ABSTRACT
Endoribonuclease (NendoU) is unique and conserved as a major genetic marker in nidoviruses that infect vertebrate hosts. Arterivirus nonstructural protein 11 (nsp11) was shown to have NendoU activity and play essential roles in the viral life cycle. Here, we report three crystal structures of porcine reproductive and respiratory syndrome virus (PRRSV) and equine arteritis virus (EAV) nsp11 mutants. The structures of arterivirus nsp11 contain two conserved compact domains: the N-terminal domain (NTD) and C-terminal domain (CTD). The structures of PRRSV and EAV endoribonucleases are similar and conserved in the arterivirus, but they are greatly different from that of severe acute respiratory syndrome (SARS) and Middle East respiratory syndrome (MERS) coronaviruses (CoV), representing important human pathogens in the Nidovirales order. The catalytic center of NendoU activity is located in the CTD, where a positively charged groove is next to the key catalytic residues conserved in nidoviruses. Although the NTD is nearly identical, the catalytic region of the arterivirus nsp11 family proteins is remarkably flexible, and the oligomerization may be concentration dependent. In summary, our structures provide new insight into this key multifunctional NendoU family of proteins and lay a foundation for better understanding of the molecular mechanism and antiviral drug development.
IMPORTANCE Porcine reproductive and respiratory syndrome virus (PRRSV) and equine arteritis virus are two major members of the arterivirus family. PRRSV, a leading swine pathogen, causes reproductive failure in breeding stock and respiratory tract illness in young pigs. Due to the lack of a suitable vaccine or effective drug treatment and the quick spread of these viruses, infected animals either die quickly or must be culled. PRRSV costs the swine industry around $644 million annually in the United States and almost €1.5 billion in Europe every year. To find a way to combat these viruses, we focused on the essential viral nonstructural protein 11 (nsp11). nsp11 is associated with multiple functions, such as RNA processing and suppression of the infected host innate immunity system. The three structures solved in this study provide new insight into the molecular mechanisms of this crucial protein family and will benefit the development of new treatments against these deadly viruses.
KEYWORDS: crystal structures, nsp11, NendoU, arterivirus, PRRSV, EAV, mutagenesis, flexibility
INTRODUCTION
The family Arteriviridae belongs to the order Nidovirales, which includes the largest currently known RNA genome viruses, such as Ronivirus, Torovirus, and Coronavirus (1). This family includes four RNA viruses: equine arteritis virus (EAV), porcine reproductive and respiratory syndrome virus (PRRSV), lactate dehydrogenase-elevating virus (LDV), and simian hemorrhagic fever virus (SHFV) (2). In 2003, the severe acute respiratory syndrome coronavirus (SARS-CoV) caused hundreds of deaths (3). Similarly, PRRSV also has caused large agricultural economic losses, especially since the outbreak of highly virulent PRRSV variants in 2006 (4). Moreover, it continues to cause great global economic losses every year due to the lack of a suitable vaccine or effective drug treatment.
EAV and PRRSV, as single-stranded, positive-sense RNA viruses, are the major members of the family Arteriviridae. Their genomes are about 13 to 15 kb and contain 10 open reading frames (ORFs), including two large ones, ORF1a and ORF1ab. The two large ORFs encode two multidomain replicase polyproteins (5), which are processed by three ORF1a-encoded proteases into several nonstructural proteins (nsps) (6), while the other ORFs encode structural proteins (7). The three proteases are nsp1, nsp2, and nsp4. Moreover, nsp1 in PRRSV has two domains, nsp1α and nsp1β. Both nsp1 and nsp2 contain a papain-like cysteine protease domain, and nsp4 is a 3C-like serine protease (8). The mature nsps form the membrane-anchored replication/transcription complex (RTC) that is essential for the synthesis of the 3′-coterminal nested set of subgenomic mRNAs and viral genome replication (9).
ORF1b encodes four proteins, nsp9 to nsp12. nsp9 (RNA-dependent RNA polymerase [RdRp]) and nsp10 (helicase) are essential for the EAV or PRRSV life cycle in the host cell (4). nsp11 (endoribonuclease [NendoU]), a major genetic marker in the nidoviruses that infect vertebrate hosts (10), is also conserved and unique. The arterivirus nsp11 has nidovirus-specific endoribonuclease activity, cleaving both single-stranded RNA (ssRNA) and double-stranded RNA (dsRNA) at a 3′ uridylate-specific site (11). The NendoU active-site mutants of EAV were shown to be crippled but replication competent, revealing the essential role of these sites in the virus life cycle (12). Based on the solved crystal structure of SARS-CoV nsp15 (13, 14), the catalytic residues likely are two histidines and a lysine in the C terminus, similar to that of RNase A. The importance of these three putative catalytic residues was confirmed by site-directed mutagenesis experiments with EAV nsp11 (11).
Although arterivirus nsp11 is known to have NendoU activity and mutations of the NendoU active sites were shown to impact the viral replication cycle, its function in the virus remains unclear (12). In EAV, the key role of NendoU in viral subgenomic RNA synthesis has been confirmed (12). Meanwhile, research on PRRSV nsp11 is mainly focused on its effect on the host cell immune processes, such as production of tumor necrosis factor alpha (TNF-α) (15) and proinflammatory interleukin-1β (16). According to RNA microarray analysis, nsp11 is associated with five different pathways: histone-related, mitogen-activated protein kinase signaling, cell cycle and DNA replication, complement, and the ubiquitin-proteasome pathways. In the cell cycle, PRRSV nsp11 can modulate the progression of S phase cells (17).
Arterivirus nsp11 and its counterpart nsp15 in coronaviruses harbor NendoU activity, but they share only 22% identical residues, and the protein sizes are different. Even while belonging to the same arterivirus family, EAV nsp11 and PRRSV nsp11 share only ∼52% amino acid identity (11). These analyses reveal that even though NendoUs of different nidoviruses are evolutionarily distant, the biochemical function is conserved. Moreover, functions of nsp11 other than its NendoU activity are unknown, and the related mechanism is unclear.
Despite its importance as a vital replicative enzyme and antiviral drug target, information about the three-dimensional structure of nsp11 is sparse. To characterize the structure and function of arterivirus nsp11, we describe here three crystal structures of PRRSV nsp11 and EAV nsp11. Understanding the mechanisms and structure of the essential NendoU would provide novel insights into viral pathogenesis and inform approaches to development of drugs against arteriviruses, such as PRRSV, causing large economic losses in agricultural industries.
RESULTS
Solving the structures of PRRSV and EAV nsp11 mutants.
We first attempted to express wild-type (WT) PRRSV and EAV nsp11 from bacteria, but their solubilities were extremely low (0.1 mg/liter), perhaps due to the high toxicity of arterivirus WT nsp11. According to previous studies (11, 16–19), we further generated NendoU active-site mutants, the EAV nsp11 H126A, H141A, and K170A mutants and the PRRSV nsp11 H144A and K173A mutants. These mutants were suitable for crystallization due to their high expression. After extensive trials, well-diffracting crystals of PRRSV nsp11 K173A and EAV nsp11 H141A and K170A mutants were finally obtained.
BLAST searches of the arterivirus nsp11 sequence among the solved structures in the PDB database revealed that the shared identities with known sequences were very low, with the highest value being only 22%. Moreover, neither PRRSV nor EAV nsp11 could be solved using the closest homolog with a known structure as the model after extensive trials by molecular replacement. Finally, we soaked the crystals with Hg2+ and used the single-wavelength anomalous diffraction (SAD) method to determine the coordinates of heavy atoms in the PRRSV nsp11 K173A mutant crystal. The number and the position of six Hg2+ atoms were determined, and native data were finally solved at 2.16 Å in the space group of P41212, with an Rwork of 18.4% and an Rfree of 21.0%. The final model contains two PRRSV nsp11 K173A mutant molecules (residues 1 to 222 of protomer A and 1 to 221 of protomer B missing residues 137 to 141 and 166 to 173), one sulfate, one chloride, one polyethylene glycol (PEG) fragment, and 440 water molecules (Table 1 and Fig. 1 and 2A).
TABLE 1.
Data collection and refinement statistics of arterivirus nsp11 mutantsa
| Parameter | Value for: |
||
|---|---|---|---|
| PRRSV K173A mutant (PDB code 5EYI) | EAV K170A mutant (PDB code 5HBZ) | EAV H141A mutant (PDB code 5HC1) | |
| Data collection statistics | |||
| Wavelength | 0.9793 | 1.0000 | 1.0000 |
| Space group | P41212 | R32 | I222 |
| Cell dimensions | |||
| a, b, c (Å) | 75.3, 75.3, 199.5 | 248.3, 248.3, 226.3 | 129.1, 133.7, 145.3 |
| α, β, γ (°) | 90, 90, 90 | 90, 90, 120 | 90, 90, 90 |
| Resolution (Å) | 50.0–2.16 (2.20–2.16) | 50.0–3.10 (3.15–3.10) | 50.0–3.10 (3.15–3.10) |
| Rsym (%)b | 9.9 | 10 | 26.6 |
| I/σ | 18.4 (1.5) | 15.7 (1.5) | 6.2 (1.6) |
| Completeness (%) | 99.6 (99.9) | 99.2 (99.5) | 90.7 (93.3) |
| Total no. of reflections | 1,657,775 | 1,174,148 | 793,92 |
| Unique reflections | 31,952 | 48,540 | 23,202 |
| Redundancy | 7.7 (7.9) | 5.5 (5.7) | 7.7 (7.7) |
| Refinement statistics | |||
| Resolution (Å) | 50.0–2.16 (2.21–2.16) | 50.0–3.10 (3.18–3.10) | 50.0–3.10 (3.18–3.10) |
| No. of reflections | 297,12 (1863) | 44,766 (2699) | 19,651 (1397) |
| Rwork/Rfree (%) | 18.4/21.0 | 19.5/19.9 | 20.8/25.2 |
| No. of atoms | |||
| Protein | 3,369 | 9,946 | 6,658 |
| Ligand/ions | 14 | 0 | 0 |
| Water | 440 | 0 | 125 |
| B-factors (Å2) | |||
| Protein | 29.1 | 73.9 | 66.3 |
| Ligand/ion | 50.4 | 0 | 0 |
| Water | 42.2 | 0 | 39.4 |
| RMSDs | |||
| Bond lengths (Å) | 0.007 | 0.009 | 0.007 |
| Bond angles (°) | 1.34 | 1.438 | 1.28 |
| Ramachandran plot (%)c | 91.5/8.5/0/0 | 84.8/14.7/0.5/0 | 83.8/15.3/0.9/0 |
Three crystal experiments for each structure.
Rsym = ΣhΣi|Ih,I − Ih|/ΣhΣlIh,i, where Ih is the mean intensity of the i observations of symmetry-related reflections of h.
Residues in most favored, additional allowed, generously allowed, and disallowed regions of the Ramachandran plot.
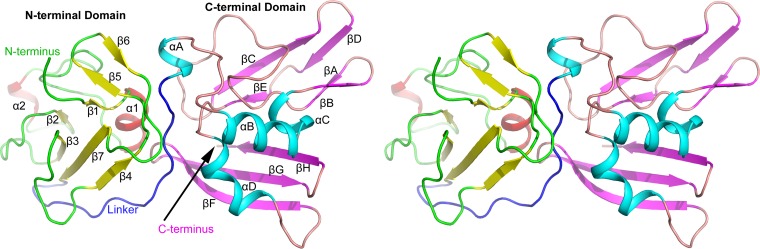
FIG 1.
Stereoview of the PRRSV nsp11 K173A mutant structure. The whole structure consists of an NTD, a CTD, and a long linker (blue). The β-sheets and α-helices are colored yellow and red, respectively, in the NTD and magenta and cyan, respectively, in the CTD.
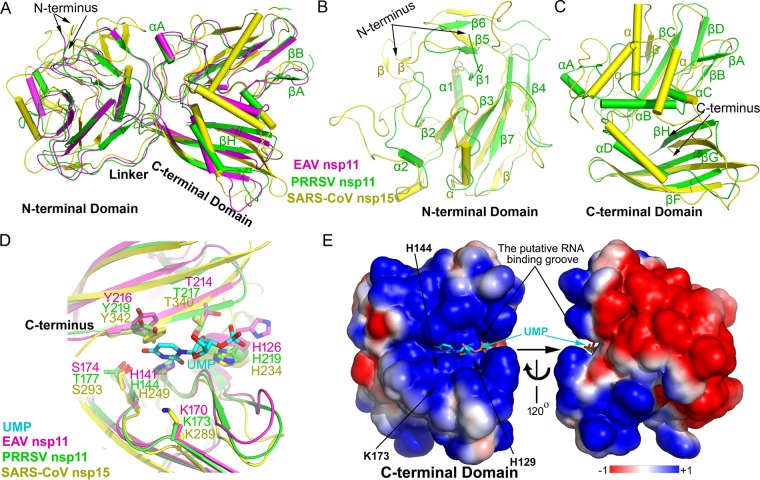
FIG 2.
Structural superimposition, catalytic site, and electrostatic potential of PRRSV nsp11, EAV nsp11, and SARS-CoV nsp15. Shown is a structural alignment of the whole protein (A), NTD (B), and CTD (C). EAV nsp11, magenta; PRRSV nsp11, green; SARS-CoV nsp15, yellow. (D) Active-site grooves in the CTD of NendoU from different nidoviruses. (E) Electrostatic potential of PRRSV nsp11 and a 120°-rotated view (right). The surface potential is displayed as a color gradient ranging from red (negative) to blue (positive). Note that three putative catalytic residues involving Lys173, His144, and His129 are located in a groove formed between β-sheet βH and a four-stranded β-sheet (βA, βB, βD, and βC). The modeled uridine 3′-phosphate (UMP) molecule is shown in a ball-and-stick representation. Note that both complete protomers of PRRSV nsp11 and EAV nsp11 are used.
The structures of EAV nsp11 mutants were all determined by molecular replacement using the PRRSV nsp11 K173A mutant structure as a model. The final model of the EAV nsp11 K170A mutant solved at 3.1 Å contains six protein molecules (residues 1 to 219, with three of the protomers missing residues 163 to 170, 163 to 172, or 162 to 171) (Table 1). Although the Rmerge of the EAV nsp11 H141A mutant was as high as 26%, the structure was able to be solved at 3.1 Å, fortunately. It contains four protein molecules (residues 1 to 219, with two of the protomers missing residues 140 to 141 or 138 to 139) and 125 water molecules (Table 1).
Arterivirus nsp11 mutants adopt a conserved compact, two-domain fold, which is greatly different from the case with SARS-CoV nsp15.
The arterivirus nsp11 mutants were found to adopt a compact fold (Fig. 1). The PRRSV nsp11 K173A mutant protomer contains two domains, the N-terminal domain (NTD; residues 1 to 90) and C-terminal domain (CTD; residues 107 to 222), which are connected with a long linker (residues 91 to 106). The NTD is formed by a three-stranded antiparallel β-sheet (β1-β5-β6) and a four-stranded antiparallel β-sheet (β2-β3-β7-β4), with two small α-helices (α1-α2) packing against both sides of the latter β-sheet. Four α-helices (αA to αD) and eight β-sheets (βA to βH) are present in the CTD. The CTD contains an antiparallel three-stranded β-sheet (βF-βH) and two two-stranded antiparallel β-sheets (βA-βB and βC-βE). Sheets βA to βE and βH form a positively electrostatic catalytic groove that is supported by α-helices αB to αD, while αA is located at the N terminus of the CTD (Fig. 1).
The EAV nsp11 H141A or K170A mutant protomer also consists of two domains, the NTD (residues 1 to 88) and the CTD (residues 104 to 219), which are connected with a long linker (residues 89 to 103) (Fig. 2A). A comparison between the PRRSV and EAV nsp11 H141A or K170A mutant structures described in this study shows that the core conformations of the CTD and NTD are nearly identical, with an overall root mean square deviation (RMSD) of 1.0 Å (Fig. 2A). However, their overall conformations slightly differ, with an RMSD of 1.4 Å, and the differences mostly occur at loops and the long linker between the NTD and CTD (Fig. 2A to C). The high similarity between the PRRSV and EAV nsp11 structures reveals that the structures of arterivirus nsp11 are conserved.
A DALI search (20) of the PRRSV nsp11 K173A mutant structure showed SARS-CoV nsp15 (PDB code 2RHB; RMSD = 3.3 Å) (14) as the closest homolog. When these two structures were superimposed, both the CTD and the NTD were found to differ significantly, with RMSDs of 3.3 Å (Fig. 2A to C). The overall folds between the β-sheets of the arterivirus nsp11 mutants and SARS-CoV nsp15 appear to be similar, but the arrangement, length, and positions of the α-helices and loops differ greatly. These structural analyses are consistent with the functional determination that arterivirus nsp11 has distinct features from its related orthologs from other nidovirus lineages, such as SARS-CoV nsp15 (11).
Active sites of NendoU are conserved in nidoviruses.
Previous mutational studies confirmed that the putative active sites, including residues His126, His141, Lys170, and Tyr216 in EAV nsp11 (His129, His144, Lys173, and Tyr219 in PRRSV nsp11), are essential for nsp11 endonuclease activity (11). In this study, the sequence alignment of arterivirus nsp11 and coronavirus nsp15 revealed that the catalytic center is conserved. We also compared our arterivirus nsp11 structures with that of SARS-CoV nsp15 (PDB code 2H85) (13). Interestingly, although the CTD of the arterivirus nsp11 mutants and SARS-CoV nsp15 could not be well superimposed (Fig. 2C), the corresponding catalytic residues of the two enzymes show similar spatial arrangements, which may explain the conserved catalytic mechanisms of nidoviruses (Fig. 2D). Furthermore, a positively charged groove was found in the vicinity of the key catalytic residues. Meanwhile, the electrostatic potential is largely negative in the other positions of the CTD (Fig. 2E). Therefore, a catalytic center involving His126, His141, and Lys170 in EAV nsp11 (His129, His144, and Lys173 in PRRSV nsp11) as well as the C-terminal β-sheet (βH in PRRSV or βI in EAV) form a positively charged groove and may thus bind the negatively charged RNA (Fig. 2D).
Catalytic regions of arterivirus nsp11 mutants differ significantly.
Surprisingly, we observed that the conformations of the CTDs in protomers of the arterivirus nsp11 mutants differ significantly (Fig. 3). This observation is unlike that with SARS-CoV nsp15 (Fig. 2A to C), in which all CTDs from different protomers have the same conformation (13, 14, 21).
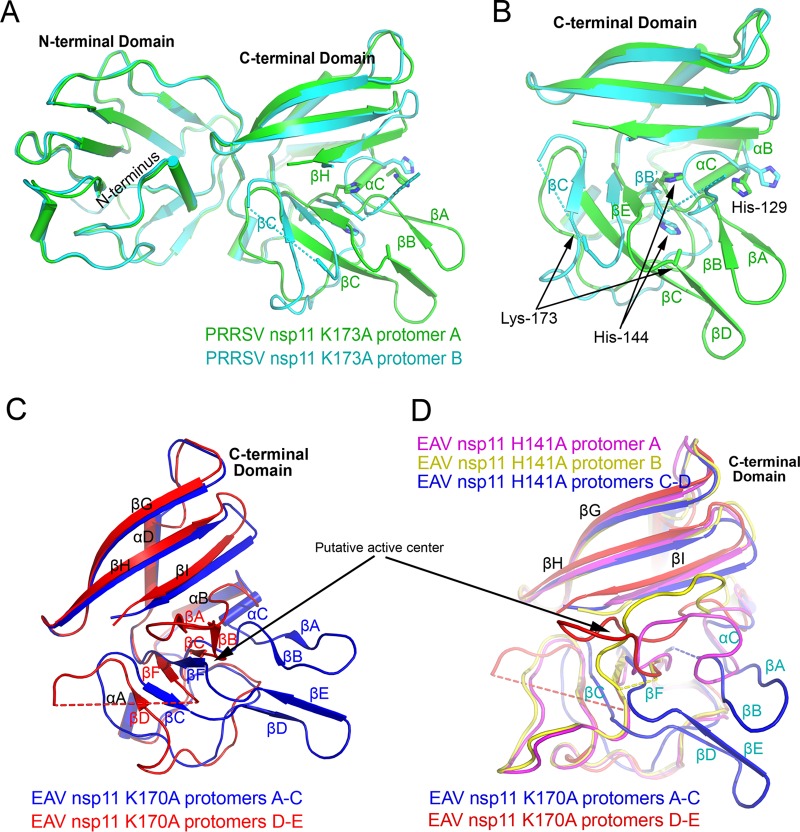
FIG 3.
Differences between nsp11 protomers. (A and B) Structural difference between the two protomers of the PRRSV nsp11 K173A mutant, especially in the CTD. The active sites His129, His144, and Lys173 are shown, and dashed lines indicate residues that have not been resolved in the structures. (C) Two different types of conformations in the CTD of EAV nsp11 K170A mutant protomers. (D) All conformations in the CTDs of EAV nsp11 mutants. Major differences occur in the putative active center.
For PRRSV nsp11 K173A, the two CTD conformations in the asymmetric unit were found to differ remarkably. The RMSD between these two PRRSV nsp11 protomers was 1.8 Å, and the major structural variation was in the catalytic region of the CTD (Fig. 3A and B). The positions of its αC, β, and βE and the loops between these two β-sheets (βA-βB and βC-βE) differ significantly. Intriguingly, βC is remarkably shorter and in the reversed orientation, while the antiparallel β-sheet βC-βE is changed to a parallel β-sheet in protomer B. Moreover, residues 137 to 141 and 166 to 173 in the catalytic region are disordered in protomer B.
Similarly, there are two types of EAV nsp11 K170A mutant conformations which differ significantly (Fig. 3C). The differences between these two types also are mainly present in the catalytic region, similar to that in PRRSV. The position of the sheet βA-βB changes greatly, and the sheet βD-βE is disordered and not solved in the three protomers D-E. For the EAV nsp11 H141A mutant, the catalytic regions have three types of conformations in the asymmetric unit (Fig. 3D). One major conformation is the same as that of the EAV nsp11 K170A mutant, as well as that of PRRSV nsp11 (complete protomer; blue in Fig. 3D). Other than this conformation, there are two additional conformations for the loops corresponding to the sheets βA-βB. Overall, the structural comparison between all arterivirus nsp11 protomers described here shows that the NTD and the noncatalytic region of CTD are nearly identical, with an overall RMSD of 1.0 Å. However, the conformations of the catalytic regions are quite diverse (Fig. 3E).
PRRSV nsp11 and its mutant exist as several types of oligomers.
SARS-CoV nsp15 has been reported to form a hexamer in solution, and its NTD mediates most of the intermolecular interactions (13, 14, 21). Hexamerization has been proposed to be required for optimal enzymatic activity and substrate binding (11). However, no ortholog corresponding to the NTD of the SARS-CoV nsp15 exists in the arterivirus nsp11 family.
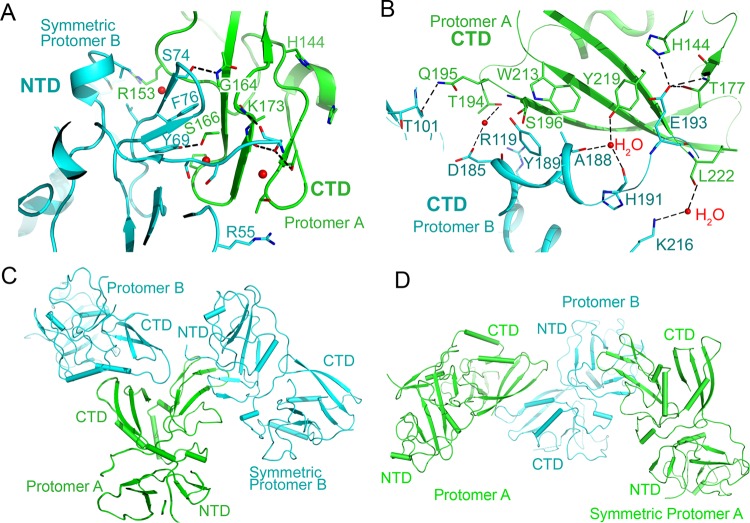
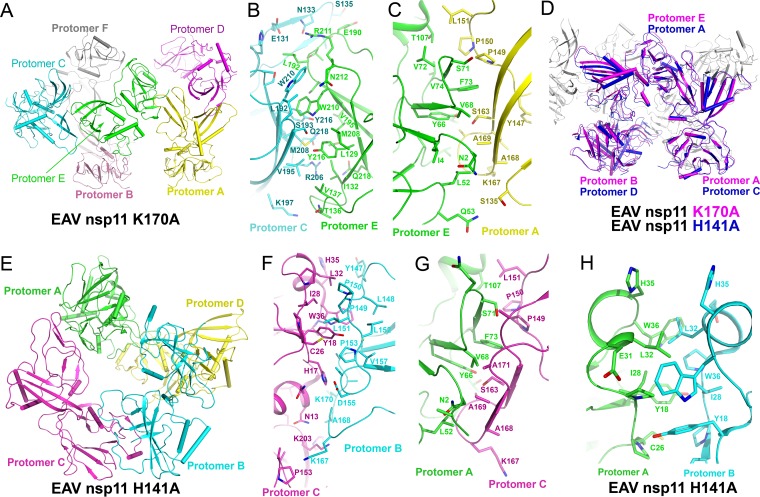
To investigate the oligomeric state of arterivirus nsp11 mutants, we first performed structural analysis of the PRRSV nsp11 K173A mutant. Interestingly, two major molecular interfaces exist according to the results calculated by the Proteins, Interfaces, Structures and Assemblies (PISA) server. The first interface is formed by the CTD and the NTD, with an interface area of 734.1 Å2. The interacting residues are Y69, S74, and F76 in the NTD of the symmetric protomer B and R153, G164, S166, and K173 in the CTD of protomer A (Fig. 4A). The second interface is formed by two CTDs in the same asymmetric unit, with an interface area of 694.9 Å2; the interacting residues are T101, R119, D185, A188, Y189, H191, E193, and K216 in one protomer, while they are H144, T177, T194, Q195, S196, W213, Y219, and L222 in the other (Fig. 4B). Several hydrogen bonds and water-mediated hydrogen bonds are present in both interfaces (Fig. 4A and B). Moreover, the catalytic region of protomer A is directly involved in the interaction of both interfaces. In particular, the CTD of protomer A is stabilized by two protomers B, with one in the same asymmetric unit and the other from a symmetry-related molecule (Fig. 4C). The observation can explain why only protomer A has a complete electron density in the active center (Fig. 3A and B), while for protomer B, the CTD is stabilized by only one protomer (Fig. 4D). These analyses also imply that PRRSV nsp11 K173A exists in solution in several different forms, such as monomers, dimers, trimers (Fig. 4), or higher-order oligomers.
FIG 4.
Interaction analysis of the PRRSV nsp11 K173A mutant. (A and B) Interfaces of PRRSV nsp11 K173A formed by protomer A or symmetry-related protomer A (green) and protomer B (cyan). The key residues involved in the interaction are shown. (C) One putative trimer. The CTD of protomer A (green, complete protomer) is stabilized by protomer B and symmetric protomer B (cyan). (D) Another putative trimer.
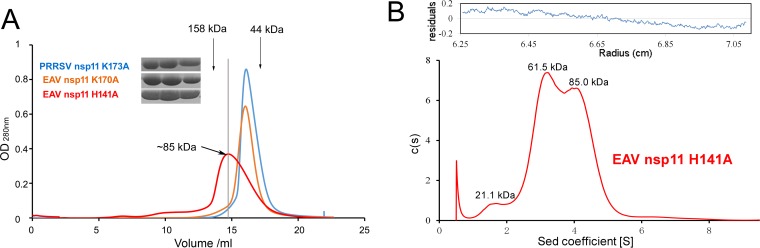
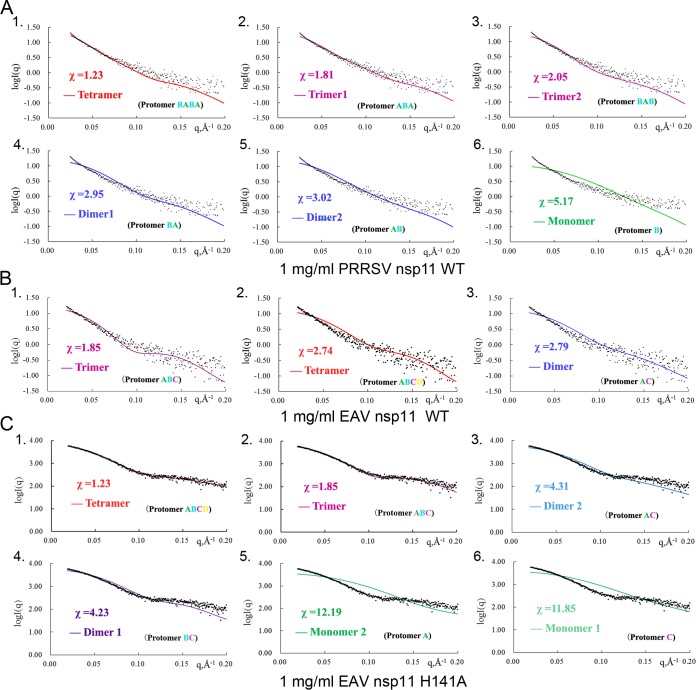
To further uncover the oligomeric states of PRRSV nsp11 in solution, we performed gel filtration and small-angle X-ray scattering (SAXS) analyses. The broad peak of the PRRSV K173A mutant in the sieve column suggests that it exists as different types of oligomers in solution (Fig. 5A). Moreover, SAXS was conducted to analyze the components in solution. SAXS is a powerful tool for structure validation and the quantitative analysis of flexible systems, and it is highly complementary to the high-resolution methods of X-ray crystallography and nuclear magnetic resonance (NMR). Due to the low yield from bacterial expression, only 1 mg/ml of WT protein was used. Among the six fitted SAXS profiles (Fig. 6A), both trimers and tetramers fit best for the WT PRRSV nsp11. The results were similar for the PRRSV nsp11 K173A mutant. Furthermore, the radius of gyration, maximum linear dimension, and molecular mass observed at different concentrations of the PRRSV nsp11 K173A mutant by SAXS (data not shown) were also found to be concentration dependent. Taken together, the results suggested that the oligomerization of PRRSV nsp11 is diverse and concentration dependent.
FIG 5.
Arterivirus nsp11 mutants exist as several types of oligomers in solution. (A) Size exclusion chromatography of arterivirus nsp11 mutants. (B) Analytical ultracentrifugation of the EAV nsp11 H141A mutant. Shown is a sedimentation coefficient [C(s)] distribution from SV analytical ultracentrifugation performed with 0.1 mM protein in 20 mM Tris-HCl (pH 8.0) and 500 mM NaCl.
FIG 6.
Comparison of SAXS experimental data and calculated scattering profiles for nsp11 at a 1-mg/ml concentration. Experimental data are represented by black dots. The theoretical scattering curves of tetramers, trimers, dimers, and monomers are shown. (A) WT PRRSV nsp11; (B) WT EAV nsp11; (C) EAV nsp11 H141A mutant.
EAV nsp11 and its mutants also exist as several types of oligomers.
For the EAV nsp11 K170A mutant structure, one asymmetric unit contains six protein molecules that belong to three dimers (Fig. 7A). These three dimers are nearly identical. The interfaces formed by the two CTDs are nearly identical in three dimers, with interface areas ranging between 700 Å2 and 750 Å2 (Fig. 7B). Moreover, there are also several other types of interfaces, such as the CTD-NTD interfaces, between different dimers, with interface areas ranging between 400 Å2 and 580 Å2 (Fig. 7C). Interestingly, residues in the interfaces are different from those observed in the PRRSV nsp11 K173A mutant, implying that the interaction between protomers is diverse and dynamic within the arterivirus endoribonuclease subfamily.
FIG 7.
Interaction analysis of EAV nsp11 H141A and K170A mutants. Shown are protomers in an asymmetric unit of the EAV nsp11 K170A (A) and H141A (E) mutants and interfaces of the EAV nsp11 K170A (B and C) and H141A (F to H) mutants. Key residues involved in the interaction are shown. Similar interfaces were found in the aligned EAV nsp11 K170A and H141A mutant structures (D); for example, the interface in panel G is similar to that in panel C.
The EAV nsp11 H141A mutant structure is a tetramer in the solved structure (Fig. 7E). The major interface between each protomer involves one CTD and one NTD, with the interface areas ranging between 600 Å2 and 700 Å2 (Fig. 7F to H). Interestingly, it has two types of interfaces which are nearly identical to the observed CTD-NTD interfaces in the PRRSV nsp11 K173A and EAV nsp11 K170A mutants. These results imply that several interactions between protomers are conserved (Fig. 7D).
The small interface areas between protomers imply that the oligomeric states are dynamic. To further characterize the oligomeric states, we performed gel filtration, sedimentation velocity (SV), and SAXS analyses. Gel filtration analyses of these mutants suggest that they exist as different types of oligomers in solution (Fig. 5A). Surprisingly, the observed peak of EAV nsp11 H141A in the sieve column suggested that it has more higher-order oligomers. To further assess the states in solution, we performed an analytical ultracentrifugation experiment. The SV analysis showed that EAV nsp11 H141A mainly exists as an equilibrium mixture of a tetramer and a trimer at a 0.1 mM concentration (Fig. 5B). Additionally, we also performed SAXS to analyze the oligomerization of WT EAV nsp11. The three theoretically calculated SAXS profiles (trimer, dimer, and tetramer) fit the experimental data similarly (χ = 1.85, 2.79, and 2.74) (Fig. 6B). This result implied that WT EAV nsp11 also exists as several types of oligomers in solution. Interestingly, both trimer and tetramer models of EAV nsp11 H141A fit best among the six fitted SAXS profiles at 1 mg/ml (Fig. 6C). However, at the 4.5-mg/ml protein concentration (data not shown), the theoretically calculated SAXS profile from any monomer, dimer, or trimer did not agree well with the experimental data, and only the tetramer model fit well. The results reveal that the oligomerization may be dependent on the concentration (data not shown). In addition to the aforementioned experiments, the other SAXS parameters observed at different concentrations of EAV nsp11 K170A or H141A were consistent with the above-described results (data not shown). Thus, the oligomerization of EAV nsp11 mutants is also concentration dependent.
DISCUSSION
Arterivirus nsp11 has been shown to have multifunctional activities and play essential roles in the virus life cycle (12, 16, 19, 22). When we initiated this work, no arterivirus nsp11 structure had been reported. In this study, we solved the first EAV nsp11 structures and found that they are conserved and the catalytic region is remarkably flexible. The findings provide a structural basis for further studies of the nsp11 family. In the final resubmission phase of the manuscript for publication, the crystal structure of WT PRRSV nsp11 was reported by another group (23). The overall structure of that WT PRRSV nsp11 is similar to the PRRSV nsp11 K173A mutant structure in this study.
Interestingly, the catalytic region of arterivirus nsp11 showed an obvious conformational flexibility (Fig. 2 and 3), greatly different from those in other nidoviruses, such as coronavirus nsp15. Although the positions of NTD are highly similar in each of the subunits, the catalytic region is especially flexible and adopts different conformations in the different subunits. The heterogeneity is most obvious in the catalytic center. In addition, several regions are disordered in some subunits and not visible in the electron density map. One reason for this observation may be that all of our solved structures are apo-structures. In an attempt to obtain a complex, we soaked all of the crystals with nucleotide uridylate or short RNA with Mn2+ or screened crystals in the buffer with 2 mM MnCl2. However, no complex structure was obtained. Another reason is that the catalytic sites are peripheric (Fig. 4) and exposed to solvents directly, making the active center region more flexible and dynamic. Therefore, the active center may be stabilized with the addition of substrates or other molecules. These analyses indicate that the arterivirus NendoU family has a flexible catalytic region with a fundamental conformational difference in the CTD. These observations suggest that the arterivirus NendoU may be poised between several different conformations, with only subtle environmental changes needed for a particular conformation to be favored. Not surprisingly, a similar observation was also reported in a recently solved structure of the RdRp from influenza C virus (24), in which the endonuclease domain of P3 is remarkably flexible.
Ser174 in EAV nsp11 has been demonstrated to be essential for the specificity of uridylates by mutagenesis (11). The mutation in EAV nsp11 only slightly impaired the NendoU activity but remarkably affected its NendoU specificity, which rendered nsp11 unable to discriminate between cytidylates and uridylates. EAV nsp11 Ser174 and PRRSV nsp11 Thr177 are the counterparts of residue Ser293 in SARS-CoV nsp15, which has been proposed to be a major determinant of the pyrimidine specificity of the enzyme (reference 11 and references therein). In our structures, both of these residues are located on the surface of the positively charged groove (Fig. 2D), which may be directly involved in binding the substrate and thus determining the NendoU specificity.
Hexamerization of coronavirus nsp15 proteins has been proposed to be required for optimal enzyme activity (13, 14, 21). The formation of the hexamer is mediated by the NTD of SARS-CoV nsp15. According to the structure-based sequence alignment, the NTD of arterivirus nsp11 is small and related to the central domain (residues 63 to 191) of SARS-CoV nsp15. Moreover, no counterpart corresponding to the NTD of SARS-CoV nsp15 exists in arterivirus nsp 11. A recent study has demonstrated the deubiquitinase (DUB) activity of PRRSV nsp11 on the K48-linked polyubiquitin chains of the protein (19). The finding suggests that PRRSV nsp11 also has a catalytic center of DUB and a binding site for ubiquitin. However, the DALI search of the NTD returned only similarities with NendoU from other species, rather than DUBs. The NTD also was found to lack a catalytic dyad such as a conserved cysteine and a downstream histidine residue. In addition, it cannot bind ubiquitin in a pulldown assay (data not shown). Therefore, we propose that nsp11 is not involved in the deubiquitination activity directly.
In this study, we expressed the WT arterivirus nsp11 and mutants bacterially. Functional studies such as gel filtration, analytical ultracentrifugation, and SAXS experiments with these proteins suggested that these proteins exist as several types of oligomers in solution. The oligomeric states of the PRRSV nsp11 K173A or the EAV nsp11 K170A or H141A mutant from the structure analysis are consistent with the observations in solution (Fig. 5 and 6). Therefore, the oligomeric states observed in the structures are not results of crystal packing and are greatly different from the hexamer in SARS-CoV nsp15. Moreover, the ratio of the oligomers of arterivirus nsp11 is concentration dependent, a novel feature in the arterivirus nsp11 family. The lack of strong hydrophobic interaction, relatively small interface area, and existence of several similar interfaces may be the major reasons for arterivirus nsp11 existing in solution as an equilibrium mixture of dimers, trimers, tetramers, and other higher-order oligomers.
Arterivirus nsp11 may form higher-order oligomers as a function of increasing protein concentration. The finding that the oligomerization of arterivirus nsp11 is dependent on concentration may be related to its enzymatic activity, especially during the rapid synthesis of viral RNA. However, further detailed study is needed to determine and clarify the function of the higher-order oligomers.
Interestingly, both sequence and structure analyses of nsp11 proteins revealed no sequence or structural homologs in the pig or horse. Therefore, NendoU may be a novel and promising drug target for arteriviruses, such as PRRSV and EAV. Our structures provide a foundation for future studies on the molecular mechanism and development of drugs against PRRSV, which remains a tremendous threat to the swine industry worldwide and causes significant economic losses every year. The structure of the arterivirus nsp11 family of proteins allows us to propose a mechanism for the enzymatic activity. In conclusion, our study demonstrates the remarkable flexibility of the essential arterivirus NendoU family both in the protomer conformation and in the oligomer composition, and it aids our understanding of the mechanisms controlling RNA processing and other functions.
MATERIALS AND METHODS
Cloning, expression, and purification of EAV and PRRSV nsp11.
Using the EAV 211 (25) (GenBank accession no. NC_002532, a gift of Eric J. Snijder, Leiden University Medical Center, The Netherlands) and PRRSV BJ-4 (26) (GenBank accession no. JX462645, a gift of Hanchun Yang, China Agricultural University, China) cDNAs as templates, the nsp11 genes were cloned into a modified pET-28a (N-terminal His tag) vector in which the thrombin recognition site was replaced by a tobacco etch virus (TEV) protease recognition site (6). The fragments were cloned between BamH I and XhoI. The nsp11 mutants (such as the K170A, H141A, or K173A mutant) were engineered using overlapping PCR. All final clones were verified by DNA sequencing.
All proteins were expressed in Escherichia coli BL21(DE3) cells (Merck). Cells were grown in LB at 37°C until the optical density at 600 nm (OD600) reached 0.6 and then induced for 6 h at 18°C with 0.2 mM isopropyl β-d-thiogalactoside. The cultures were harvested by centrifugation at 4,000 × g for 10 min at 4°C, resuspended in lysis buffer (20 mM Tris-HCl [pH 8.0], 500 mM NaCl, and 50 mM imidazole), and disrupted by sonication. The lysate was centrifuged at 47,000 × g for 30 min to remove cell debris. The soluble fraction was applied to a Ni2+-chelating column (GE Healthcare). After sample loading, the column was washed with lysis buffer, and the protein was eluted with elution buffer (20 mM Tris-HCl [pH 8.0], 500 mM NaCl, and 200 mM imidazole). Total yields of the PRRSV nsp11 K173A and EAV nsp11 K170A mutants per liter of bacterial culture were estimated to be 30 mg, and that of the EAV nsp11 H141A mutant was 80 mg. The His tag was cleaved using His-TEV protease at 4°C for 8 h and removed by an Ni2+-chelating column. Further purification was performed by size exclusion chromatography using a Superdex 200 (GE Healthcare) with buffer (20 mM Tris-HCl [pH 8.0], 500 mM NaCl). Peak fractions were collected and analyzed by SDS-PAGE. All purified proteins were concentrated by ultrafiltration using Amicon Ultra-15 centrifugal filter units (Millipore) to 10 to 12 mg/ml for crystallization.
Crystallization and data collection.
Initial crystal screening was performed at 18°C and 4°C using the sitting-drop vapor diffusion method, in which 1 μl of protein solution was mixed with 1 μl of reservoir solution. For the PRRSV nsp11 K173A mutant, small crystals appeared in several solutions containing PEG, but they were fragile. The conditions were optimized, and high-quality crystals were finally obtained in 0.1 M Tris-HCl (pH 7.5), 15% PEG 3350, and 0.1 M (NH4)2SO4. For the EAV nsp11 K170A mutant, small crystals also grew in many reservoir solutions containing PEG. However, the diffraction ability of these crystals was poor. The diffraction qualities were not improved after extensive optimization of crystallization conditions or various postcrystallization treatments, such as annealing and dehydration. Fortunately, the crystals for the EAV nsp11 K170A fragment were finally obtained in 0.1 M sodium citrate (pH 6.6) and 1 M (NH4)3PO4. The crystals of the EAV nsp11 H141A fragment were obtained in 20% PEG 3350 and 0.1 M lithium citrate.
For data collection, crystals were cryoprotected in mother liquor containing 25% (vol/vol) ethylene glycol and flash cooled in liquid nitrogen. Native data for the PRRSV nsp11 K173A and EAV nsp11 K170A and H141A mutants were collected on beamlines BL18U and BL17U at Shanghai Synchrotron Radiation Facility (SSRF) and beamline BL1A at the Photon Factory (Tsukuba, Japan), respectively. Data were indexed, integrated, and scaled using HKL2000 (27). Data collection and processing statistics are shown in Table 1.
To solve the structure, PRRSV nsp11 K173A crystals were soaked in the crystallizing condition with 5 mM HgCl2, cryoprotected in the above-described mother liquor containing 25% (vol/vol) ethylene glycol, and flash cooled in liquid nitrogen. The data were collected on beamline 3W1A at the Beijing Synchrotron Radiation Facility (BSRF).
Structure determination.
The structure of the PRRSV nsp11 K173A protein was determined by the single-wavelength anomalous diffraction (SAD) method in the space group P41212. Because the Rmerge of the Hg2+-soaked crystal was 16.6%, the large margin of error in the data made it difficult to find the phasing. After extensive trials of different resolution ranges and a number of heavy atoms, a final solution of six Hg2+ ions was found by the program SHELEX C (28) at 2.6 Å. Using the identified Hg2+ atomic coordinates, the positions were optimized by SOLVE, and the phases were subsequently improved using RESOLVE (29) with a figure of merit of 0.16. Many β-strand bundles and α-helices could be automatically modeled into the electron density map by AUTOBUILD (30). The best solution stopped at an Rwork/Rfree value of 0.33/0.41. A polypeptide model was then built using the program COOT (31) and refined using REFMAC5 (32) in iterative cycles. The structure was finally refined to 2.16 Å with an Rwork of 18.4% and an Rfree of 21.0%. All structural figures were prepared by using the PyMOL program (33). The structures of EAV nsp11 were all determined by molecular replacement using the PRRSV nsp11 K173A mutant structure as a model.
Size exclusion chromatography.
Proteins (2 to 4 mg) were applied to a Superdex-200 10/300 column (GE Healthcare) equilibrated with a buffer containing 20 mM Tris-HCl (pH 8.0) and 500 mM NaCl. Peak fractions were visualized by SDS-PAGE, followed by Coomassie blue staining.
SAXS experiments.
Small-angle X-ray scattering (SAXS) data were collected at the beamline BL19U2 of the SSRF using our previously published methods (34, 35). Briefly, all proteins were subjected to size exclusion chromatography with a buffer containing 20 mM Tris-HCl (pH 8.0) and 500 mM NaCl. Various concentrations of protein (40 μl) were used, and the data were collected at 1.03 Å with a distance of 1 m from the detector. Individual data were processed by RAW (36). The scattering data from the buffer alone was measured before and after each sample measurement, and the average of the scattering data was used for background subtraction. The theoretical scattering curves from the possible configurations of all proteins were fitted to the experimental scattering curve using the Minimal Ensemble Search (MES) algorithm (37).
Accession number(s).
Coordinates and structure factors have been deposited in the Protein Data Bank under accession codes 5EYI, 5HBZ, and 5HC1.
ACKNOWLEDGMENTS
We thank Eric J. Snijder, Wenhai Feng, and Hanchun Yang for kindly providing cDNA. We thank Jiafu Long for help with analytical ultracentrifugation experiments. We thank Peng Gong for critical reading. We thank the staff at beamlines BL19U2, BL18U, and BL17U at the Shanghai Synchrotron Radiation Facility, beamline BL1A at the Photon Factory in Tsukuba, Japan, and beamline 3W1A at the Beijing Synchrotron Radiation Facility for the excellent technical assistance during data collection.
This work was supported financially by the National Key Research and Development Program of China (2016YFC1200400) and the National Natural Science Foundation of China (31370720, 91519332, and 31570725).
M.Z. performed gene cloning, protein expression, purification, crystallization, diffraction data collection, and structure determination. All authors contributed to experimental design, data analyses, and discussions. M.Z. and Z.C. conceived the project and wrote the manuscript.
We declare that we have no conflicts of interest.
REFERENCES
- 1.Gorbalenya AE, Enjuanes L, Ziebuhr J, Snijder EJ. 2006. Nidovirales: evolving the largest RNA virus genome. Virus Res 117:17–37. doi: 10.1016/j.virusres.2006.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Snijder EJ, Meulenberg JJ. 1998. The molecular biology of arteriviruses. J Gen Virol 79:961–979. doi: 10.1099/0022-1317-79-5-961. [DOI] [PubMed] [Google Scholar]
- 3.Selmi C, Ansari AA, Invernizzi P, Podda M, Gershwin ME. 2002. The search for a practical approach to emerging diseases: the case of severe acute respiratory syndrome (SARS). Dev Immunol 9:113–117. doi: 10.1080/1044667031000137575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Li Y, Zhou L, Zhang J, Ge X, Zhou R, Zheng H, Geng G, Guo X, Yang H. 2014. Nsp9 and Nsp10 contribute to the fatal virulence of highly pathogenic porcine reproductive and respiratory syndrome virus emerging in China. PLoS Pathog 10:e1004216. doi: 10.1371/journal.ppat.1004216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.den Boon JA, Snijder EJ, Chirnside ED, de Vries AA, Horzinek MC, Spaan WJ. 1991. Equine arteritis virus is not a togavirus but belongs to the coronaviruslike superfamily. J Virol 65:2910–2920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Deng Z, Lehmann KC, Li X, Feng C, Wang G, Zhang Q, Qi X, Yu L, Zhang X, Feng W, Wu W, Gong P, Tao Y, Posthuma CC, Snijder EJ, Gorbalenya AE, Chen Z. 2014. Structural basis for the regulatory function of a complex zinc-binding domain in a replicative arterivirus helicase resembling a nonsense-mediated mRNA decay helicase. Nucleic Acids Res 42:3464–3477. doi: 10.1093/nar/gkt1310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Thaa B, Kabatek A, Zevenhoven-Dobbe JC, Snijder EJ, Herrmann A, Veit M. 2009. Myristoylation of the arterivirus E protein: the fatty acid modification is not essential for membrane association but contributes significantly to virus infectivity. J Gen Virol 90:2704–2712. doi: 10.1099/vir.0.011957-0. [DOI] [PubMed] [Google Scholar]
- 8.Li Y, Tas A, Sun Z, Snijder EJ, Fang Y. 2015. Proteolytic processing of the porcine reproductive and respiratory syndrome virus replicase. Virus Res 202:48–59. doi: 10.1016/j.virusres.2014.12.027. [DOI] [PubMed] [Google Scholar]
- 9.Snijder EJ, Kikkert M, Fang Y. 2013. Arterivirus molecular biology and pathogenesis. J Gen Virol 94:2141–2163. doi: 10.1099/vir.0.056341-0. [DOI] [PubMed] [Google Scholar]
- 10.Nga PT, Parquet Mdel C, Lauber C, Parida M, Nabeshima T, Yu F, Thuy NT, Inoue S, Ito T, Okamoto K, Ichinose A, Snijder EJ, Morita K, Gorbalenya AE. 2011. Discovery of the first insect nidovirus, a missing evolutionary link in the emergence of the largest RNA virus genomes. PLoS Pathog 7:e1002215. doi: 10.1371/journal.ppat.1002215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nedialkova DD, Ulferts R, van den Born E, Lauber C, Gorbalenya AE, Ziebuhr J, Snijder EJ. 2009. Biochemical characterization of arterivirus nonstructural protein 11 reveals the nidovirus-wide conservation of a replicative endoribonuclease. J Virol 83:5671–5682. doi: 10.1128/JVI.00261-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Posthuma CC, Nedialkova DD, Zevenhoven-Dobbe JC, Blokhuis JH, Gorbalenya AE, Snijder EJ. 2006. Site-directed mutagenesis of the Nidovirus replicative endoribonuclease NendoU exerts pleiotropic effects on the arterivirus life cycle. J Virol 80:1653–1661. doi: 10.1128/JVI.80.4.1653-1661.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ricagno S, Egloff MP, Ulferts R, Coutard B, Nurizzo D, Campanacci V, Cambillau C, Ziebuhr J, Canard B. 2006. Crystal structure and mechanistic determinants of SARS coronavirus nonstructural protein 15 define an endoribonuclease family. Proc Natl Acad Sci U S A 103:11892–11897. doi: 10.1073/pnas.0601708103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bhardwaj K, Palaninathan S, Alcantara JM, Yi LL, Guarino L, Sacchettini JC, Kao CC. 2008. Structural and functional analyses of the severe acute respiratory syndrome coronavirus endoribonuclease Nsp15. J Biol Chem 283:3655–3664. doi: 10.1074/jbc.M708375200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.He Q, Li Y, Zhou L, Ge XN, Guo X, Yang HC. 2015. Both Nsp1 beta and Nsp11 are responsible for differential TNF-alpha production induced by porcine reproductive and respiratory syndrome virus strains with different pathogenicity in vitro. Virus Res 201:32–40. doi: 10.1016/j.virusres.2015.02.014. [DOI] [PubMed] [Google Scholar]
- 16.Beura LK, Sarkar SN, Kwon B, Subramaniam S, Jones C, Pattnaik AK, Osorio FA. 2010. Porcine reproductive and respiratory syndrome virus nonstructural protein 1beta modulates host innate immune response by antagonizing IRF3 activation. J Virol 84:1574–1584. doi: 10.1128/JVI.01326-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sun Y, Li D, Giri S, Prasanth SG, Yoo D. 2014. Differential host cell gene expression and regulation of cell cycle progression by nonstructural protein 11 of porcine reproductive and respiratory syndrome virus. Biomed Res Int 2014:430508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shi XB, Wang L, Li XW, Zhang GP, Guo JQ, Zhao D, Chai SJ, Deng RG. 2011. Endoribonuclease activities of porcine reproductive and respiratory syndrome virus nsp11 was essential for nsp11 to inhibit IFN-beta induction. Mol Immunol 48:1568–1572. doi: 10.1016/j.molimm.2011.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang D, Fan J, Fang L, Luo R, Ouyang H, Ouyang C, Zhang H, Chen H, Li K, Xiao S. 2015. The nonstructural protein 11 of porcine reproductive and respiratory syndrome virus inhibits NF-kappaB signaling by means of its deubiquitinating activity. Mol Immunol 68:357–366. doi: 10.1016/j.molimm.2015.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Holm L, Rosenstrom P. 2010. Dali server: conservation mapping in 3D. Nucleic Acids Res 38:W545–W549. doi: 10.1093/nar/gkq366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Joseph JS, Saikatendu KS, Subramanian V, Neuman BW, Buchmeier MJ, Stevens RC, Kuhn P. 2007. Crystal structure of a monomeric form of severe acute respiratory syndrome coronavirus endonuclease nsp15 suggests a role for hexamerization as an allosteric switch. J Virol 81:6700–6708. doi: 10.1128/JVI.02817-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yoo D, Song C, Sun Y, Du Y, Kim O, Liu HC. 2010. Modulation of host cell responses and evasion strategies for porcine reproductive and respiratory syndrome virus. Virus Res 154:48–60. doi: 10.1016/j.virusres.2010.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shi Y, Li Y, Lei Y, Ye G, Shen Z, Sun L, Luo R, Wang D, Fu ZF, Xiao S, Peng G. 2016. A dimerization-dependent mechanism drives the endoribonuclease function of porcine reproductive and respiratory syndrome virus nsp11. J Virol 90:4579–4592. doi: 10.1128/JVI.03065-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hengrung N, El Omari K, Serna Martin I, Vreede FT, Cusack S, Rambo RP, Vonrhein C, Bricogne G, Stuart DI, Grimes JM, Fodor E. 2015. Crystal structure of the RNA-dependent RNA polymerase from influenza C virus. Nature 527:114–117. doi: 10.1038/nature15525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ziebuhr J, Snijder EJ, Gorbalenya AE. 2000. Virus-encoded proteinases and proteolytic processing in the Nidovirales. J Gen Virol 81:853–879. doi: 10.1099/0022-1317-81-4-853. [DOI] [PubMed] [Google Scholar]
- 26.Yan Y, Xin A, Zhu G, Huang H, Liu Q, Shao Z, Zang Y, Chen L, Sun Y, Gao H. 2013. Complete genome sequence of a novel natural recombinant porcine reproductive and respiratory syndrome virus isolated from a pig farm in Yunnan province, southwest China. Genome Announc 1:e00003-12. doi: 10.1128/genomeA.00003-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Otwinowski Z, Minor W. 1997. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol 276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 28.Sheldrick GM. 2008. A short history of SHELX. Acta Crystallogr A 64:112–122. doi: 10.1107/S0108767307043930. [DOI] [PubMed] [Google Scholar]
- 29.Terwilliger TC, Berendzen J. 1999. Automated MAD and MIR structure solution. Acta Crystallogr D Biol Crystallogr 55:849–861. doi: 10.1107/S0907444999000839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Terwilliger TC, Grosse-Kunstleve RW, Afonine PV, Moriarty NW, Zwart PH, Hung LW, Read RJ, Adams PD. 2008. Iterative model building, structure refinement and density modification with the PHENIX AutoBuild wizard. Acta Crystallogr D Biol Crystallogr 64:61–69. doi: 10.1107/S090744490705024X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Emsley P, Lohkamp B, Scott WG, Cowtan K. 2010. Features and development of Coot. Acta Crystallogr D Biol Crystallogr 66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Murshudov GN, Skubak P, Lebedev AA, Pannu NS, Steiner RA, Nicholls RA, Winn MD, Long F, Vagin AA. 2011. REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr D Biol Crystallogr 67:355–367. doi: 10.1107/S0907444911001314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schrödinger LLC. 2010. PyMOL molecular graphics system, version 1.3. Schrödinger LLC. [Google Scholar]
- 34.Zhang X, Zhang Q, Xin Q, Yu L, Wang Z, Wu W, Jiang L, Wang G, Tian W, Deng Z, Wang Y, Liu Z, Long J, Gong Z, Chen Z. 2012. Complex structures of the abscisic acid receptor PYL3/RCAR13 reveal a unique regulatory mechanism. Structure 20:780–790. doi: 10.1016/j.str.2012.02.019. [DOI] [PubMed] [Google Scholar]
- 35.Hu Y, Chen Z, Fu Y, He Q, Jiang L, Zheng J, Gao Y, Mei P, Chen Z, Ren X. 2015. The amino-terminal structure of human fragile X mental retardation protein obtained using precipitant-immobilized imprinted polymers. Nat Commun 6:6634. doi: 10.1038/ncomms7634. [DOI] [PubMed] [Google Scholar]
- 36.Nielsen SS, Toft KN, Snakenborg D, Jeppesen MG, Jacobsen JK, Vestergaard B, Kutter JP, Arleth L. 2009. BioXTAS RAW, a software program for high-throughput automated small-angle X-ray scattering data reduction and preliminary analysis. J Appl Crystallogr 42:959–964. doi: 10.1107/S0021889809023863. [DOI] [Google Scholar]
- 37.Pelikan M, Hura GL, Hammel M. 2009. Structure and flexibility within proteins as identified through small angle X-ray scattering. Gen Physiol Biophys 28:174–189. doi: 10.4149/gpb_2009_02_174. [DOI] [PMC free article] [PubMed] [Google Scholar]