and XbaI enzymes.
The pp28-mCherry virus was made by PCR amplification of galK with the forward primer 5′-AACGTCCACCCACCCCCGGGACAAAAAAGCCCGCCGCCTCCTTGCCCTTTCCTGTTGACAATTAATCATCGGCA and reverse primer 5′-TGTCCCATTCCCGACTCGCGAATCGTACGCGAGACCTGAAAGTTTATGAGTCAGCACTGTCCTGCTCCTT. mCherry was amplified and inserted into the galK BAC using the forward primer 5′-AACGTCCACCCACCCCCGGGACAAAAAAGCCCGCCGCCTCCTTGCCCTTTCGGGATCCCGCCACCATGGTG and reverse primer 5′-TGTCCCATTCCCGACTCGCGAATCGTACGCGAGACCTGAAAGTTTATGAGTTACTTGTACAGCTCGTCCA.
The pp150-GFP virus was made by PCR amplification of galK with the forward primer 5′-GCCGTGCAGAACATCCTCCAAAAGATCGAGAAGATTAAGAACACGGAGGAACCTGTTGACAATTAATCATCGGCA and reverse primer 5′-ACACGTCACTATCCGATGGTTTCATTAAAAAGTACGTCTGCGTGTGTGTTTCTTAATCAGCACTGTCCTGCTCCTT. EGFP was amplified and inserted into the galK BAC using the forward primer 5′-GCCGTGCAGAACATCCTCCAAAAGATCGAGAAGATTAAGAACACGGAGGAACCACCGGTCGCCACCATGGTGAGC and reverse primer 5′-ACACGTCACTATCCGATGGTTTCATTAAAAAGTACGTCTGCGTGTGTGTTTCTTAATTACTTGTACAGCTCGTCCATGCC.
To make tetracycline (Tet)-inducible miRNA-expressing viruses, multiple recombineering steps were performed. First, the EF1a-Tet3G construct was inserted into the TRS1 area of the BAC genome. Since pEF1a-Tet3G and pTRE3G-Bi both contain simian virus 40 (SV40) termination sequences, the SV40 terminator on EF1a-Tet3G was replaced with the bovine growth hormone (BGH) termination sequence through a PCR sewing step to avoid additional sites for potential recombination to occur. To insert EF1a-Tet3G-BGH, the GalK sequence was inserted upstream of TRS1 using the forward primer 5′-CGTTTGCCGTTGGGCGTACGCTACGTTTGTATTTCTGGCTATAATATGTGCCTGTTGACAATTAATCATCGGCA and reverse primer 5′-ATACACCCTACAGTCACACCCTTCCCAATAGGAACATCGACACATGACCGTCAGCACTGTCCTGCTCCTT. EF1a-Tet3G-BGH was amplified and inserted into the galK BAC using the forward primer 5′-CGTTTGCCGTTGGGCGTACGCTACGTTTGTATTTCTGGCTATAATATGTGGAGTAATTCATACAAAAG and reverse primer 5′-ATACACCCTACAGTCACACCCTTCCCAATAGGAACATCGACACATGACCGCCATAGAGCCCACCGCATCC. To insert constructs under the control of the bidirectional promoter, the GalK sequence was first inserted downstream of US34a area using the forward primer 5′-AGGGTGGCGAGGTGTGAGGATGAAACATATGCAGATACGCAGTGTTGTTACCTGTTGACAATTAATCATCGGCA and reverse primer 5′-GACTTTCATACTGAAGTACCGTTGTACGCATTACACGGGTTTCGTTCGGATCAGCACTGTCCTGCTCCTT. The GFP-Tet promoter miRNA sequence was then inserted by amplifying the miRNA-containing plasmid with the forward primer 5′-AGGGTGGCGAGGTGTGAGGATGAAACATATGCAGATACGCAGTGTTGTTAAAGTGCCACCTGACGTCG and reverse primer 5′-GACTTTCATACTGAAGTACCGTTGTACGCATTACACGGGTTTCGTTCGGAGTGAGCGAGGAAGCTCGG-3′.
Recombinant viruses were made by BAC mutagenesis using Escherichia coli strain SW105 and galK selection as described previously (26). Briefly, overnight cultures grown in YENB (0.75% yeast extract, 0.8% nutrient broth) medium with 12.5 μg/ml chloramphenicol at 32.5°C were expanded and grown to mid-log phase. Following a heat shock at 42°C, 50 μl of cells was then electroporated (2.5 kV/25 μFD, 200 Ω) (Bio-Rad Gene-Pulser) with purified PCR DNA products. Following a recovery period, bacteria were washed in M9 salts and plated on selection medium. For galK selection, cells were plated on M63 minimal plates containing galactose and replated on MacConkey agar for confirmation of presence of GalK. For gene-of-interest selection, cells were grown on plates containing 2-deoxygalactose. BAC DNA was isolated, and insertion sites were amplified and verified by Sanger sequencing (Eurofins or Genewiz).
Virus preparation, titration, infections, and growth curves.
Titered MCMV (Smith; ATCC VR-1399) and HCMV TB40-mCherry, Toledo, Towne, and Merlin stocks were provided by David Spector (20, 23). AD169 and its above-named derivatives were generated from BAC stocks. To produce virus stocks, BAC DNA was electroporated into MRC5 cells according to previously published protocols (23). Virus-producing cells were saved at passage zero (P0) in liquid nitrogen and used to infect cells in roller bottles to produce larger stocks of virus. Virus stocks were concentrated by high-speed centrifugation on a 20% sorbitol cushion at 20,000 rpm for 1 h at 20°C in a Beckman SW32 rotor.
Infections were done at an MOI of 0.05 for multistep growth curves and at an MOI of 3 for single-step growth curves. Infections were done on NHDFs unless otherwise noted. Virus was incubated with cells for 2 h before addition of fresh medium. For growth curve analyses, virus was harvested at the time points indicated in the figure legends for each experiment by scraping the cells into the medium, sonicating the samples 10 times with 1-s pulses, vortexing them for 15 s, and centrifuging them at 13,000 rpm for 10 min. Supernatants were collected, aliquoted, and flash-frozen in liquid nitrogen before storage at −80°C.
For MCMV growth curve analysis, samples were titrated on NIH 3T3 cells, and titers were calculated using the 50% tissue culture infective dose method. Both HCMV virus stocks and samples for growth curves were titrated by serial dilutions on MRC5 cells and quantified by the immunological detection of immediate early proteins, as previously described (27). The numbers of stained nuclei were acquired on a Nikon Eclipse Ti inverted microscope, and fluorescent nuclei were quantified using NIS Elements software. Images of viral foci from infection of ARPE-19 cells were also acquired on an Eclipse Ti inverted microscope. The number and size of foci were quantified using NIS Elements software.
Reagents and antibodies.
Doxycycline (Enzo Life Sciences) was added to the medium at 2 h postinfection at 10 μg/ml (22.5 μM), and the medium was supplemented every 3 days. Acyclovir was purchased from EMD Millipore and used at a concentration of 100 μg/ml (445 μM). Cytomegalovirus gB late antigen antibody (C58G) and beta-actin antibody (BA3R) were purchased from ThermoFisher Scientific. Anti-pp28 (5C3) was purchased from Abcam, Inc., and an antibody against STX5 (Western analysis; B-8) was purchased from Santa Cruz. Antibodies against GM130 and p230 were purchased from BD Transduction Laboratories. Antibodies against STX18 and STX5 (immunofluorescence) were from Synaptic Systems. Antibodies against syntaxin 7 (Bethyl Laboratories), syntaxin 16 (Proteintech), and α-tubulin (Sigma-Aldrich) were also purchased. A rabbit polyclonal antibody that detects exons 2 and 3 of the HCMV major immediate early proteins was a kind gift from Jim Alwine (28). Horseradish peroxidase-conjugated secondary antibodies for Western blotting detection were purchased from Santa Cruz (goat anti-rabbit) and Jackson Laboratories (goat anti-mouse). Fluorescein isothiocyanate (FITC)- and rhodamine-conjugated secondary antibodies for immunofluorescence staining were purchased from Santa Cruz Biotechnology, Inc.
Retro94 synthesis and use.
1-Methyl-2-(5-(2-methylthiazol-4-yl)thiophen-2-yl)-3phenyl-2,3-dihydroquinazolin-4(1H)-one (Retro94) was synthesized as reported in the literature with minor modifications (16). Briefly, to a solution of 2-aminobenzanilide (1 mmol) in methanol (3 ml) was added 5-(2-methyl-3-thiazol-4-yl)-2-thiophencarboxaldehyde (1 mmol), and the reaction mixture was stirred at room temperature overnight. The precipitated yellow imino compound was filtered and washed with a small quantity of methanol to give a 62% yield.
The imino compound (0.75 mmol) was dissolved in anhydrous tetrahydrofuran (THF) (10 ml) and reacted with NaH (60% in oil washed with hexane; 2.5 mmol) at 0°C for 4 to 6 h, and iodomethane (3 mmol) was added dropwise at 0°C. After the addition, the reaction mixture was allowed to warm to room temperature and stirred overnight. The reaction mixture was carefully quenched with saturated NaHCO3. The aqueous layer was extracted with CH2Cl2 (four times at 25 ml), washed with 1N HCl (two times at 10 ml), dried over MgSO4, filtered, and evaporated to dryness. The crude product was purified over silica gel column by eluting with ethyl acetate (EtOAc)-hexane (2:8) to give Retro94 in an overall 71% yield. 1H nuclear magnetic resonance (NMR) (500 mHz, CDCl3) δ (ppm) 8.12 (dd, 1H, J = 6.5 Hz, J′ = 1.5 Hz), 7.46 to 7.40 (m, 1H), 7.40 to 7.24 (m, 5H), 7.12 (s, 1H), 6.99 (dt, 1H, J = 8.5 Hz, j′ = 1 Hz), 6.84 (d, 1H, J = 3.0 Hz), 6.69 (d, 1H, j = 7.0 Hz), 5.99 (s, 1H), 3.02 (s, 3H, NCH3), 2.74 (s, 3H, CH3); mass spectrum (electrospray ionization) [M = H]+, 417.8.
1H NMR spectra were recorded on a 500 MHz Bruker spectrometer (Billerica, MA) using tetramethylsilane (TMS) as the internal reference. The following abbreviations were used to designate chemical shift multiplicities: s, singlet; d, doublet; dd, double doublet; t, triplet; dt, doublet of triplet; m, multiplet. Mass spectrometry (MS) analysis was performed on 4000 Q trap hybrid triple quadrupole/linear ion trap instrument (Applied Biosystems/MDS Sciex) at the proteomic facility at Penn State Hershey Cancer Institute, Hershey, PA. Thin-layer chromatography (TLC) was on aluminum-supported, precoated silica gel plates (EM Industries, Gibbstown, NJ). 5-(2-Methyl-3-thiazol-4-yl)-2-thiophencarboxaldehyde was purchased from Maybridge-Thermo Fisher (Cornwall, United Kingdom). All other starting materials were obtained from Aldrich Chemical Co. (Milwaukee, WI) and used without further purification. Retro94 was added to cells at 2 hpi at a concentration of 10 μM unless otherwise indicated.
Western blotting.
Total cell lysates were prepared from uninfected or HCMV-infected cells (MOI of 3) at the time points indicated in the figure legends by harvesting lysates in radioimmunoprecipitation assay buffer (1% NP-40, 1% sodium deoxycholate, 0.1% sodium dodecyl sulfate [SDS], 0.15 M NaCl, 10 mM sodium phosphate [pH 7.2], 2 mM EDTA, 50 mM NaF, 1 mM phenylmethylsulfonyl fluoride, 1 mM aprotinin, 0.2 mM Na3VO4, 1 μg/ml leupeptin). After centrifugation, the supernatant was collected and stored at −80°C. The protein was quantified by a Bradford assay (Bio-Rad), and equal protein amounts (15 or 20 μg) were prepared for SDS-PAGE and immunoblotting on nitrocellulose or polyvinylidene difluoride (PVDF) membrane blocked with 5% milk or 5% bovine serum albumin (BSA) in Tris-buffered saline (0.1% Tween). Antibody dilutions were in accordance with the manufacturer's instructions.
DNA replication.
For DNA replication analysis, cells were infected at an MOI of 3 and harvested at 24, 48, 72, and 96 h postinfection. DNA was isolated using a DNeasy blood and tissue kit (Qiagen). Quantitative PCR (qPCR) was performed on the extracted DNA using the following primers: HCMV UL99, 5′-GTGTCCCATTCCCGACTCG-3′ (forward) and 5′-TTCACAACGTCCACCCACC-3′ (reverse); β-globin, 5′-ACGTGGATGAAGTTGGTGGT (forward) and 5′-GCCCAGTTTCTATTGGTCTCC (reverse). To generate a standard curve for UL99, 10-fold dilutions (2.77 × 109 to 2.77 × 103 genome copies) of the HCMV XbaI plasmid C (provided by David Spector) was subjected to qPCR analysis. A standard curve for β-globin was obtained by diluting the β-globin sequence from 8.86 × 1010 to 8.86 × 105 genome copies. UL99 genomic equivalents were normalized to the β-globin level.
Immunofluorescence microscopy and imaging.
Coverslips or chamber slides containing either uninfected or HCMV-infected cells were washed in phosphate-buffered saline (PBS) and fixed in 2% paraformaldehyde for 15 min at room temperature. Cells were blocked in PBS containing 10% human serum, 0.5% Tween 20, and 5% glycine. Triton X-100 (0.1%) was added for permeabilization. Primary and secondary antibodies as specified in the antibody section were diluted in blocking buffer. Nuclei were stained with 4′,6′-diamidino-2-phenylindole (DAPI) prior to mounting with ProLong Diamond Antifade Mountant (ThermoScientific). All images were acquired using a C2+ Confocal Microscope System (Nikon). Images were processed using NIS Elements software. Images shown are volume renderings of z-stacks. Images of monolayers were acquired on a Nikon Eclipse TS100 microscope with Infinity3-1URFC camera and Infinity software.
EM.
Cells were seeded on 60-mm Permanox tissue culture dishes (Nalge Nunc International). Infections were done at an MOI of 3 for 96 h. Cells were first washed with PBS three times, followed by fixation for 1 h at 4°C in fixation buffer (0.5% [vol/vol] glutaraldehyde, 0.04% [wt/vol] paraformaldehyde, 0.1 M sodium cacodylate). Cells were then processed for electron microscopy (EM) by the Microscopy Imaging Facility (Pennsylvania State University College of Medicine). Briefly, the fixed samples were washed three times with 0.1 M sodium cacodylate, followed by postfixation in 1% osmium–1.5% potassium ferrocyanide overnight at 4°C. Samples were then washed three times in 0.1 M sodium cacodylate, dehydrated with ethanol, and embedded in Epon 812 for staining and sectioning. Images were acquired using a JEOL JEM-1400 Digital Capture transmission electron microscope.
ACKNOWLEDGMENTS
We thank Alex Buchkovich for assistance in generating reagents and viruses. We thank Roland Myers and the Penn State Hershey imaging core for the preparation of EM samples. We particularly thank David Spector for sharing CMV strains, cell types, and other reagents and for many helpful discussions and insights. We thank Aron Lukacher and Saumya Maru for discussions on Retro94.
This work was supported by the Pennsylvania State University College of Medicine and NIH Training Grant T32 CA60395 (L.C.).
REFERENCES
- 1.Das S, Vasanji A, Pellett PE. 2007. Three-dimensional structure of the human cytomegalovirus cytoplasmic virion assembly complex includes a reoriented secretory apparatus. J Virol 81:11861–11869. doi: 10.1128/JVI.01077-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sanchez V, Greis KD, Sztul E, Britt WJ. 2000. Accumulation of virion tegument and envelope proteins in a stable cytoplasmic compartment during human cytomegalovirus replication: characterization of a potential site of virus assembly. J Virol 74:975–986. doi: 10.1128/JVI.74.2.975-986.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bughio F, Umashankar M, Wilson J, Goodrum F. 2015. Human cytomegalovirus UL135 and UL136 genes are required for postentry tropism in endothelial cells. J Virol 89:6536–6550. doi: 10.1128/JVI.00284-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hook LM, Grey F, Grabski R, Tirabassi R, Doyle T, Hancock M, Landais I, Jeng S, McWeeney S, Britt W, Nelson JA. 2014. Cytomegalovirus miRNAs target secretory pathway genes to facilitate formation of the virion assembly compartment and reduce cytokine secretion. Cell Host Microbe 15:363–373. doi: 10.1016/j.chom.2014.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Buchkovich NJ, Maguire TG, Yu Y, Paton AW, Paton JC, Alwine JC. 2008. Human cytomegalovirus specifically controls the levels of the endoplasmic reticulum chaperone BiP/GRP78, which is required for virion assembly. J Virol 82:31–39. doi: 10.1128/JVI.01881-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Xie M, Xuan B, Shan J, Pan D, Sun Y, Shan Z, Zhang J, Yu D, Li B, Qian Z. 2015. Human cytomegalovirus exploits interferon-induced transmembrane proteins to facilitate morphogenesis of the virion assembly compartment. J Virol 89:3049–3061. doi: 10.1128/JVI.03416-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Das S, Ortiz DA, Gurczynski SJ, Khan F, Pellett PE. 2014. Identification of human cytomegalovirus genes important for biogenesis of the cytoplasmic virion assembly complex. J Virol 88:9086–9099. doi: 10.1128/JVI.01141-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Buchkovich NJ, Maguire TG, Paton AW, Paton JC, Alwine JC. 2009. The endoplasmic reticulum chaperone BiP/GRP78 is important in the structure and function of the human cytomegalovirus assembly compartment. J Virol 83:11421–11428. doi: 10.1128/JVI.00762-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Buchkovich NJ, Maguire TG, Alwine JC. 2010. Role of the endoplasmic reticulum chaperone BiP, SUN domain proteins, and dynein in altering nuclear morphology during human cytomegalovirus infection. J Virol 84:7005–7017. doi: 10.1128/JVI.00719-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Indran SV, Ballestas ME, Britt WJ. 2010. Bicaudal D1-dependent trafficking of human cytomegalovirus tegument protein pp150 in virus-infected cells. J Virol 84:3162–3177. doi: 10.1128/JVI.01776-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Krzyzaniak MA, Mach M, Britt WJ. 2009. HCMV-encoded glycoprotein M (UL100) interacts with Rab11 effector protein FIP4. Traffic 10:1439–1457. doi: 10.1111/j.1600-0854.2009.00967.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu ST, Sharon-Friling R, Ivanova P, Milne SB, Myers DS, Rabinowitz JD, Brown HA, Shenk T. 2011. Synaptic vesicle-like lipidome of human cytomegalovirus virions reveals a role for SNARE machinery in virion egress. Proc Natl Acad Sci U S A 108:12869–12874. doi: 10.1073/pnas.1109796108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cepeda V, Fraile-Ramos A. 2011. A role for the SNARE protein syntaxin 3 in human cytomegalovirus morphogenesis. Cell Microbiol 13:846–858. doi: 10.1111/j.1462-5822.2011.01583.x. [DOI] [PubMed] [Google Scholar]
- 14.Fasshauer D, Sutton RB, Brunger AT, Jahn R. 1998. Conserved structural features of the synaptic fusion complex: SNARE proteins reclassified as Q- and R-SNAREs. Proc Natl Acad Sci U S A 95:15781–15786. doi: 10.1073/pnas.95.26.15781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stechmann B, Bai SK, Gobbo E, Lopez R, Merer G, Pinchard S, Panigai L, Tenza D, Raposo G, Beaumelle B, Sauvaire D, Gillet D, Johannes L, Barbier J. 2010. Inhibition of retrograde transport protects mice from lethal ricin challenge. Cell 141:231–242. doi: 10.1016/j.cell.2010.01.043. [DOI] [PubMed] [Google Scholar]
- 16.Noel R, Gupta N, Pons V, Goudet A, Garcia-Castillo MD, Michau A, Martinez J, Buisson DA, Johannes L, Gillet D, Barbier J, Cintrat JC. 2013. N-Methyldihydroquinazolinone derivatives of Retro-2 with enhanced efficacy against Shiga toxin. J Med Chem 56:3404–3413. doi: 10.1021/jm4002346. [DOI] [PubMed] [Google Scholar]
- 17.Carney DW, Nelson CD, Ferris BD, Stevens JP, Lipovsky A, Kazakov T, DiMaio D, Atwood WJ, Sello JK. 2014. Structural optimization of a retrograde trafficking inhibitor that protects cells from infections by human polyoma- and papillomaviruses. Bioorg Med Chem 22:4836–4847. doi: 10.1016/j.bmc.2014.06.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gupta N, Pons V, Noël R, Buisson DA, Michau A, Johannes L, Gillet D, Barbier J, Cintrat JC. 2014. (S)-N-Methyldihydroquinazolinones are the active enantiomers of retro-2 derived compounds against toxins. ACS Med Chem Lett 5:94–97. doi: 10.1021/ml400457j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Parlati F, Varlamov O, Paz K, McNew JA, Hurtado D, Söllner TH, Rothman JE. 2002. Distinct SNARE complexes mediating membrane fusion in Golgi transport based on combinatorial specificity. Proc Natl Acad Sci U S A 99:5424–5429. doi: 10.1073/pnas.082100899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Miyazaki K, Wakana Y, Noda C, Arasaki K, Furuno A, Tagaya M. 2012. Contribution of the long form of syntaxin 5 to the organization of the endoplasmic reticulum. J Cell Sci 125:5658–5666. doi: 10.1242/jcs.105304. [DOI] [PubMed] [Google Scholar]
- 21.Weinberger A, Kamena F, Kama R, Spang A, Gerst JE. 2005. Control of Golgi morphology and function by Sed5 t-SNARE phosphorylation. Mol Biol Cell 16:4918–4930. doi: 10.1091/mbc.E05-02-0101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rendón WO, Martínez-Alonso E, Tomás M, Martínez-Martínez N, Martínez-Menárguez JA. 2013. Golgi fragmentation is Rab and SNARE dependent in cellular models of Parkinson's disease. Histochem Cell Biol 139:671–684. doi: 10.1007/s00418-012-1059-4. [DOI] [PubMed] [Google Scholar]
- 23.Rabouille C, Kondo H, Newman R, Hui N, Freemont P, Warren G. 1998. Syntaxin 5 is a common component of the NSF- and p97-mediated reassembly pathways of Golgi cisternae from mitotic Golgi fragments in vitro. Cell 92:603–610. doi: 10.1016/S0092-8674(00)81128-9. [DOI] [PubMed] [Google Scholar]
- 24.Nelson CD, Carney DW, Derdowski A, Lipovsky A, Gee GV, O'Hara B, Williard P, DiMaio D, Sello JK, Atwood WJ. 2013. A retrograde trafficking inhibitor of ricin and Shiga-like toxins inhibits infection of cells by human and monkey polyomaviruses. mBio 4:e00729-13. doi: 10.1128/mBio.00729-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lipovsky A, Popa A, Pimienta G, Wyler M, Bhan A, Kuruvilla L, Guie MA, Poffenberger AC, Nelson CD, Atwood WJ, DiMaio D. 2013. Genome-wide siRNA screen identifies the retromer as a cellular entry factor for human papillomavirus. Proc Natl Acad Sci U S A 110:7452–7457. doi: 10.1073/pnas.1302164110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Suga K, Hattori H, Saito A, Akagawa K. 2005. RNA interference-mediated silencing of the syntaxin 5 gene induces Golgi fragmentation but capable of transporting vesicles. FEBS Lett 579:4226–4234. doi: 10.1016/j.febslet.2005.06.053. [DOI] [PubMed] [Google Scholar]
- 27.Amessou M, Fradagrada A, Falguières T, Lord JM, Smith DC, Roberts LM, Lamaze C, Johannes L. 2007. Syntaxin 16 and syntaxin 5 are required for efficient retrograde transport of several exogenous and endogenous cargo proteins. J Cell Sci 120:1457–1468. doi: 10.1242/jcs.03436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Harel NY, Alwine JC. 1998. Phosphorylation of the human cytomegalovirus 86-kilodalton immediate-early protein IE2. J Virol 72:5481–5492. [DOI] [PMC free article] [PubMed] [Google Scholar]