ABSTRACT
Middle East respiratory syndrome coronavirus (MERS-CoV) binds to cellular receptor dipeptidyl peptidase 4 (DPP4) via the spike (S) protein receptor-binding domain (RBD). The RBD contains critical neutralizing epitopes and serves as an important vaccine target. Since RBD mutations occur in different MERS-CoV isolates and antibody escape mutants, cross-neutralization of divergent MERS-CoV strains by RBD-induced antibodies remains unknown. Here, we constructed four recombinant RBD (rRBD) proteins with single or multiple mutations detected in representative human MERS-CoV strains from the 2012, 2013, 2014, and 2015 outbreaks, respectively, and one rRBD protein with multiple changes derived from camel MERS-CoV strains. Like the RBD of prototype EMC2012 (EMC-RBD), all five RBDs maintained good antigenicity and functionality, the ability to bind RBD-specific neutralizing monoclonal antibodies (MAbs) and the DPP4 receptor, and high immunogenicity, able to elicit S-specific antibodies. They induced potent neutralizing antibodies cross-neutralizing 17 MERS pseudoviruses expressing S proteins of representative human and camel MERS-CoV strains identified during the 2012-2015 outbreaks, 5 MAb escape MERS-CoV mutants, and 2 live human MERS-CoV strains. We then constructed two RBDs mutated in multiple key residues in the receptor-binding motif (RBM) of RBD and demonstrated their strong cross-reactivity with anti-EMC-RBD antibodies. These RBD mutants with diminished DPP4 binding also led to virus attenuation, suggesting that immunoevasion after RBD immunization is accompanied by loss of viral fitness. Therefore, this study demonstrates that MERS-CoV RBD is an important vaccine target able to induce highly potent and broad-spectrum neutralizing antibodies against infection by divergent circulating human and camel MERS-CoV strains.
IMPORTANCE MERS-CoV was first identified in June 2012 and has since spread in humans and camels. Mutations in its spike (S) protein receptor-binding domain (RBD), a key vaccine target, have been identified, raising concerns over the efficacy of RBD-based MERS vaccines against circulating human and camel MERS-CoV strains. Here, we constructed five vaccine candidates, designated 2012-RBD, 2013-RBD, 2014-RBD, 2015-RBD, and Camel-RBD, containing single or multiple mutations in the RBD of representative human and camel MERS-CoV strains during the 2012-2015 outbreaks. These RBD-based vaccine candidates maintained good functionality, antigenicity, and immunogenicity, and they induced strong cross-neutralizing antibodies against infection by divergent pseudotyped and live MERS-CoV strains, as well as antibody escape MERS-CoV mutants. This study provides impetus for further development of a safe, highly effective, and broad-spectrum RBD-based subunit vaccine to prevent MERS-CoV infection.
KEYWORDS: MERS, MERS-CoV, spike protein, receptor-binding domain, multiple strains, antibody escape mutants, cross-neutralization
Middle East respiratory syndrome (MERS) is caused by a newly emerged coronavirus, MERS coronavirus (MERS-CoV) (1). This new coronavirus was first identified in Saudi Arabia in June 2012 and has since infected at least 1,806 individuals worldwide, with 643 deaths (mortality rate, 36%), as of 29 September 2016, in 27 countries (http://www.who.int/emergencies/mers-cov/en/). MERS-CoV is derived from zoonotic sources, with bats as its probable original reservoirs and dromedary camels as key intermediate hosts. Thus, animal-to-human transmission of MERS-CoV is believed to be the major route for primary MERS-CoV infection (2–10). Nevertheless, MERS-CoV has gained the ability to infect humans via human-to-human transmission, particularly in health care facilities where patients are concentrated and infection prevention control is limited, as seen in the recent “MERS-CoV superspreading” event in South Korea (11–15). Thus, rapid development of broad-spectrum, effective, and safe vaccines is urgently needed to prevent MERS-CoV infection.
MERS-CoV spike (S) protein plays a major role in virus infection and pathogenesis by binding to the cellular receptor dipeptidyl peptidase 4 (DPP4) through the receptor-binding domain (RBD) in the S1 subunit, followed by fusion between virus and cell membranes through the S2 subunit (16–19). Our previous studies have demonstrated that the MERS-CoV RBD contains a critical neutralizing domain (CND; residues 377 to 588), which is able to induce highly potent neutralizing antibodies that protect vaccinated human DPP4-transgenic (hDPP4-Tg)- and adenovirus 5 (Ad5)-hDPP4-transduced mice from challenge with MERS-CoV EMC2012, a prototypic virus strain, suggesting that the MERS-CoV RBD, particularly the fragment containing the identified CND, can be a major target for MERS vaccine development (20–28).
As the virus evolved during its spread in humans, key mutations, including L506F, D509G, and D510G, were detected in the RBD of various MERS-CoV strains isolated from different regions and at different times throughout the course of the MERS outbreak from 2012 to 2015 (Table 1) (29, 30). In addition, the RBD sequences of MERS-CoV from dromedary camels are distinct from those of infected humans (Table 1), suggesting that the key residues in RBD susceptible to mutation may play critical roles in MERS-CoV transmission. Furthermore, using RBD-specific neutralizing monoclonal antibodies (MAbs), several important antibody escape mutations, such as those at residues 511 and 513, have been identified in the MERS-CoV RBD (31–34). This tendency of MERS-CoV RBD to mutate over time may facilitate viral evasion of cross-neutralizing antibodies present in humans and camels previously infected with MERS-CoV or immunized with S protein-based vaccines. In specific, it is essential to investigate whether RBD-based MERS vaccines under development can be effective against MERS-CoV strains now in circulation in humans and camels.
TABLE 1.
Representative MERS-CoV strains isolated in the 2012-2015 outbreaks and their mutations in MERS-CoV RBD containing residues 377 to 588a
| GenBank protein ID | Yr isolated | Host | Region | Mutation in MERS-CoV RBD residue: |
No. of mutations | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 400 | 424 | 431 | 434 | 457 | 460 | 482 | 506 | 509 | 510 | 520 | 522 | 529 | 530 | 534 | 582 | |||||
| AFS88936.1 | 2012 | Human | EMC | K | T | A | A | S | S | A | L | D | D | A | Q | I | V | V | N | 0 |
| AFY13307.1 | 2012 | Human | England | F | 1 | |||||||||||||||
| AGG22542.1 | 2012 | Human | England | F | 1 | |||||||||||||||
| AGV08379.1 | 2012 | Human | KSA | G | 1 | |||||||||||||||
| AGV08584.1 | 2012 | Human | KSA | A | 1 | |||||||||||||||
| AHI48528.1 | 2013 | Human | KSA | P | V | 2 | ||||||||||||||
| AHI48733.1 | 2013 | Human | KSA | V | 1 | |||||||||||||||
| AHC74088.1 | 2013 | Human | Qatar | F | 1 | |||||||||||||||
| AGV08438.1 | 2013 | Human | KSA | I | 1 | |||||||||||||||
| AID55090.1 | 2014 | Human | KSA | I | 1 | |||||||||||||||
| AID55095.1 | 2014 | Human | KSA | I | 1 | |||||||||||||||
| AID55087.1 | 2014 | Human | KSA | H | 1 | |||||||||||||||
| AKL59401.1 | 2015 | Human | Korea | L | 1 | |||||||||||||||
| ALB08322.1 | 2015 | Human | Korea | G | 1 | |||||||||||||||
| ALB08289.1 | 2015 | Human | Korea | T | 1 | |||||||||||||||
| AHY22545.1 | 2013 | Camel | KSA | N | 1 | |||||||||||||||
| AHL18090.1 | 2013 | Camel | Egypt | S | 1 | |||||||||||||||
| AHX00711.1 | 2013 | Camel | KSA | G | 1 | |||||||||||||||
| AHX00721.1 | 2013 | Camel | KSA | G | 1 | |||||||||||||||
| AHY22555.1 | 2013 | Camel | KSA | S | 1 | |||||||||||||||
EMC, Erasmus Medical Center; KSA, Saudi Arabia. Key residues in the RBM of the RBD are in bold.
The RBD of MERS-CoV is composed of a core subdomain and an external receptor-binding motif (RBM; residues 484 to 567). The RBM is the main domain interacting with the DPP4 receptor, and it is defined by a number of key residues, including L506, D509, D510, R511, and E513, which, in the aggregate, determine receptor-binding properties and subsequent viral entry into target cells (19, 35, 36). However, only some of these key residues in the RBM of current MERS-CoV strains have been identified (29, 37). Although these key mutations are not fixed in isolated MERS-CoV strains, it is important to note that S protein, particularly the RBM, continues to undergo strong positive selection during virus transmission (38). Consequently, it is possible that key mutations in the RBM of RBD might accumulate in one single virus in the course of viral evolution, resulting in the emergence of immune escape virus strains. Thus, to improve our understanding of virus escape mutants relative to viral fitness, it is important to establish whether simultaneous changes of multiple key residues in the RBM of RBD will alter the antigenicity, functionality, and immunogenicity of the RBD.
In this study, we initially constructed five recombinant RBD proteins, designated 2012-RBD, 2013-RBD, 2014-RBD, 2015-RBD, and Camel-RBD. They contain single or multiple mutations in the RBD of representative human MERS-CoV strains circulating in the 2012, 2013, 2014, and 2015 outbreaks or several mutations noted in the camel RBD. We evaluated whether RBDs with these scattered mutations would maintain their antigenicity, functionality, and immunogenicity. We also evaluated the cross-neutralizing activity of the antibodies induced by these RBDs against divergent human and camel MERS-CoV strains, as well as antibody escape mutants of MERS-CoV. Two additional RBDs, RBD-FGG and RBD-FGGAA, which contain mutations of 3 and 5 key residues in the RBM of RBD, respectively, were constructed. Our results demonstrate strong cross-reactivity when mice were immunized with wild-type or variant RBDs. They demonstrate that RBD mutations with diminished DPP4 binding also led to virus attenuation, suggesting that immunoevasion after RBD immunization may result only in the context of loss of viral fitness.
RESULTS
Recombinant RBD proteins of representative human and camel MERS-CoV strains in 2012 to 2015 maintained good conformation and antigenicity.
The RBD sequences of MERS-CoVs isolated from various infection regions, different time periods (2012 to 2015), and different hosts (humans and camels) are slightly different from the RBD sequence of EMC2012, the prototype strain. The mutations are summarized in Table 1.
Accordingly, we initially constructed 5 recombinant RBD (rRBD) proteins (2012-RBD, 2013-RBD, 2014-RBD, 2015-RBD, and Camel-RBD) containing single and multiple natural mutations in the critical neutralizing domain (CND) of RBD of representative human MERS-CoV strains isolated from 2012 to 2015 and representative camel MERS-CoV strains, respectively (Table 2; Fig. 1A and B). These proteins, which were fused with a C-terminal human Fc tag, were characterized by SDS-PAGE and Western blot analysis. Similar to the wild-type RBD (EMC-RBD), the five RBD mutants of native (nonboiled) proteins had twice the molecular mass of those that were boiled (denatured) proteins (Fig. 1C, top), suggesting that the Fc tags promoted dimer formation. In addition, all RBD proteins of human and camel MERS-CoVs reacted strongly with antibodies targeting the RBD of MERS-CoV EMC2012 (Fig. 1C, bottom).
TABLE 2.
Constructed MERS-CoV RBD fragments containing single or multiple mutations in the RBD of representative MERS-CoV strains isolated in 2012 to 2015 and multiple key mutations in the RBM of the RBD (residues 377 to 588)a
| Protein | Yr isolated | Host | Mutation in MERS-CoV RBD residue: |
No. of mutations | ||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 400 | 424 | 431 | 434 | 457 | 460 | 482 | 506 | 509 | 510 | 511 | 513 | 520 | 522 | 530 | 534 | 582 | ||||
| EMC-RBD | 2012 | Human | K | T | A | A | S | S | A | L | D | D | R | E | A | Q | V | V | N | |
| 2012-RBD | 2012 | Human | F | G | A | 3 | ||||||||||||||
| 2013-RBD | 2013 | Human | P | V | F | V | I | 5 | ||||||||||||
| 2014-RBD | 2014 | Human | I | H | 2 | |||||||||||||||
| 2015-RBD | 2015 | Human | L | 1 | ||||||||||||||||
| Camel-RBD | 2013 | Camel | N | S | G | S | 4 | |||||||||||||
| RBD-FGG | 2012 | Human | F | G | 3 | |||||||||||||||
| 2015 | Human | G | ||||||||||||||||||
| RBD-FGGAA | 2012 | Human | F | G | 5 | |||||||||||||||
| 2015 | Human | G | ||||||||||||||||||
| Escape mutants | A | |||||||||||||||||||
| A | ||||||||||||||||||||
EMC-RBD, RBD fragment constructed based on the sequence of MERS-CoV EMC2012 (prototype) strain. Key residues in the RBM of the RBD are in bold.
FIG 1.
Construction, characterization, and antigenicity of human and camel MERS-CoV RBD proteins. (A) Schematic diagram of MERS-CoV S1 subunit. Residues 1 to 18, signal peptide. RBD, receptor-binding domain, which contains the identified critical neutralizing domain covering residues 377 to 588. (B) Construction of RBDs of divergent human and camel MERS-CoV strains fused with Fc of human IgG. Residues represent single or multiple mutations in the RBD of representative human MERS-CoV strains in 2012 to 2015, designated 2012-RBD, 2013-RBD, 2014-RBD, and 2015-RBD, or MERS-CoV from camels (Camel-RBD) in comparison with the RBD of prototype strain EMC2012 (EMC-RBD). (C) SDS-PAGE and Western blot analysis of purified rRBD proteins. Nonboiled (nondenatured) or boiled (denatured) samples (5 μg) were subjected to SDS-PAGE (top) or Western blotting (bottom), and the binding was tested using MERS-CoV RBD-specific antibody (1:1,000). The molecular mass markers (in kilodaltons) are indicated on the left. (D) Detection of antigenicity of rRBD proteins by ELISA. ELISA plates were coated with respective human and camel RBD proteins or human Fc (hIgG-Fc) control and then incubated with neutralizing mouse MAb Mersmab1 and human MAbs m336, m337, and m338 (1.25 μg/ml), which recognize conformational epitopes in the RBD of MERS-CoV EMC2012. The data are presented as mean A450 ± standard deviation (SD) (n = 4) of RBDs binding to MAbs.
To investigate whether the above-mentioned rRBD proteins of divergent human and camel MERS-CoV strains maintained good antigenicity, we performed an enzyme-linked immunosorbent assay (ELISA) to test the binding activity of these proteins to EMC2012 RBD-specific neutralizing MAbs (33, 34). All mutant and wild-type RBDs bound strongly to mouse MAb Mersmab1 and human MAbs m336, m337, and m338 (Fig. 1D), demonstrating good antigenicity.
Variant rRBD proteins bound strongly to human DPP4 receptor.
A coimmunoprecipitation (Co-IP) assay was initially performed to identify whether the rRBD proteins of representative human and camel MERS-CoV strains circulating in 2012 to 2015 could bind to DPP4, the receptor of MERS-CoV. Strong reactivity to both proteins was observed in the immunoprecipitated samples containing RBD and hDPP4 or RBD and hDPP4-expressing Huh-7 cell lysates. However, hDPP4 in the absence of RBD was recognized only by anti-hDPP4 antibody, not by anti-MERS-CoV-RBD antibody (Fig. 2A). These data suggest that rRBD proteins of representative human and camel MERS-CoV strains in 2012 to 2015 bound efficiently to soluble and cell-associated hDPP4 receptors.
FIG 2.
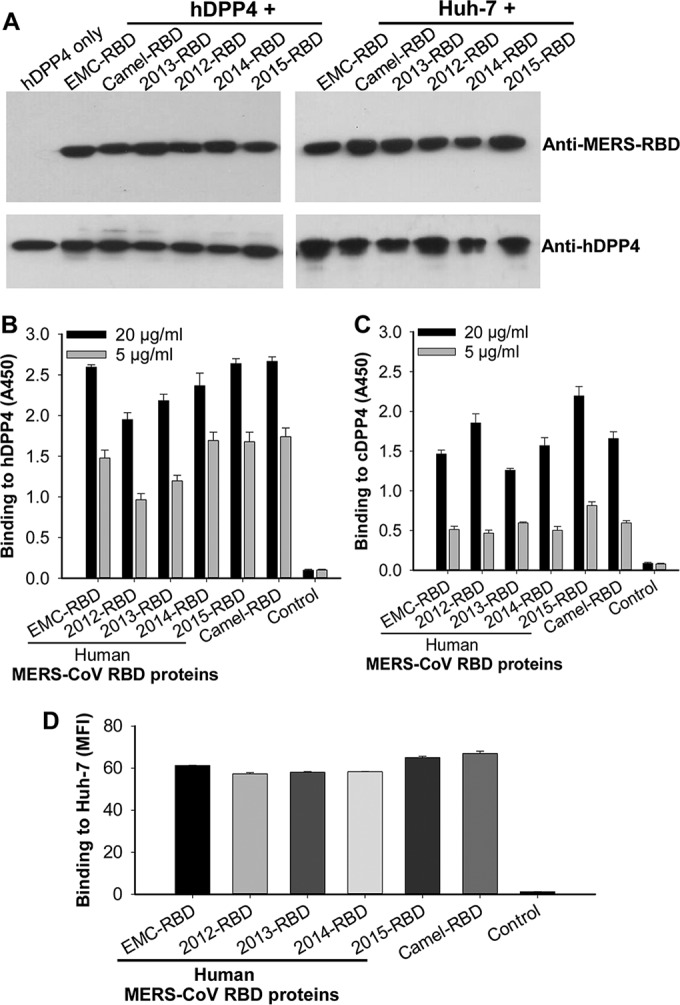
Detection of binding of human and camel MERS-CoV RBD proteins to DPP4 receptor. (A) Co-IP followed by Western blotting of binding between human and camel RBD proteins and soluble hDPP4 protein or cell-associated hDPP4 in Huh-7 cells. Recombinant RBD proteins were incubated with hDPP4 protein (left) or Huh-7 cell lysates (right) plus protein A beads and then detected for binding using MERS-CoV RBD-specific (1:1,000, top) or DPP4-specific (0.5 μg/ml, bottom) antibodies. The hDPP4 protein only was included as a control. Quantification of binding between rRBD proteins and hDPP4 (B) or cDPP4 (C) protein by ELISA. ELISA plates were coated with hDPP4 or cDPP4 protein (2 μg/ml) and then incubated with dilutions of MERS-CoV RBD proteins or hIgG-Fc control. The data are presented as means ± SD (n = 4) of RBD binding to hDPP4 or cDPP4 protein. (D) Quantification of binding between rRBD proteins and cell-associated hDPP4 receptor by flow cytometry analysis. Huh-7 cells were sequentially incubated with rRBD proteins (40 μg/ml) or hIgG-Fc control and FITC-labeled anti-human IgG antibody, followed by analysis for binding. The data are presented as means ± SD (n = 4) of RBD binding to Huh-7-expressed hDPP4 receptor. MFI, median fluorescence intensity.
ELISA and flow cytometry assays were then carried out to quantify the binding between RBD and DPP4. ELISA results demonstrated that the RBDs from multiple human and camel MERS-CoVs bound to both hDPP4 (Fig. 2B) and camel DPP4 (cDPP4) (Fig. 2C) proteins in a dose-dependent manner and that the binding to hDPP4 was much stronger than that of cDPP4 protein. In contrast, no binding was observed between human Fc and hDPP4 or cDPP4 (Fig. 2B and C). Results from flow cytometry analysis also revealed strong binding of these rRBD proteins to Huh-7 cell-associated hDPP4 receptor (Fig. 2D). Taken together, these results confirm the binding specificity and potency between human or camel MERS-CoV RBDs and the hDPP4 receptor.
Recombinant RBD proteins of representative MERS-CoV strains in 2012 to 2015 induced highly potent cross-reactive antibody responses.
The purified rRBD proteins were then evaluated for their capacity to induce cross-reactive antibody responses in immunized mouse sera. All RBDs elicited similarly high titers of IgG antibodies that cross-reacted potently with S1 protein of MERS-CoV EMC2012 (Fig. 3A). Similar to EMC-RBD, 2012-RBD, 2014-RBD, 2015-RBD, and Camel-RBD all induced potent S1-specific IgG1 (Th2) and IgG2a (Th1) antibody responses. In addition, 2013-RBD, which contained 5 mutations spread throughout the RBD, still elicited strong IgG1 and IgG2a antibodies specific to S1 of the EMC2012 strain (Fig. 3B and C). In contrast, phosphate-buffered saline (PBS) control induced only background levels of MERS-CoV-specific antibody (Fig. 3). These data suggest that RBD proteins of representative human and camel MERS-CoV strains in 2012 to 2015 are highly immunogenic in inducing cross-reactive antibody responses.
FIG 3.

Human and camel MERS-CoV RBD proteins induced highly potent cross-reactive antibody responses in immunized mice. PBS was included as a control. Sera from 10 days after the third immunization were tested for IgG (A), IgG1 (B), and IgG2a (C) antibody responses specific to S1 of MERS-CoV prototype strain EMC2012. The antibody titers are expressed as the endpoint dilutions that remain positively detectable, and they are presented as mean antibody titers ± SD for five mice in each group. 2012-RBD, 2013-RBD, 2014-RBD, 2015-RBD, and Camel-RBD represent MERS-CoV strains isolated from humans in 2012 to 2015 and from camels, respectively. EMC-RBD, RBD of MERS-CoV prototype strain EMC2012.
Recombinant RBD proteins of 2012-2015 MERS-CoV strains induced highly potent cross-neutralizing antibodies.
An ideal MERS vaccine should induce strong neutralizing antibodies against divergent MERS-CoV strains. Therefore, we generated a series of pseudoviruses expressing S proteins of human and camel MERS-CoV isolates in 2012 to 2015 with single or multiple natural mutations in the RBD (Table 1). We then tested the ability of the aforementioned RBD-immunized mouse sera to prevent infection of these pseudoviruses in Huh-7 cells. All pseudoviruses efficiently expressed MERS-CoV RBD and HIV-1 p24, which are recognized by anti-RBD antibody (Fig. 4A, top) and p24-specific antibody (Fig. 4A, bottom), and had sufficient infectivity in hDPP4-expressing Huh-7 cells (Fig. 4B). As expected, EMC-RBD, the RBD of EMC2012, the prototypic MERS-CoV, induced highly potent neutralizing antibodies that cross-neutralized all 17 pseudoviruses of MERS-CoV strains tested, including those isolated from humans in Saudi Arabia and South Korea in the 2012-2015 outbreaks and those from infected camels (Fig. 5A). The RBDs of human MERS-CoV isolates, including 2012-RBD, 2014-RBD, and 2015-RBD, as well as camel MERS-CoV (Camel-RBD), elicited similarly high titers of neutralizing antibodies against these pseudoviruses (Fig. 5B, D, E, and F). Although 2013-RBD, which contains 5 mutations in multiple sites of RBD from 4 human MERS-CoV strains in 2013, induced slightly lower titers of neutralizing antibodies than did the other RBDs, these antibodies could still efficiently cross-neutralize all MERS pseudoviruses tested (Fig. 5C). In contrast, no specific neutralizing antibody was induced in PBS-treated (control) mice (Fig. 5A). These results confirm the ability of the test rRBD proteins in inducing strong and cross-neutralizing antibodies against divergent MERS-CoV strains isolated from humans and camels.
FIG 4.

Detection of target proteins and infectivity of MERS pseudoviruses. (A) Packaged MERS pseudoviruses were tested for expression of MERS-CoV S and HIV-1 p24 proteins by Western blotting using anti-MERS-CoV RBD (1:1,000, top) and anti-HIV-1 p24 (183-H12-5C, 1:50, bottom) antibodies, respectively. (B) Detection of infectivity of MERS pseudoviruses in DPP4-expressing Huh-7 cells. Vesicular stomatitis virus G glycoprotein (VSV-G) was included as a positive control.
FIG 5.
Human and camel MERS-CoV RBD proteins induced highly potent cross-neutralizing antibodies against divergent human and camel MERS pseudoviruses. MERS pseudoviruses expressing S proteins of divergent human and camel MERS-CoV strains isolated from 2012 to 2015 with single or multiple mutations in the RBD were tested for the ability to cross-neutralize MERS-CoV RBD proteins in Huh-7 cells. Sera of mice immunized with EMC-RBD (A), 2012-RBD (B), 2013-RBD (C), 2014-RBD (D), 2015-RBD (E), Camel-RBD (F), or PBS control (A) were collected at 10 days after the third immunization and analyzed. Neutralizing activity was expressed as 50% neutralizing antibody titers (NT50). The data are presented as means ± SD for five mice in each group.
Recombinant RBD proteins of 2012-2015 MERS-CoV strains induced highly potent cross-neutralizing antibodies against MERS-CoV MAb escape variants.
Polyclonal anti-MERS-CoV antibody is expected to neutralize MAb escape variants, since many sites on the RBD are targeted in such a preparation. To assess this, we generated 5 pseudoviruses expressing S proteins of MAb escape mutants with single or multiple mutations in RBD (Fig. 4) (31–34) and examined their sensitivity to the antibodies generated in the aforementioned RBD-immunized mice. Except for 2013-RBD, which induced a slightly lower level of neutralizing antibodies, as previously noted, all other RBDs, including EMC-RBD, 2012-RBD, 2014-RBD, 2015-RBD, and Camel-RBD, elicited similarly high titers of antibodies able to cross-neutralize all MERS-CoV pseudoviruses tested (Fig. 6). Thus, rRBD proteins of MERS-CoV strains isolated from humans and camels in the 2012-2015 outbreaks induce strong, broad-spectrum antibodies capable of cross-neutralizing pseudoviruses of MAb escape strains of MERS-CoV.
FIG 6.
Human and camel MERS-CoV RBD proteins induced highly potent cross-neutralizing antibodies against MAb escape mutants of MERS pseudoviruses. MERS pseudoviruses expressing RBD MAb escape variants were generated as specified above and tested for cross-neutralizing ability of human and camel RBD proteins in Huh-7 cells. Sera of mice immunized with EMC-RBD (A), 2012-RBD (B), 2013-RBD (C), 2014-RBD (D), 2015-RBD (E), Camel-RBD (F), or PBS control (A) were collected at 10 days after the third immunization and analyzed. Neutralizing activity was expressed as NT50, and the data are presented as means ± SD for five mice in each group.
Antibodies induced by rRBD proteins of 2012-2015 MERS-CoV strains cross-neutralized live human MERS-CoV strains EMC2012 and London1-2012.
To determine whether RBD immunization also provided protection against infection with infectious MERS-CoV, we infected cells with two representative MERS-CoVs, EMC2012 and London1-2012, isolated in 2012 (1, 39). Notably, all RBDs, including EMC-RBD, 2012-RBD, 2013-RBD, 2014-RBD, 2015-RBD, and Camel-RBD, induced cross-neutralizing antibodies against both human MERS-CoV strains, among which EMC-RBD elicited the highest levels of neutralizing antibodies. Even though relatively low titers of neutralizing antibodies were induced by 2012-RBD and 2013-RBD compared with those induced by other RBD proteins, levels were sufficient to neutralize both test MERS-CoV strains. In contrast, PBS control induced no neutralizing antibodies against MERS-CoV (Fig. 7). These data show that infectious viruses, as well as pseudoviruses, were neutralized after immunization with a panel of RBDs.
FIG 7.

Human and camel MERS-CoV RBD proteins induced cross-neutralizing antibodies against different human MERS-CoVs. Mice were immunized with the indicated RBD or PBS as a control, and sera were collected at 10 days after the third immunization and examined for the presence of antibodies that neutralized MERS-CoV strains EMC2012 and London1-2012 in Vero E6 cells. Neutralizing antibody titers are presented as the reciprocal of the highest dilution of sera that resulted in a complete inhibition of virus infectivity in at least 50% of the wells (NT50). The data are from pooled sera of five mice in each group.
The MERS-CoV RBD with simultaneous mutations of multiple key residues in the RBM displayed significantly reduced activity of receptor binding and viral entry.
The data reported above demonstrated that RBDs with single or multiple natural mutations derived from representative strains in 2012 to 2015, which contain scattered mutations in the RBM, had no significant changes in antigenicity, functionality, and neutralizing immunogenicity. To test whether deliberate mutation of multiple key residues in the RBM of the RBD would affect hDPP4-binding ability, we expressed two additional Fc-tagged RBD mutant proteins, RBD-FGG and RBD-FGGAA in the RBM that either occurred naturally, but sporadically (L506F, D509G, or D510G), or were detected in MAb escape variants (R511A or E513A) (Table 2). Similar to wild-type RBD (EMC-RBD WT), both mutant proteins had high purity, formed conformational dimeric structures (Fig. 8A, top), and were recognized by RBD-specific antibodies (Fig. 8A, bottom). Then, we tested their binding activity to DPP4 by ELISA and flow cytometry analyses. The results revealed that RBD-FGG and RBD-FGGAA exhibited significantly reduced binding activity to recombinant hDPP4 (Fig. 8B), cDPP4 (Fig. 8C), and cell-associated hDPP4 (Fig. 8D) proteins, with effects most obvious when 5 residues were mutated (Fig. 8B to D).
FIG 8.

MERS-CoV RBD with multiple mutations of key residues in the RBM exhibited significantly reduced activity of receptor binding and viral entry. (A) Characterization of mutant MERS-CoV RBD proteins. SDS-PAGE (top) and Western blot (bottom) analyses of the purified mutant RBD proteins containing 3 (RBD-FGG) and 5 (RBD-FGGAA) key mutations, respectively, in the RBM. Nonboiled and boiled protein samples (5 μg) were subjected to SDS-PAGE or Western blotting, followed by detection by MERS-CoV RBD-specific antibody (1:1,000). EMC-RBD wild-type (WT) was included as a control. The molecular mass markers (in kilodaltons) are indicated on the left. (B and C) Detection of binding affinity between mutant MERS-CoV RBD proteins and hDPP4 (B) or cDPP4 (C) protein by ELISA. The ELISA plates were coated with hDPP4 or cDPP4 protein (2 μg/ml) and then incubated with different RBDs. The data are presented as means ± SD (n = 4) of RBD binding to hDPP4 or cDPP4 protein. (D) Detection of binding between mutant RBD proteins and Huh-7 cells expressing hDPP4 by flow cytometry analysis. EMC-RBD WT was included as a control. The data are presented as means ± SD (n = 4) of each RBD (40 μg/ml) binding to hDPP4 in Huh-7 cells. MFI, median fluorescence intensity. In panels B to D, three asterisks (***) indicate P values of <0.001 between mutant and WT RBDs. (E) Detection of entry of MERS pseudoviruses expressing S proteins with 3 (L506F, D509G, D510G) or 5 (L506F, D509G, D510G, R511A, E513A) mutations in the RBM. The infectivity of EMC2012 WT pseudovirus in Huh-7 cells was set as 100% entry, and the infectivity of the corresponding mutant pseudovirus was expressed as the percentage of entry (%). ***, P < 0.001 between mutant and WT pseudoviruses.
To evaluate the effects of these mutations on S-mediated viral entry, we constructed 2 additional MERS pseudoviruses expressing S proteins with the 3 or 5 aforementioned mutations in RBD and used them to infect Huh-7 cells. These MERS-CoV mutant pseudoviruses were significantly inhibited from entering Huh-7 cells, with the greatest inhibition observed after infection with pseudovirus carrying 5 mutations (Fig. 8E).
The MERS-CoV RBD with mutations of multiple key residues in the RBM exhibited significantly reduced antigenicity and neutralizing immunogenicity.
To determine whether simultaneous mutations of key residues in the RBM affected antigenicity, we initially evaluated the binding affinity of mutant RBD proteins (RBD-FGG and RBD-FGGAA) to wild-type RBD-specific neutralizing monoclonal and polyclonal antibodies by ELISA. Compared to EMC-RBD WT, the two mutant RBDs exhibited significantly reduced binding to neutralizing MAbs Mersmab1 and m336 (Fig. 9A). Both mutant RBDs bound less well than wild-type RBD to polyclonal sera, with the greatest reduction observed when RBD-FGGAA was assayed (Fig. 9B).
FIG 9.

MERS-CoV RBD with multiple mutations of key residues in the RBM showed reduced antigenicity and neutralizing immunogenicity. (A and B) Detection of the binding between mutant RBD proteins and RBD-specific neutralizing antibodies by ELISA. ELISA plates were precoated with rRBD proteins (1 μg/ml), and binding was detected using RBD-specific neutralizing MAbs Mersmab1 and m336 (1.25 μg/ml) (A), as well as polyclonal antibodies from sera of mice immunized with EMC-RBD wild-type (WT) protein (B). Serum IgG antibody titers are expressed as the endpoint dilutions that remain positively detectable, and the data are presented as means ± SD (n = 4) of each RBD binding to the antibodies. EMC-RBD WT protein was included as a control. **, P < 0.01; ***, P < 0.001 between mutant and WT RBD proteins. (C and D) Detection of neutralizing activity of MERS-CoV RBD-specific neutralizing MAbs Mersmab1 and m336 (C), as well as polyclonal antibodies from sera of mice immunized with EMC-RBD WT protein (D), against the above-described mutant and WT pseudoviruses. ND50 and NT50 represent the 50% neutralizing dose (for MAbs) and 50% neutralizing antibody titers (for sera), respectively. **, P < 0.01; ***, P < 0.001 between mutant and WT pseudoviruses. (E and F) Detection of IgG (E) and neutralizing antibodies (F) induced by MERS-CoV RBD mutant proteins, or EMC-RBD WT protein control, by ELISA and MERS pseudovirus neutralization assay, respectively. Sera from 10 days after the second immunization were tested for IgG antibodies specific to EMC-RBD and neutralizing antibodies against EMC2012 WT pseudovirus. The antibody titers are presented as means ± SD for five mice in each group. The neutralizing antibody titers are expressed as mean NT50 ± SD for five mice in each group. *, P < 0.05; **, P < 0.01 between mutant and WT RBD proteins.
Then, we further investigated whether RBD-FGG and RBD-FGGAA were as immunogenic as wild-type RBD. First, we examined whether MAbs Mersmab1 and m336 efficiently neutralized pseudoviruses with RBD-FGG and RBD-FGGAA mutations. The results demonstrated (by comparing 50% neutralizing doses [ND50]) that the pseudoviruses with these mutations were significantly less sensitive to neutralization than wild-type pseudoviruses (Fig. 9C). Similar results were obtained when the pseudoviruses were exposed to mouse sera containing polyclonal neutralizing antibodies (Fig. 9D). After immunization, RBD-FGG and RBD-FGGAA elicited significantly decreased levels of IgG (Fig. 9E) and neutralizing antibodies in mouse sera against MERS pseudovirus (EMC2012 WT) (Fig. 9F). These results suggest that simultaneous mutations of multiple key residues in the RBM of MERS-CoV RBD resulted in significantly reduced antigenicity and neutralizing immunogenicity, but at the cost of reduced ability to enter cells.
DISCUSSION
Development of safe, effective, and broad-spectrum vaccines against MERS-CoV infection is still urgently needed to combat the continuing threat posed by MERS-CoV. Compared with other vaccine types, including those based on viruses and viral vectors, subunit vaccines are safer since viral genomic components are absent (25, 28). We previously identified the RBD in the S protein of MERS-CoV as a critical vaccine target and demonstrated that RBD-based MERS vaccines induce highly potent neutralizing antibodies that protect immunized animals against MERS-CoV challenge (20, 21, 23, 24, 40).
Studies have revealed the presence of a number of single and multiple mutations in the RBDs of MERS-CoV strains isolated from humans and camels at different time periods during the 2012-2015 outbreaks (Table 1) (29, 30). Also, analysis of RBD-specific neutralizing MAbs has identified a number of mutations in the RBDs of MERS-CoV mutants that escaped neutralization by these MAbs (31–34). The presence of both natural and antibody escape mutations in the RBD of MERS-CoV has raised concerns about the capacity of RBDs to induce cross-neutralizing antibodies against different mutant strains of MERS-CoV. Therefore, this study aimed to design and develop RBD subunit vaccines based on different human and camel MERS-CoV strains isolated from 2012 through 2015 and evaluate their cross-neutralizing ability against divergent MERS-CoV strains and MAb escape mutants.
Using the RBD sequence of MERS-CoV strain EMC2012 (EMC-RBD) as a prototype, we constructed five mutant RBD proteins, designated 2012-RBD, 2013-RBD, 2014-RBD, 2015-RBD, and Camel-RBD, that contain single or multiple mutations in the RBD of representative MERS-CoV strains isolated from humans and camels in 2012 to 2015 (Tables 1 and 2). Our data indicated that all five mutant RBDs maintained good conformation and antigenicity, reacting strongly with polyclonal and MAb neutralizing antibodies that recognize neutralizing epitopes in the RBD of strain EMC2012 (33, 34, 41). In addition, these RBDs bound strongly to the hDPP4 receptor in soluble and cell-associated forms, suggesting good functionality. It should be noted that while the binding between these RBDs and hDPP4 protein was stronger than that between RBDs and cDPP4 protein, the binding between Camel-RBD and hDPP4-expressing Huh-7 cells was enhanced, not reduced. These results suggest that the camel RBD retains its high binding activity to human receptor during evolution, indicating that camels will remain an important reservoir for sporadic human infection.
MERS vaccines are expected to have broad-spectrum neutralizing ability against different MERS-CoV strains. Indeed, we have found that similar to the prototype EMC-RBD, all five mutant RBDs containing scattered key mutations elicited high-titer antibody responses in immunized mice as assessed by their ability to strongly cross-react with MERS-CoV S protein from the prototype strain EMC2012. Most importantly, these RBD-induced antibodies could cross-neutralize infections by all MERS-CoV strains tested, including 17 pseudotyped human and camel MERS-CoV strains isolated in the 2012-2015 outbreaks, 5 MAb escape MERS-CoV mutants, and 2 live MERS-CoV strains isolated in the early stage of the 2012 outbreak, thus confirming their ability to induce cross-neutralizing antibodies against divergent circulating MERS-CoV strains. Our other studies have demonstrated that neutralizing antibody titers of ≥1:119 completely protect highly susceptible hDPP4-transgenic (hDPP4-Tg) mice from lethal MERS-CoV challenge (our unpublished data). It is thus expected that immunization with vaccine candidates containing the individual mutant RBDs (2012-RBD, 2013-RBD, 2014-RBD, 2015-RBD, or Camel-RBD), as well as prototype EMC2012-RBD, will protect hDPP4-Tg mice from MERS-CoV infection since all of them induced neutralizing antibody titers of ≥1:120 against two live MERS-CoV strains (EMC2012 and London1-2012). Therefore, irrespective of these scattered mutations at single or multiple sites of MERS-CoV RBD, the data presented here suggest that RBD-based MERS vaccines will be able to induce sufficient cross-neutralizing antibodies for protection against current circulating strains, as well as other strains that might occur in the future.
The tertiary structure of MERS-CoV S trimer was modeled on the basis of the recently solved cryo-electron microscopy (cryo-EM) structure of mouse hepatitis virus (MHV) S trimer (Fig. 10) (42). Mapping of these naturally occurring scattered mutations in RBD of MERS-CoV on the modeled structure of MERS-CoV S trimer revealed that eight of these residues (506, 509, 510, 520, 522, 529, 530, and 534) are located in the RBM region whereas the rest are located in the core region of the RBD. Among these eight RBM residues, three (506, 509, 510) are directly involved in DPP4 binding (35, 36). The epitopes covering these three residues have been shown to be critical for the binding of neutralizing monoclonal antibodies (32–34). Two additional key residues (511 and 513) in the RBM are also responsible for virus-DPP4 binding and play a role in inducing MAb escape mutant virus strains (31, 32, 34). Thus, simultaneous mutations of the above-mentioned three (506, 509, 510) or five (506, 509, 510, 511, 513) key residues in a single viral strain led to significant changes in the neutralizing immunogenicity of MERS-CoV RBD, facilitating escape of the virus from host immune surveillance. Several reasons explain why this has not happened in nature. First, the chance for simultaneous mutations of these three or five key residues in the RBM of the RBD is significantly lower than that for single mutations. Second, we found that mutating all three or five residues simultaneously significantly reduced viral binding to the DPP4 receptor and, hence, reduced the ability of the virus to enter and infect target cells, a hefty price that the virus cannot afford to pay. Consequently, only scattered mutations of these residues were detected in different viral strains, which led to less significant changes in the neutralizing immunogenicity of each RBD. The other mutated residues play less important roles in receptor binding and in overall neutralizing immunogenicity of the RBD and are also inconsistently detected in different viral strains. Since these binding and inhibition assays were based on viral RBD protein or pseudoviruses expressing MERS-CoV S protein with the test mutations in the RBD, there exists the possibility that the results might be different when mutations are identified in live MERS-CoV. It is also possible that some live MERS-CoV strains that contain the mutations of key residues in RBD might become resistant to neutralizing antibodies without causing significant reduction of infectivity. Nevertheless, the results presented in this study suggest that it might take much longer for MERS-CoV to acquire immune escape mutations in the RBD than in other regions of the viral S protein since decreased neutralization is accompanied by reduced binding to DPP4. Therefore, the RBD remains a major target site for development of MERS vaccines.
FIG 10.
Distribution of RBD mutation residues in the structural model of MERS-CoV S trimer. Based on the structural homology between MERS-CoV RBD (PDB access code 4L3N) and the corresponding domain in the trimeric MHV S (PDB access code 3JCL), the crystal structure of the former was modeled into the cryo-EM structure of the latter. The core structure of MERS-CoV RBD is in cyan, the RBM is in red, and the MERS-CoV RBD residues that have undergone mutations are in blue. The trimeric MHV S protein contains three copies of this domain, with two colored in magenta and the third replaced by MERS-CoV RBD.
MERS-CoV RBD contains multiple conformational neutralizing epitopes encompassing key residues that include L506, D509, D510, R511, E513, W535, E536, D539, Y540, and R542 (33, 34, 41, 43–45); thus, vaccines targeting the RBD are effective against a virus with mutations in one or more epitopes. In contrast, other target sites in the S protein may contain only one single neutralizing epitope. Therefore, vaccines targeting such an epitope would become ineffective if a single mutation occurred. In addition, the RBD is also a critical functional domain, and antibodies targeting the RBD can also block the binding between RBD and viral receptor, in addition to their virus-neutralizing activity (26).
To summarize, we constructed five rRBD proteins respectively covering different mutations in the RBD of MERS-CoV that circulated during the course of the 2012-2015 outbreaks, as well as two mutant RBDs with simultaneous mutations of multiple key residues in the RBM of the RBD. Their antigenicity to bind MERS-CoV RBD-specific neutralizing antibodies, as well as their functionality to bind the DPP4 receptor of MERS-CoV, was demonstrated. This study also explored the broad-spectrum capability of the RBDs containing naturally scattered mutations in inducing cross-neutralizing antibodies against human and camel strains isolated from the 2012-2015 outbreaks, as well as antibody escape mutant strains. Taken together, this study confirms the feasibility of developing an RBD-based MERS vaccine that is safe, effective, and broad-spectrum, with the added ability to cross-neutralize antibodies against infection of current and future divergent MERS-CoV strains.
MATERIALS AND METHODS
Ethics statement.
Female BALB/c mice aged 6 to 8 weeks were used in this study. The animal studies were carried out in strict accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health (NIH), and the protocol was approved by the Committee on the Ethics of Animal Experiments of the New York Blood Center (permit number 194.17).
Sources of sequences.
The MERS-CoV S sequences from years 2012 through 2015 were obtained from the GenBank database at the NCBI website (https://www.ncbi.nlm.nih.gov) and aligned with the S protein sequence of MERS-CoV EMC2012 strain to identify key mutations within residues 377 to 588 of the RBD (Table 1).
Construction, expression, and purification of recombinant proteins.
Construction, expression, and purification of recombinant proteins was performed as previously described with some modifications (21, 40). Briefly, the MERS-CoV EMC-RBD plasmid was constructed by fusing residues 377 to 588 of EMC2012 RBD with human IgG Fc. This plasmid was used as the template to generate 2012-RBD, 2013-RBD, 2014-RBD, 2015-RBD, and Camel-RBD with point or multiple natural mutations, as well as to generate RBD-FGG and RBD-FGGAA with simultaneous multiple mutations of key RBM residues (Table 2), using a QuikChange site- or multisite-directed mutagenesis kit (Agilent Technologies). Recombinant S1 of MERS-CoV (EMC2012) (residues 18 to 725) and cDPP4 with a C-terminal His6 were constructed using the pJW4303 expression vector (26). The aforementioned proteins were harvested from 293T cell culture supernatants. Recombinant hDPP4 protein (residues 39 to 766) containing a C-terminal His6 was expressed in the culture medium of insect cells using the Bac-to-Bac expression system (Invitrogen) (19). The Fc- and His-tagged proteins were purified by protein A affinity chromatography (GE Healthcare) and nickel-nitrilotriacetic acid (Ni-NTA) Superflow (Qiagen), respectively.
SDS-PAGE and Western blot analysis.
Purified MERS-CoV RBD proteins were subjected to SDS-PAGE and Western blot analysis as previously described (21). Briefly, proteins (boiled or nonboiled) were separated by 10% Tris-glycine SDS-PAGE and then stained directly by Coomassie brilliant blue or transferred to nitrocellulose membranes. The blots were blocked with 5% nonfat milk–PBST (where PBST is PBS with Tween 20) at 4°C overnight, followed by sequential incubation with MERS-CoV RBD-specific antibody (1:1,000) and horseradish peroxidase (HRP)-conjugated goat anti-mouse IgG (1:3,000) (GE Healthcare) for 1 h at room temperature. Binding signals were visualized using ECL Western blot substrate reagents and Amersham Hyperfilm (GE Healthcare).
Expression of MERS-CoV spike and HIV-1 p24 in the generated pseudoviruses was identified by Western blotting in lysed pseudoviruses using MERS-CoV RBD-specific antibody (1:1,000) and anti-HIV-1 p24 (183-H12-5C, 1:50) MAb as described above (46).
Coimmunoprecipitation assay.
Binding between MERS-CoV RBD proteins and hDPP4 receptor was performed using a coimmunoprecipitation (Co-IP) assay and Western blotting as previously described (21). Briefly, RBDs (10 μg) were incubated with hDPP4 protein (10 μg) or hDPP4-expressing Huh-7 cell lysates (5 × 107/ml) at 4°C for 1 h in the presence of protein A beads. Protein A beads were washed with lysis buffer and PBS. Proteins were eluted from the beads and assessed for RBD and hDPP4 by Western blotting using MERS-CoV RBD-specific antibody (1:1,000) and hDPP4-specific MAb (0.5 μg/ml; R&D Systems), as described above.
ELISA.
ELISA was performed to detect binding between MERS-CoV RBD proteins and RBD-specific neutralizing MAbs (21). Briefly, ELISA plates were precoated with RBD proteins (1 μg/ml) overnight at 4°C, blocked with 2% nonfat milk–PBST for 2 h at 37°C, and then incubated sequentially with MAbs (1.25 μg/ml) and HRP-conjugated anti-mouse IgG (1:3,000, for mouse MAb) (GE Healthcare) or anti-human IgG-Fab (1:5,000, for human MAbs) (Sigma) for 1 h at 37°C. The reaction was visualized by addition of 3,3′,5,5′-tetramethylbenzidine (TMB) substrate (Invitrogen) and stopped by 1 N H2SO4. Absorbance at 450 nm (A450) was measured using an ELISA plate reader (Tecan).
Binding between MERS-CoV RBD and DPP4 proteins was performed using an ELISA protocol similar to that described above, except that the plates were coated with hDPP4 or cDPP4 proteins (2 μg/ml) prior to addition of serially diluted rRBD proteins. Binding was detected using RBD-specific mouse antibody (1:1,000), followed by HRP-conjugated anti-mouse IgG (1:3,000).
Assays to detect MERS-CoV S-specific antibody responses in mouse sera were performed by coating ELISA plates with MERS-CoV S1 or rRBD protein (1 μg/ml), followed by sequential incubation with serially diluted mouse sera and HRP-conjugated anti-mouse IgG (1:3,000), IgG1 (1:2,000), or IgG2a (1:5,000) (Invitrogen) antibodies.
Flow cytometry.
To detect the binding between MERS-CoV RBD proteins and hDPP4-expressing Huh-7 cells, cells were incubated with the indicated RBDs (40 μg/ml) for 30 min at room temperature, followed by the addition of fluorescein isothiocyanate (FITC)-labeled anti-human IgG antibody for 30 min. Cells were analyzed by flow cytometry (26, 27).
Animal vaccination and sample collection.
Animal vaccination and sample collection were performed as previously described with some modifications (24). Briefly, mice were subcutaneously (s.c.) immunized with MERS-CoV RBD proteins (10 μg/mouse) or PBS control, each with MF59 adjuvant, and boosted once or twice at 3 weeks with the same immunogens and adjuvant. Sera were collected at 10 days after the last immunization.
Generation of wild-type and mutant MERS pseudoviruses and pseudovirus neutralization assay.
MERS pseudoviruses were generated and pseudovirus-based neutralization assays performed as previously described with some modifications (22, 46). Briefly, 293T cells were cotransfected with a plasmid encoding an Env-defective, luciferase-expressing HIV-1 genome (pNL4-3.luc.RE) and each of the plasmids encoding the indicated S proteins (Table 1) using the calcium phosphate method. The medium was replaced with fresh Dulbecco's modified Eagle medium (DMEM) 8 h later, and pseudovirus-containing supernatants were collected 72 h after transfection for single-cycle infection. Wild-type and mutant MERS pseudoviruses were incubated with serially diluted mouse sera at 37°C for 1 h and added to Huh-7 cells, followed by addition of fresh medium 24 h later. The cells were lysed 72 h later in cell lysis buffer (Promega), incubated with luciferase substrate (Promega), and assessed for relative luciferase activity using an Infinite 200 PRO Luminator (Tecan). The 50% MERS pseudovirus neutralizing antibody titer (NT50) was calculated as previously described (47).
Measurement of neutralizing antibody titers.
A virus plaque reduction assay was carried out to determine neutralizing antibody titers in sera as previously described (43, 48). Briefly, sera were serially diluted and incubated with 100 PFU of MERS-CoV strain EMC2012 or London1-2012 at 37°C for 30 min before transferring to Vero cell monolayers. Cultured cells were overlaid with 1% agar medium, and plaques were counted.
Statistical analysis.
Statistical significance among different groups was calculated by Student's t test using GraphPad Prism statistical software. Asterisks in the figures indicate significance (*, **, and *** denote P values of <0.05, <0.01, and <0.001, respectively).
ACKNOWLEDGMENTS
We thank Dimiter S. Dimitrov and Tianlei Ying at the National Institutes of Health for providing m336, m337, and m338 MAbs.
This study was supported by NIH grants R01AI098775, U01AI124260, and R21AI109094 to S.J. and L.D., R01AI089728 and R01AI110700 to F.L., and PO1AI060699 to S.P. G.Z. and Y.Z. received funding from the China National Program of Infectious Disease (2014ZX10004001004). The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
We declare no conflict of interest.
REFERENCES
- 1.Zaki AM, van Boheemen S, Bestebroer TM, Osterhaus AD, Fouchier RA. 2012. Isolation of a novel coronavirus from a man with pneumonia in Saudi Arabia. N Engl J Med 367:1814–1820. doi: 10.1056/NEJMoa1211721. [DOI] [PubMed] [Google Scholar]
- 2.Wang Q, Qi J, Yuan Y, Xuan Y, Han P, Wan Y, Ji W, Li Y, Wu Y, Wang J, Iwamoto A, Woo PC, Yuen KY, Yan J, Lu G, Gao GF. 2014. Bat origins of MERS-CoV supported by bat coronavirus HKU4 usage of human receptor CD26. Cell Host Microbe 16:328–337. doi: 10.1016/j.chom.2014.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lu G, Wang Q, Gao GF. 2015. Bat-to-human: spike features determining ‘host jump’ of coronaviruses SARS-CoV, MERS-CoV, and beyond. Trends Microbiol 23:468–478. doi: 10.1016/j.tim.2015.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yang Y, Du L, Liu C, Wang L, Ma C, Tang J, Baric RS, Jiang S, Li F. 2014. Receptor usage and cell entry of bat coronavirus HKU4 provide insight into bat-to-human transmission of MERS coronavirus. Proc Natl Acad Sci U S A 111:12516–12521. doi: 10.1073/pnas.1405889111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Munster VJ, Adney DR, van Doremalen N, Brown VR, Miazgowicz KL, Milne-Price S, Bushmaker T, Rosenke R, Scott D, Hawkinson A, de Wit E, Schountz T, Bowen RA. 2016. Replication and shedding of MERS-CoV in Jamaican fruit bats (Artibeus jamaicensis). Sci Rep 6:21878. doi: 10.1038/srep21878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sabir JS, Lam TT, Ahmed MM, Li L, Shen Y, Abo-Aba SE, Qureshi MI, Abu-Zeid M, Zhang Y, Khiyami MA, Alharbi NS, Hajrah NH, Sabir MJ, Mutwakil MH, Kabli SA, Alsulaimany FA, Obaid AY, Zhou B, Smith DK, Holmes EC, Zhu H, Guan Y. 2016. Co-circulation of three camel coronavirus species and recombination of MERS-CoVs in Saudi Arabia. Science 351:81–84. doi: 10.1126/science.aac8608. [DOI] [PubMed] [Google Scholar]
- 7.Hemida MG, Chu DK, Poon LL, Perera RA, Alhammadi MA, Ng HY, Siu LY, Guan Y, Alnaeem A, Peiris M. 2014. MERS coronavirus in dromedary camel herd, Saudi Arabia. Emerg Infect Dis 20:1231–1234. doi: 10.3201/eid2007.140571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yusof MF, Eltahir YM, Serhan WS, Hashem FM, Elsayed EA, Marzoug BA, Abdelazim AS, Bensalah OK, Al Muhairi SS. 2015. Prevalence of Middle East respiratory syndrome coronavirus (MERS-CoV) in dromedary camels in Abu Dhabi Emirate, United Arab Emirates. Virus Genes 50:509–513. doi: 10.1007/s11262-015-1174-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yang Y, Liu C, Du L, Jiang S, Shi Z, Baric RS, Li F. 2015. Two mutations were critical for bat-to-human transmission of Middle East respiratory syndrome coronavirus. J Virol 89:9119–9123. doi: 10.1128/JVI.01279-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hemida MG, Elmoslemany A, Al-Hizab F, Alnaeem A, Almathen F, Faye B, Chu DK, Perera RA, Peiris M. 2015. Dromedary camels and the transmission of Middle East respiratory syndrome coronavirus (MERS-CoV). Transbound Emerg Dis doi: 10.1111/tbed.12401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Korea Centers of Disease Control and Prevention. 2015. Middle East respiratory syndrome coronavirus outbreak in the Republic of Korea, 2015. Osong Public Health Res Perspect 6:269–278. doi: 10.1016/j.phrp.2015.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Oh MD, Choe PG, Oh HS, Park WB, Lee SM, Park J, Lee SK, Song JS, Kim NJ. 2015. Middle East respiratory syndrome coronavirus superspreading event involving 81 persons, Korea 2015. J Korean Med Sci 30:1701–1705. doi: 10.3346/jkms.2015.30.11.1701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ki M. 2015. 2015 MERS outbreak in Korea: hospital-to-hospital transmission. Epidemiol Health 37:e2015033. doi: 10.4178/epih/e2015033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Oboho IK, Tomczyk SM, Al-Asmari AM, Banjar AA, Al-Mugti H, Aloraini MS, Alkhaldi KZ, Almohammadi EL, Alraddadi BM, Gerber SI, Swerdlow DL, Watson JT, Madani TA. 2015. 2014 MERS-CoV outbreak in Jeddah—a link to health care facilities. N Engl J Med 372:846–854. doi: 10.1056/NEJMoa1408636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Memish ZA, Al-Tawfiq JA, Alhakeem RF, Assiri A, Alharby KD, Almahallawi MS, Alkhallawi M. 2015. Middle East respiratory syndrome coronavirus (MERS-CoV): a cluster analysis with implications for global management of suspected cases. Travel Med Infect Dis 13:311–314. doi: 10.1016/j.tmaid.2015.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lu L, Liu Q, Zhu Y, Chan KH, Qin L, Li Y, Wang Q, Chan JF, Du L, Yu F, Ma C, Ye S, Yuen KY, Zhang R, Jiang S. 2014. Structure-based discovery of Middle East respiratory syndrome coronavirus fusion inhibitor. Nat Commun 5:3067. doi: 10.1038/ncomms4067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li F. 2015. Receptor recognition mechanisms of coronaviruses: a decade of structural studies. J Virol 89:1954–1964. doi: 10.1128/JVI.02615-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Raj VS, Mou H, Smits SL, Dekkers DH, Muller MA, Dijkman R, Muth D, Demmers JA, Zaki A, Fouchier RA, Thiel V, Drosten C, Rottier PJ, Osterhaus AD, Bosch BJ, Haagmans BL. 2013. Dipeptidyl peptidase 4 is a functional receptor for the emerging human coronavirus-EMC. Nature 495:251–254. doi: 10.1038/nature12005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen Y, Rajashankar KR, Yang Y, Agnihothram SS, Liu C, Lin YL, Baric RS, Li F. 2013. Crystal structure of the receptor-binding domain from newly emerged Middle East respiratory syndrome coronavirus. J Virol 87:10777–10783. doi: 10.1128/JVI.01756-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang N, Tang J, Lu L, Jiang S, Du L. 2015. Receptor-binding domain-based subunit vaccines against MERS-CoV. Virus Res 202:151–159. doi: 10.1016/j.virusres.2014.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ma C, Wang L, Tao X, Zhang N, Yang Y, Tseng CT, Li F, Zhou Y, Jiang S, Du L. 2014. Searching for an ideal vaccine candidate among different MERS coronavirus receptor-binding fragments—the importance of immunofocusing in subunit vaccine design. Vaccine 32:6170–6176. doi: 10.1016/j.vaccine.2014.08.086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tang J, Zhang N, Tao X, Zhao G, Guo Y, Tseng CT, Jiang S, Du L, Zhou Y. 2015. Optimization of antigen dose for a receptor-binding domain-based subunit vaccine against MERS coronavirus. Hum Vaccin Immunother 11:1244–1250. doi: 10.1080/21645515.2015.1021527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tao X, Garron T, Agrawal AS, Algaissi A, Peng BH, Wakamiya M, Chan TS, Lu L, Du L, Jiang S, Couch RB, Tseng CK. 2015. Characterization and demonstration of the value of a lethal mouse model of Middle East respiratory dyndrome coronavirus infection and disease. J Virol 90:57–67. doi: 10.1128/JVI.02009-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang N, Channappanavar R, Ma C, Wang L, Tang J, Garron T, Tao X, Tasneem S, Lu L, Tseng CT, Zhou Y, Perlman S, Jiang S, Du L. 2016. Identification of an ideal adjuvant for receptor-binding domain-based subunit vaccines against Middle East respiratory syndrome coronavirus. Cell Mol Immunol 13:180–190. doi: 10.1038/cmi.2015.03. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang N, Jiang S, Du L. 2014. Current advancements and potential strategies in the development of MERS-CoV vaccines. Expert Rev Vaccines 13:761–774. doi: 10.1586/14760584.2014.912134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Du L, Kou Z, Ma C, Tao X, Wang L, Zhao G, Chen Y, Yu F, Tseng CT, Zhou Y, Jiang S. 2013. A truncated receptor-binding domain of MERS-CoV spike protein potently inhibits MERS-CoV infection and induces strong neutralizing antibody responses: implication for developing therapeutics and vaccines. PLoS One 8:e81587. doi: 10.1371/journal.pone.0081587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Du L, Zhao G, Kou Z, Ma C, Sun S, Poon VK, Lu L, Wang L, Debnath AK, Zheng BJ, Zhou Y, Jiang S. 2013. Identification of a receptor-binding domain in the S protein of the novel human coronavirus Middle East respiratory syndrome coronavirus as an essential target for vaccine development. J Virol 87:9939–9942. doi: 10.1128/JVI.01048-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Du L, Jiang S. 2015. Middle East respiratory syndrome: current status and future prospects for vaccine development. Expert Opin Biol Ther 15:1647–1651. doi: 10.1517/14712598.2015.1092518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kim Y, Cheon S, Min CK, Sohn KM, Kang YJ, Cha YJ, Kang JI, Han SK, Ha NY, Kim G, Aigerim A, Shin HM, Choi MS, Kim S, Cho HS, Kim YS, Cho NH. 2016. Spread of mutant Middle East respiratory syndrome coronavirus with reduced affinity to human CD26 during the South Korean outbreak. mBio 7:e00019-15. doi: 10.1128/mBio.00019-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kim DW, Kim YJ, Park SH, Yun MR, Yang JS, Kang HJ, Han YW, Lee HS, Man KH, Kim H, Kim AR, Heo DR, Kim SJ, Jeon JH, Park D, Kim JA, Cheong HM, Nam JG, Kim K, Kim SS. 2016. Variations in spike glycoprotein gene of MERS-CoV, South Korea, 2015. Emerg Infect Dis 22:100–104. doi: 10.3201/eid2201.151055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jiang L, Wang N, Zuo T, Shi X, Poon KM, Wu Y, Gao F, Li D, Wang R, Guo J, Fu L, Yuen KY, Zheng BJ, Wang X, Zhang L. 2014. Potent neutralization of MERS-CoV by human neutralizing monoclonal antibodies to the viral spike glycoprotein. Sci Transl Med 6:234ra59. doi: 10.1126/scitranslmed.3008140. [DOI] [PubMed] [Google Scholar]
- 32.Tang XC, Agnihothram SS, Jiao Y, Stanhope J, Graham RL, Peterson EC, Avnir Y, Tallarico AS, Sheehan J, Zhu Q, Baric RS, Marasco WA. 2014. Identification of human neutralizing antibodies against MERS-CoV and their role in virus adaptive evolution. Proc Natl Acad Sci U S A 111:E2018–E2026. doi: 10.1073/pnas.1402074111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ying T, Du L, Ju TW, Prabakaran P, Lau CC, Lu L, Liu Q, Wang L, Feng Y, Wang Y, Zheng BJ, Yuen KY, Jiang S, Dimitrov DS. 2014. Exceptionally potent neutralization of middle East respiratory syndrome coronavirus by human monoclonal antibodies. J Virol 88:7796–7805. doi: 10.1128/JVI.00912-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Du L, Zhao G, Yang Y, Qiu H, Wang L, Kou Z, Tao X, Yu H, Sun S, Tseng CT, Jiang S, Li F, Zhou Y. 2014. A conformation-dependent neutralizing monoclonal antibody specifically targeting receptor-binding domain in Middle East respiratory syndrome coronavirus spike protein. J Virol 88:7045–7053. doi: 10.1128/JVI.00433-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lu G, Hu Y, Wang Q, Qi J, Gao F, Li Y, Zhang Y, Zhang W, Yuan Y, Bao J, Zhang B, Shi Y, Yan J, Gao GF. 2013. Molecular basis of binding between novel human coronavirus MERS-CoV and its receptor CD26. Nature 500:227–231. doi: 10.1038/nature12328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang N, Shi X, Jiang L, Zhang S, Wang D, Tong P, Guo D, Fu L, Cui Y, Liu X, Arledge KC, Chen YH, Zhang L, Wang X. 2013. Structure of MERS-CoV spike receptor-binding domain complexed with human receptor DPP4. Cell Res 23:986–993. doi: 10.1038/cr.2013.92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kossyvakis A, Tao Y, Lu X, Pogka V, Tsiodras S, Emmanouil M, Mentis AF, Tong S, Erdman DD, Antoniadis A. 2015. Laboratory investigation and phylogenetic analysis of an imported Middle East respiratory syndrome coronavirus case in Greece. PLoS One 10:e0125809. doi: 10.1371/journal.pone.0125809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang Z, Shen L, Gu X. 2016. Evolutionary dynamics of MERS-CoV: potential recombination, positive selection and transmission. Sci Rep 6:25049. doi: 10.1038/srep25049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bermingham A, Chand MA, Brown CS, Aarons E, Tong C, Langrish C, Hoschler K, Brown K, Galiano M, Myers R, Pebody RG, Green HK, Boddington NL, Gopal R, Price N, Newsholme W, Drosten C, Fouchier RA, Zambon M. 2012. Severe respiratory illness caused by a novel coronavirus, in a patient transferred to the United Kingdom from the Middle East, September 2012. Euro Surveill 17:20290 http://www.eurosurveillance.org/ViewArticle.aspx?ArticleId=20290. [PubMed] [Google Scholar]
- 40.Ma C, Li Y, Wang L, Zhao G, Tao X, Tseng CT, Zhou Y, Du L, Jiang S. 2014. Intranasal vaccination with recombinant receptor-binding domain of MERS-CoV spike protein induces much stronger local mucosal immune responses than subcutaneous immunization: implication for designing novel mucosal MERS vaccines. Vaccine 32:2100–2108. doi: 10.1016/j.vaccine.2014.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ying T, Prabakaran P, Du L, Shi W, Feng Y, Wang Y, Wang L, Li W, Jiang S, Dimitrov DS, Zhou T. 2015. Junctional and allele-specific residues are critical for MERS-CoV neutralization by an exceptionally potent germline-like antibody. Nat Commun 6:8223. doi: 10.1038/ncomms9223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Walls AC, Tortorici MA, Bosch BJ, Frenz B, Rottier PJ, DiMaio F, Rey FA, Veesler D. 2016. Cryo-electron microscopy structure of a coronavirus spike glycoprotein trimer. Nature 531:114–117. doi: 10.1038/nature16988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang L, Shi L, Joyce MG, Modjarrad K, Zhang Y, Leung K, Lees CR, Zhou T, Yassine HM, Kanekiyo M, Yang ZY, Chen X, Becker MM, Freeman M, Vogel L, Johnson JC, Olinger G, Todd JP, Bagci U, Solomon J, Mollura DJ, Hensley L, Jahrling P, Denison MR, Rao SS, Subbarao K, Kwong PD, Mascola JR, Kong WP, Graham BS. 2015. Evaluation of candidate vaccine approaches for MERS-CoV. Nat Commun 6:7712. doi: 10.1038/ncomms8712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Corti D, Zhao J, Pedotti M, Simonelli L, Agnihothram S, Fett C, Fernandez-Rodriguez B, Foglierini M, Agatic G, Vanzetta F, Gopal R, Langrish CJ, Barrett NA, Sallusto F, Baric RS, Varani L, Zambon M, Perlman S, Lanzavecchia A. 2015. Prophylactic and postexposure efficacy of a potent human monoclonal antibody against MERS coronavirus. Proc Natl Acad Sci U S A 112:10473–10478. doi: 10.1073/pnas.1510199112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yu X, Zhang S, Jiang L, Cui Y, Li D, Wang D, Wang N, Fu L, Shi X, Li Z, Zhang L, Wang X. 2015. Structural basis for the neutralization of MERS-CoV by a human monoclonal antibody MERS-27. Sci Rep 5:13133. doi: 10.1038/srep13133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhao G, Du L, Ma C, Li Y, Li L, Poon VK, Wang L, Yu F, Zheng BJ, Jiang S, Zhou Y. 2013. A safe and convenient pseudovirus-based inhibition assay to detect neutralizing antibodies and screen for viral entry inhibitors against the novel human coronavirus MERS-CoV. Virol J 10:266. doi: 10.1186/1743-422X-10-266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chou TC. 2006. Theoretical basis, experimental design, and computerized simulation of synergism and antagonism in drug combination studies. Pharmacol Rev 58:621–681. doi: 10.1124/pr.58.3.10. [DOI] [PubMed] [Google Scholar]
- 48.Fett C, DeDiego ML, Regla-Nava JA, Enjuanes L, Perlman S. 2013. Complete protection against severe acute respiratory syndrome coronavirus-mediated lethal respiratory disease in aged mice by immunization with a mouse-adapted virus lacking E protein. J Virol 87:6551–6559. doi: 10.1128/JVI.00087-13. [DOI] [PMC free article] [PubMed] [Google Scholar]






