Abstract
Abciximab, a derivative of the murine mAb 7E3, protects against ischemic complications of percutaneous coronary interventions by inhibiting ligand binding to the αIIbβ3 receptor. In this study we identified regions on integrin β3 that control 7E3 binding. Murine/human amino acid substitutions were created in two regions of the βA domain that previous studies found to influence 7E3 binding: the C177–C184 loop and K125–N133. The T182N substitution and a K125Q mutation reduced 7E3 binding to human β3 in complex with αIIb. The introduction of both the human C177–C184 region and human W129 into murine β3 was necessary and sufficient to permit 7E3 binding to the human αIIb/murine β3 complex. Although we cannot exclude allosteric effects, we propose that 7E3 binds between C177–C184 and W129, which are within 15 Å of each other in the crystal structure and close to the β3 metal ion-dependent adhesion site. We previously demonstrated that 7E3 binds more rapidly to activated than unactivated platelets. Because it has been proposed that αIIbβ3 changes from a bent to an extended conformation upon activation, we hypothesized that 7E3 binds less well to the bent than the extended conformation. In support of this hypothesis we found that 7E3 bound less well to an αIIbβ3 construct locked in a bent conformation, and unlocking the conformation restored 7E3 binding. Thus, our data are consistent with αIIbβ3 existing in variably bent conformations in equilibrium with each other on unactivated platelets, and activation resulting in αIIbβ3 adopting a more extended conformation.
The integrin αIIbβ3 and αVβ3 receptors are important in a number of physiologic and pathologic phenomena, including hemostasis, thrombosis, tumor angiogenesis, and bone resorption (1, 2). The murine mAb 7E3 (3) blocks ligand binding to both αIIbβ3 and αVβ3 receptors (4, 5). Abciximab is a mouse/human chimeric Fab fragment of the mAb 7E3 that inhibits αIIbβ3-mediated platelet aggregation and is approved for human use to prevent the ischemic complications associated with percutaneous coronary interventions (6).
Previous studies of 7E3 binding to cells expressing recombinant αIIbβ3 receptors demonstrated that: (i) swapping select murine for human αIIb sequences does not decrease 7E3 binding (7), (ii) removing the specificity determining loop (SDL) (K156–G189 sequence) from β3 results in loss of 7E3 binding (8), (iii) swapping the murine for human C177–C184 sequence within the SDL region in β3 results in loss of 7E3 binding (7), and (iv) swapping the murine S129–T133 sequence for the human W129–N133 sequence results in partial loss of 7E3 binding (7).
The above human/mouse swapping studies identified regions within β3 that affect 7E3 binding, but the biochemical and functional properties of these chimeric receptors were not characterized. In addition, the W129–N133 region contains two amino acid differences between human β3(β3Hu) and mouse β3 (β3M) and the C177–C184 region contains three amino acid differences (Table 1). To define further the regions on αIIbβ3 that control 7E3 binding, we assessed the effects of individual amino acid substitutions on 7E3 binding to cells expressing αIIbβ3, as well as the effects of these substitutions on receptor biochemistry and function. In addition, we studied the effect of mutating a highly conserved lysine residue (K125) that appears to link the C177–C184 loop to the α-helix containing W129–N133.
Table 1. Constructs for mammalian cell expression.
| Construct | Name | 129-133 | 177-184 |
|---|---|---|---|
| Human β3 | β3Hu | 129WSIQN133 | 177CYDMKTTC184 |
| Human β3 mouse 177-184 | β3Hu—M177-184 | 129-----133 | 177--N--NA-184 |
| Human β3 D179N | D179N | 129-----133 | 177--N-----184 |
| Human β3 T182N | T182N | 129-----133 | 177-----N--184 |
| Human β3 T183A | T183A | 129-----133 | 177------A-184 |
| Human β3 C177A + C184A | C177A-C184A | 129-----133 | 177A------A184 |
| Human β3 C177A-M-C184A | 129-----133 | 177A-N--NAA184 | |
| Human β3 W129S | W129S | 129S----133 | 177--------184 |
| Human β3 N133T | N133T | 129----T133 | 177--------184 |
| Human β3 K125A/R/Q | K125A/R/Q | 129-----133 | 177--------184 |
| Human αIIb-R320C β3-R563C | αIIbR320Cβ3R563C | 129-----133 | 177--------184 |
| Mouse β3 | β3M | 129S---T133 | 177--N--NA-184 |
| Mouse β3 human 177-184 | β3M—Hu177-184 | 129S---T133 | 177--------184 |
| Mouse β3 human 177-184 + S129W | β3M—Hu177-184+W129 | 129----T133 | 177--------184 |
Finally, in view of our localization of the region involved in 7E3 binding to the head region of β3 adjacent to the arginine-glycine-aspartic acid (RGD) binding site, our previous studies demonstrating that 7E3 IgG (but not 7E3 Fab) binds much more rapidly to activated than unactivated platelets, (3, 9), and our recently proposed model of αIIbβ3 undergoing a change from a bent to an extended conformation upon activation, (10, 11), we also assessed the binding of 7E3 IgG and 7E3 Fab to an αIIbβ3 receptor reversibly locked in a bent conformation (10).
Materials and Methods
mAbs. The mAbs used in this study were: 7E3 IgG and Fab (anti-αVβ3+αIIbβ3) (4), 10E5 (αIIbβ3-specific) (5), LM609 (αVβ3-specific) (kindly provided by David Cheresh, The Scripps Research Institute, La Jolla, CA), AP3 (β3-specific) (kindly provided by Peter Newman, Blood Center of Southeastern Wisconsin, Milwaukee) (12), FITC-labeled goat anti-mouse IgG F(ab′)2 (The Jackson Laboratory), and purified mouse IgG (The Jackson Laboratory).
Generation of Constructs. β3Hu and human αIIb cDNAs were generously provided by Peter Newman; β3M cDNA was a generous gift from Patrick Ross (Washington University School of Medicine, St. Louis). Table 1 lists the constructs used for this study. Mutations in β3Hu and β3M/pcDNA3.1 cDNA constructs were generated by using either the splice by overlap extension PCR method as described (13) or the QuikChange XL Site-Directed Mutagenesis Kit (Stratagene) as per the manufacturer's instructions. The generation of pEF–αIIbR320C and pCDNA3.1–β3R563C has been described (10).
Cell Transfections. Normal or mutant αIIbβ3 receptors were expressed in Chinese hamster ovary (CHO), human 293T, and human 293 cells. CHO cells express relatively small amounts of endogenous hamster αV, which can combine with transfected β3 to form αVβ3. The 293 and 293T cells express relatively small amounts of endogenous human αV, which can combine with transfected β3 to form αVβ3. The expression of these small amounts of αVβ3 was considered in designing the experiments and interpreting the results. Transfections were performed with Lipofectamine (GIBCO/BRL), PerFectin (Gene Therapy System, San Diego), or CaCl2/Hepes methods (14). Transfections using Lipofectamine and PerFectin were performed according to the manufacturer's instructions. For transfections using the CaCl2/Hepes method, cells were plated to ≈20% confluency in 100-mm tissue culture dishes and incubated overnight at 37°C. A DNA transfection solution [DNA (6–10 μg) and 2 M CaCl2 (62 μl) in 500 μl of double distilled H2O and 500 μl of Hepes-buffered saline] was added to the cells and incubated for an additional 48 h; the cells were then washed with PBS and harvested. For stable transfections, cells were selected in media containing 800 μg/ml G418 for 2–4 weeks. To obtain a population of cells expressing higher levels of human and chimeric receptors, cells were labeled with the mAb 10E5 (αIIbβ3-specific) and sorted with a FACSCalibur cell sorter (Becton Dickinson).
Flow Cytometry. Cells (4–6 × 105 cells per ml) were incubated with primary antibody (5–10 μg/ml) for 30 min at room temperature followed by FITC-labeled goat anti-mouse IgG F(ab′)2 (10 μg per sample) for 30 min on ice. For some experiments 7E3 and 10E5 were labeled with Alexa Fluor 488 (Molecular Probes) as per the manufacturer's protocol and then used to directly label the cells. Cells were washed, resuspended in PBS containing 2% FBS, and analyzed with a FACSCalibur flow cytometer. Background controls were transfected cells incubated with secondary FITC-labeled antibody alone or cells transfected with pCDNA3.1 and then incubated with FITC-labeled 10E5 and/or 7E3; background levels were set such that the control cells had ≤5% positive cells. In some experiments surface β3 receptor expression was normalized for total β3 integrin expression by using the anti-β3Hu-specific mAb VIPL2. In some experiments cells were pretreated with 2 mM DTT for 15 min at room temperature before adding the mAbs.
Fibrinogen Adhesion Assay. To minimize any contribution of αVβ3 receptors to fibrinogen adhesion of transfected cells, the assay was conducted in the presence of Ca2+, which preferentially supports αIIbβ3-mediated ligand binding rather than αVβ3-mediated ligand binding (15). Wells of polystyrene (Nunc) flat-bottom 96-well plates were precoated with Tris-saline buffer (50 mM Tris·HCl/100 mM NaCl, pH 7.4) (100 μl per well) containing 10 μg/ml human fibrinogen (Enzyme Research Laboratories, South Bend, IN) for 1 h at room temperature. The wells were blocked with DMEM (GIBCO/BRL) (100 μl per well) containing 1% BSA for 1 h at room temperature. Cells (106 cells per ml) were preincubated with mAbs 7E3 or 10E5, or purified mouse IgG (10 μg/ml), and 2 mM CaCl2 for 20 min at room temperature, and then added to the wells (100 μl per well) and incubated at 37°C for 1 h. Nonadherent cells were removed by gently rinsing the wells three times with Tris-saline buffer (200 μl per well). Adherent cells were fixed with 4% formaldehyde for 20 min, and then stained with a solution containing 0.5% crystal violet and 20% methanol for 30 min at room temperature. After washing, a 10% acetic acid solution was added (100 μl per well) to solubilize the dye, and the OD550 of the solution was measured on a microplate reader (Thermo Max, Biomatic Technologies, Stoughton, MA). The OD550 using untransfected cells was considered background and subtracted from the OD550 using transfected cells.
Biosynthetic Labeling and Immunoprecipitation. Cells (5 × 105 per 60-mm tissue culture well; two wells per group) were incubated at 37°C in medium containing [35S]methionine/cysteine (300 μCi per well) for 2–4 h. Cells were then incubated for 30 min on ice with lysis buffer (150 mM NaCl/50 mM Tris·HCl, pH 7.5) containing 1% Triton X-100 and PMSF (2 mM), leupeptin (10 μM), calpeptin (20 μM), and N-ethylmaleimide (10 mM). Lysates were centrifuged at 4°C at 12,000 × g for 30 min, and supernatants were precleared with protein A or protein G Sepharose (Amersham Pharmacia) at 4°C for 30 min. Using equivalent amounts of trichloroacetic acid-precipitable radioactivity (≈5–6 × 106 counts per sample), immunoprecipitations were performed with 10E5 or 7E3 (8 μg/ml). Protein G Sepharose (5%) was added and incubated for 1 h at 4°C. The beads were washed twice with lysis buffer containing 500 mM NaCl, and then the bound material was eluted with SDS/PAGE sample buffer at 100°C. Samples (reduced and nonreduced) were separated by SDS/PAGE (7.5% gel) and the dried gels were exposed to film (Blue Sensitive, Denville Scientific, Metuchen, NJ). Preliminary studies demonstrated that antibody LM609 immunoprecipitated only trace amounts of αVβ3 from stable 293 cell lines expressing either αIIbβ3 or just β3, indicating that only trace amounts of αVβ3 are produced by these cells (data not shown).
Immunoblotting. Preparation of cell lysates and immunoprecipitation with 10E5 or 7E3 were performed as above. For some experiments cells were labeled for 30 min with a sulfo-N-hydroxysuccinimide-biotin (1 mg/ml) (Pierce) on ice; the reaction was stopped by adding glycine (5 mM final concentration). Samples (reduced and nonreduced) were separated by SDS/PAGE (7.5% gel) and electro-transferred to poly(vinylidene difluoride) membranes. Membranes were blocked in 5% nonfat dry milk for 1 h at room temperature, washed, and hybridized with a primary antibody (10 μg/ml) for 2 h at room temperature or overnight at 4°C. Membranes were washed twice in 0.1% Tween-TBS and incubated with horseradish peroxidase (HRP)-conjugated AP3 (anti-β3) or HRP-avidin for 1 h at room temperature. Membranes were washed and developed by using chemiluminescence as per the manufacturer's instructions (Amersham Pharmacia).
Molecular Modeling. The structural effects of the substitutions were analyzed by using a molecular modeling approach using the charmm molecular program as described (16). Briefly, the individual amino acids were substituted into the β-A domain of β3 and then energy minimization was performed with the Adopted Basis Newton–Raphson method (17).
Results
7E3 Binding to Receptors Containing β3M C177–C184 Amino Acid Residues. Antibodies 10E5 and 7E3 reacted with similar percentages of 293T cells transfected with normal human αIIb and normal β3Hu, 42 ± 12% and 46 ± 17%, respectively (Table 2). In sharp contrast, when 293T cells were transfected with human αIIb in conjunction with β3Hu containing the murine C177–C184 sequence (β3Hu–M177-184), 16 ± 10% were positive with 10E5, but only 0.1 ± 0.02% were positive with 7E3 (P < 0.01). Similar results were obtained with CHO cells expressing endogenous hamster αV with either β3Hu or β3Hu–M177-184; both sets of cells reacted with antibody LM609, which is specific for αVβ3 (data not shown). Thus, the binding of 7E3 to both αIIbβ3 and the hamster/human hybrid αVβ3 were affected by substituting the murine C177–C184 sequence.
Table 2. Percentage (± standard deviation) of 293T cells expressing αIIb and either normal or mutated β3 that binds 10E5 or 7E3 as judged by flow cytometry.
| Construct | 10E5 | 7E3 | 7E3/10E5 | P value (10E5 vs. 7E3) |
|---|---|---|---|---|
| β3Hu | 42 ± 12 | 46 ± 17 | 1.1 | n.s. |
| β3Hu-M177-184 | 16 ± 10 | 0.1 ± 0.2 | 0.01 | <0.01 |
| β3M | 36 ± 18 | 2 ± 2 | 0.06 | <0.01 |
| β3M-Hu177-184 | 32 ± 8 | 24 ± 6 | 0.75 | <0.01 |
| β3M-Hu177-184+W129 | 32 ± 8 | 32 ± 8 | 1 | n.s. |
n.s., Not significant.
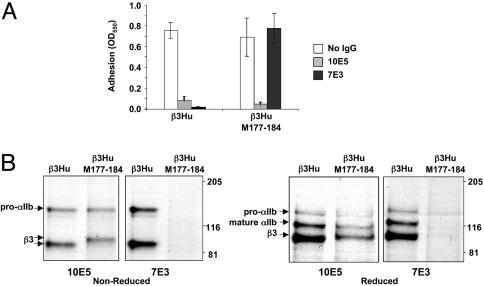
To ascertain whether the flow cytometry data correlate with antibody-induced inhibition of ligand binding, transfected CHO cells were tested for their ability to adhere to fibrinogen (Fig. 1A). Cells transfected with human αIIb and either β3Hu or β3Hu–M177-184 adhered to immobilized fibrinogen, and adhesion of both transfected cell lines was markedly inhibited by mAb 10E5 (Fig. 1 A). In contrast, although the adhesion of cells transfected with β3Hu was markedly inhibited by mAb 7E3, 7E3 did not inhibit the adhesion of cells transfected with β3Hu–M177-184.
Fig. 1.
Effects of swapping of murine sequence in β3Hu on 7E3 binding. (A) Adhesion to immobilized fibrinogen of CHO cells transiently expressing β3Hu or β3Hu–M177-184 in association with human αIIb. Cells were preincubated with 2 mM Ca2+ and different antibodies. Adhesion is expressed as OD550 of solubilized crystal violet-stained cells. Data presented are the mean ± standard deviation of three experiments. (B) Radiograph of immunoprecipitates under nonreduced and reduced conditions of 35S-labeled cells using 10E5 or 7E3. Cells were transiently transfected with β3Hu or β3Hu–M177-184 in association with human αIIb.
To assess the biochemical characteristics of the receptors, transfected cells were grown in the presence of [35S]methionine/cysteine for 3 h and lysed; the resulting lysates were then immunoprecipitated with either mAb 10E5 or mAb 7E3 (Fig. 1B). 10E5 immunoprecipitated radiolabeled receptors from the lysates of the cells transfected with human αIIb in conjunction with either β3Hu or β3Hu–M177-184. When samples were analyzed under nonreduced conditions, β3Hu–M177-184 migrated slower than did β3Hu. Under reduced conditions, however, the migration of β3Hu and β3Hu–M177-184 were similar or identical, suggesting that the difference in migration patterns under nonreduced conditions was caused by loss of, or altered, disulfide bond formation in β3Hu–M177-184. Trace amounts of pro-αIIb were also observed in reduced samples of 10E5 immunoprecipitates of both constructs. As expected, β3Hu–M177-184 was not immunoprecipitated by 7E3, confirming the data obtained in the flow cytometry and adhesion studies.
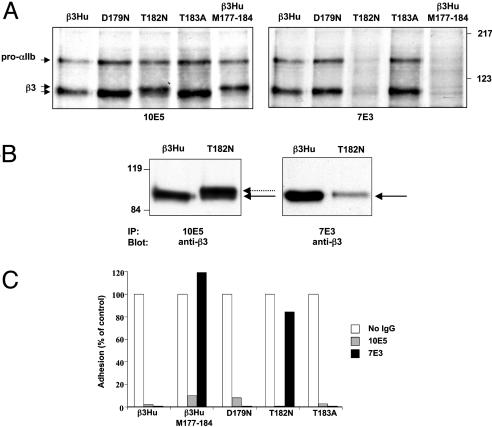
Biochemical, Functional, and Immunologic Characterization of αIIbβ3 Receptors with Single Murine/Human Amino Acid Substitutions in the β3 C177–C184 Sequence. To identify the amino acid(s) within the murine C177–C184 sequence that affect the binding of 7E3, we prepared β3 constructs containing the three single amino acid substitutions that distinguish the murine from the human sequence (D179N, T182N, T183A) (Fig. 2A). Cells were cotransfected with human αIIb and either normal or mutant β3. Receptors reactive with 10E5 were expressed at comparable levels as demonstrated by similar patterns of αIIbβ3 immunoprecipitation of biosynthetically labeled αIIbβ3 by 10E5 (Fig. 2 A). In contrast, although 7E3 and 10E5 immunoprecipitated comparable amounts of αIIbβ3 from β3Hu, D179, and T183 cells, it precipitated only a small amount of αIIbβ3 from T182N, and nearly none of the αIIbβ3 from β3Hu–M177-184. The results of the adhesion studies were consistent with the immunoprecipitation data (Fig. 2C). Thus, 10E5 inhibited adhesion with all of the constructs, indicating the dominant role of αIIbβ3 in mediating the adhesion. In sharp contrast, 7E3 did not inhibit the adhesion of the β3Hu–M177-184 cells and only slightly reduced the adhesion of the T182N cells. Under nonreduced conditions, the β3 containing the T182N substitution immunoprecipitated by 10E5 migrated as a doublet, with the less intense lower band migrating at the same Mr as β3Hu and the more intense upper band migrating at the same Mr as β3Hu–M177-184 (Fig. 2 A and B). In contrast 7E3 appeared to only immunoprecipitate the minor form that migrated at the same Mr as β3Hu (Fig. 2 A and B). These data suggest that 7E3 can bind to a subpopulation of T182N receptors that adopts the proper conformation, but we cannot rigorously exclude the possibility that these observations are caused by differences in affinity. The β3Hu–T182N doublet disappeared when the samples were reduced (data not shown).
Fig. 2.
Effects of introducing single murine substitutions in β3Hu C177–C184. (A) Radioautographs of 35S-labeled cells immunoprecipitated with 10E5 or 7E3 and electrophoresed under nonreduced conditions in SDS/PAGE. (B) Immunoblot analysis using mAb AP3 (anti-β3) of 7E3 or 10E5 immunoprecipitates of lysates of cells transfected with β3Hu or T182N. T182N was immunoprecipitated by 10E5 and migrated as a closely spaced doublet. Only the minor band migrating with the same Mr as β3Hu was immunoprecipitated by 7E3. (C) Adhesion to immobilized fibrinogen of 293T cells expressing native and mutated β3. Cells were preincubated with 2 mM Ca2+. Data are the mean of two experiments and are normalized for adhesion of cells in the absence of antibodies.
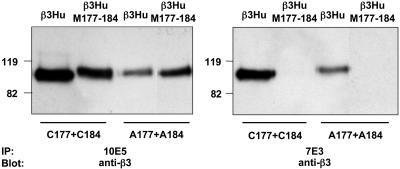
Effect of Modifying the C177–C184 Disulfide Bond on 7E3 Binding. To determine whether 7E3 binding to β3 receptors has an absolute requirement for disulfide bond formation between residues C177 and C184, constructs were generated in which alanines were substituted for the two cysteines. Cell lysates were immunoprecipitated with 10E5 or 7E3 and immunoblotted with mAb A P3. 10E5 immunoprecipitated receptors containing C177A+C184A; under nonreduced conditions this β3 mutant migrated as did β3Hu–M177-184, that is, more slowly than β3Hu (Fig. 3). Because the C177A+C184A mutant cannot form the normal C177–C184 disulfide bond, these data support the hypothesis that both the β3Hu–M177-184 and T182N substitutions also disrupt the C177–C184 disulfide bond. At least a portion of the C177A+C184A-containing receptors could also be immunoprecipitated with 7E3, suggesting that at least some of the molecules adopt a conformation(s) recognized by 7E3 even in the absence of a disulfide bond between residues 177 and C184 in β3Hu (Fig. 3). The substitution of the three murine residues (M177-184) into the C177A+C184A construct (A177–A184), however, eliminated 7E3 binding. Thus, both the conformation and the amino acid composition of the C177–C184 sequence can profoundly affect 7E3 binding.
Fig. 3.
Effect of substituting alanines for C177 and C184 on 10E5 and 7E3 binding. Lysates of constructs containing β3Hu or β3Hu–M178-183 and/or C177A+C184A residues were immunoprecipitated by 10E5 (Left) or 7E3 (Right), electrophoresed under nonreduced conditions in SDS/PAGE, and immunoblotted with anti-β3 mAb AP-3.
Effect of β3 W129S and T133N Substitutions on 7E3 Binding to αIIbβ3 Receptors. Puzon-McLaughlin et al. (7) previously showed that substituting both β3 W129S and N133T partially decreased 7E3 binding, so we assessed the effects of making the two individual single amino acid substitutions on 7E3 binding. By flow cytometry and immunoprecipitation, neither single amino acid substitutions affected 7E3 binding (data not shown), suggesting that both substitutions are required in combination to partially decrease 7E3 binding.
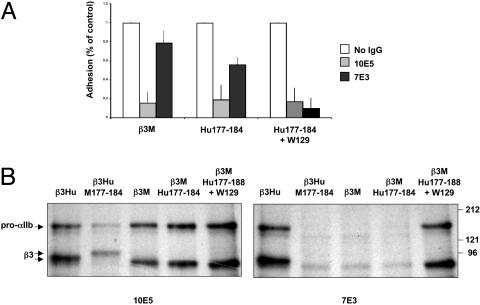
Studies to Engineer the 7E3 Epitope into Receptors Containing β3M and Human αIIb. β3M constructs were generated containing either β3Hu amino acids C177–C184 (β3M–Hu177-184) or β3Hu C177–C184 plus the β3Hu tryptophane at position 129 (β3M–Hu177-184+W129). Cells transiently or stably expressing human αIIb in combination with WT β3M or the chimeric β3 subunits were analyzed for 7E3 binding by flow cytometry, SDS/PAGE, and cell adhesion. By flow cytometry, 7E3 binding to cells expressing β3M–Hu177-184 was greater than 7E3 binding to cells expressing β3M (Table 2), but significantly less than 10E5 binding to the same cells (P < 0.01). In sharp contrast, 7E3 binding to cells expressing β3M–Hu177-184+W129 was virtually identical to 10E5 binding (P = not significant) (Table 2). Cell adhesion to immobilized fibrinogen in the absence of inhibitory antibodies was similar for cells expressing β3M, β3M–Hu177–184, or β3M–Hu177-184+W129 (Fig. 4A). 10E5 inhibited adhesion of cells expressing each of these chimeric receptors by 80% or more. In contrast, and consistent with the flow cytometry data, 7E3 did not significantly inhibit adhesion to immobilized fibrinogen of cells expressing β3M, only partially inhibited adhesion of cells expressing β3M–Hu177-184, and nearly completely inhibited adhesion of cells expressing β3M–Hu177-184+W129. These findings were supported by immunoprecipitation analyses of 35S-labeled cell lysates in which 7E3 did not immunoprecipitate the receptors from cells expressing β3M or β3M–Hu177-184, but did immunoprecipitate the receptors from cells expressing β3M–Hu177-184+W129 (Fig. 4B).
Fig. 4.
Engineering of 7E3 epitope into β3M. (A) Adhesion to immobilized fibrinogen of cells stably expressing native and mutated β3. Cells were preincubated with 2 mM Ca2+ and purified murine IgG, 10E5, or 7E3. Data are the mean of three experiments ± standard deviation and are normalized for adhesion of cells in the presence of purified murine mAb. (B) Radioautographs of immunoprecipitates using 10E5 or 7E3 of lysates of 35S-labeled cells transfected with β3Hu, β3M, or chimeric β3 and electrophoresed under nonreduced conditions in SDS/PAGE.
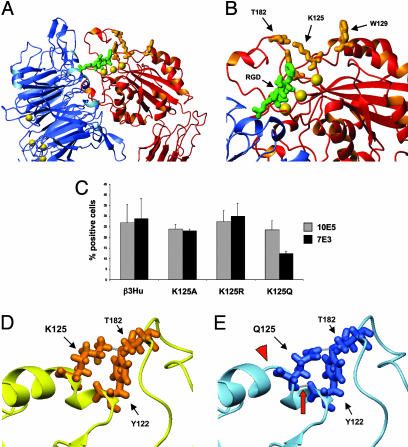
Effect of K125 Substitutions on 7E3 Binding to αIIbβ3 Receptors. We further investigated the effects of substitutions in K125, which is present in both β3Hu and β3M. K125 is on α-helix 1 in close proximity to W129; it forms hydrogen bonds with Y122, and the latter forms a hydrogen bond with T182, thus linking the α-helix 1 to the C177–C184 loop (Fig. 5 A and B) (18). Neither the K125A nor the K125R substitution altered 7E3 binding. However, when K125 was mutated to a glutamine, the binding of 7E3 was reduced by ≈50% relative to 10E5 (Fig. 5C). The structural effects of these substitutions were analyzed by using a molecular modeling approach. The alanine and arginine substitutions at K125 did not affect the structure or the hydrogen-bonding pattern in the region. In contrast, the substitution of the smaller glutamine for K125 resulted in the development of a new hydrogen bond between Q125 and Y122, as well as a 1-Å shift in T182 toward Y122 and a 1-Å shift in Y122 toward Q125 (Fig. 5 D and E, arrowhead). In addition, there was partial loss of α-helix 1 (from amino acids 125–127), increasing the flexibility of this region (Fig. 5 D and E, arrow).
Fig. 5.
The role of K125 in 7E3 binding. (A) Crystal structure of selected regions of the external domain of integrin αVβ3 in complex with the peptide RGDW as per Xiong et al. (31). The αV propeller is in blue, and the β3–βA domain is in red. The peptide is in green, and Mn2+ ions are depicted as yellow spheres. (B) Higher-power view of the top surface of the β3–βA domain in association with the peptide. The three yellow spheres represent the three Mn2+ ions in the ligand-associated metal binding site, MIDAS, and adjacent to MIDAS positions, respectively. The amino acids depicted in orange are the ones described in the text as important in 7E3 binding. (C) Effect of K125 substitutions on 10E5 and 7E3 binding. Data are the mean ± standard deviation of three independent experiments. (D and E) Molecular modeling of the effect of the Q125 substitution on C177–C184 and α-helix 1. Models are rotated 180° in comparison with A and B. Residues at positions 122, 125, and 182 are shown for β3Hu (D) and K125Q (E). Selected hydrogen bonds are depicted in gray and the new hydrogen bond between Y122 and K125Q is identified by a red arrow. Notice the reorientation of K125 as a result of the additional hydrogen bond, along with the loss of a portion of the α-helical structure of α-helix 1 (red arrowhead).
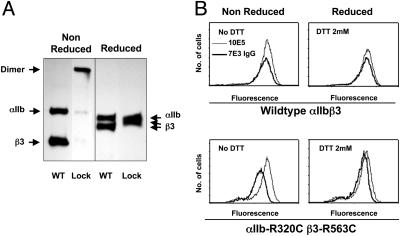
Effects of Locking αIIbβ3 in a Bent Conformation on 7E3 and 10E5 Binding. Some 293 cells were generated that stably expressed αIIbβ3 or the mutant αIIb-R320C β3-R563C receptor, which locks αIIbβ3 into a bent position by introducing a disulfide bond between the αIIb propeller and the EGF3 domain of β3 (10). In accord with our previous report, almost all of the mutant receptors formed disulfide-bonded bent-locked heterodimers (Fig. 6A). As judged by flow cytometry, the mean fluorescent intensity (MFI) binding ratio of 7E3 IgG/10E5 to αIIbβ3-expressing cells was 0.78 ± 0.04. In sharp contrast, the 7E3 IgG/10E5 MFI ratio was only 0.56 ± 0.05 with cells expressing αIIb-R320C β3-R563C (P < 0.008; Fig. 6B and Table 3). When the smaller 7E3 Fab fragment was tested, the 7E3 Fab/10E5 MFI ratios were nearly identical in cells expressing the normal and the mutated receptors (0.58 ± 0.04 and 0.56 ± 0.04). Mild reduction with DTT (2 mM for 15 min) did not affect 10E5 binding to either αIIbβ3 or mutant-expressing cells (Table 3 and Fig. 6B). DTT increased 7E3 IgG binding to normal αIIbβ3 by only 12%, but increased the 7E3 IgG binding to the cells expressing the mutant receptor such that the 7E3 IgG/10E5 MFI ratio increased by 32%. The 7E3 Fab binding increased by only 5% and 9% with DTT reduction of αIIbβ3 and mutant receptors, respectively.
Fig. 6.
7E3 binding to αIIbβ3 and αIIb-R320C β3-R563C. (A) Immunoprecipitation with 10E5 of lysates of surface biotin-labeled 293 cells expressing αIIbβ3 or αIIb-R320C β3-R563C. Samples were electrophoresed under nonreduced and reduced conditions. (B) Fluorescent intensity of cells expressing αIIbβ3 or αIIb-R320C β3-R563C after incubation with Alexa647-7E3 IgG or Alexa647-10E5, with or without treatment with 2 mM DTT.
Table 3. MFI of cells expressing αIIbβ3 or αIIb-R320C β3-R563C with presence or absence of 2 mM DTT.
| 10E5 | 7E3 IgG | 7E3 Fab | |
|---|---|---|---|
| αIIbβ3 | 435 ± 85 | 336 ± 49 | 255 ± 64 |
| αIIbβ3 + DTT | 440 ± 96 (1%) | 379 ± 55 (12%) | 267 ± 77 (5%) |
| αIIb-R320C β3-R563C | 389 ± 63 | 218 ± 43 | 218 ± 53 |
| αIIb-R320C β3-R563C + DTT | 387 ± 64 (-1%) | 290 ± 53 (32%)* | 237 ± 78 (9%) |
Data are the mean ± SD of four separate experiments. In parentheses is shown the change in MFI after DTT treatment.
P < 0.008 with DTT treatment.
Discussion
In the present study we provide immunologic, biochemical, and functional data on the binding of 7E3 and the mechanism of integrin activation. Because the binding properties of 7E3 IgG and related forms of 7E3 have been well characterized (3, 5, 9), these data have implications for 7E3's mechanism of inhibition of ligand binding as well as the structural changes in β3 integrins that accompany platelet activation. In accord with the study of Puzon-McLaughlin et al. (7) we found that swapping the murine for the β3Hu C177–C184 sequence (β3Hu–M177-184) greatly reduced 7E3 binding. This chimeric β3 subunit migrated slower than β3Hu on SDS/PAGE under nonreduced conditions, suggesting that the insertion of the murine residues in the human backbone reduces the likelihood of forming the C177–C184 bond. This hypothesis is strengthened by the observation that β3Hu–M177-184 migrated at the same Mr as a construct in which the C177–C184 bond could not be formed because alanines were substituted for C177 and C184. However, it is not the loss of the disulfide bond itself that causes the loss of the 7E3 epitope, because at least some of the receptors with the C177A and C184A substitutions could still be immunoprecipitated by 7E3. Only when the β3M 179–183 sequence was swapped into the C177A+C184A construct was 7E3 binding lost.
Of the three differences in amino acids in the human and murine C177–C184 sequences, only the T182N substitution reduced the binding of 7E3 as judged by immunoprecipitation. In accord with these binding data, adhesion to immobilized fibrinogen of cells expressing this mutated receptor was not inhibited by 7E3. Remarkably, the β3 subunit containing the T182N substitution migrated in SDS/PAGE gels as a closely spaced doublet. The minor band migrated as did β3Hu and was immunoprecipitated by 7E3. The major band migrated more slowly, like the β3 subunit containing alanines in place of cysteines at positions 177 and 184, and could not be immunoprecipitated by 7E3. Our hypothesis is that the T182N substitution decreases the likelihood of the formation of the C177–C184 disulfide loop, thus resulting in the presence of two different conformations, one in which the C177–C184 disulfide bond forms (faster migrating) and the other in which it does not (slower migrating). 7E3 can react with the former, but not the latter, indicating that 7E3 binding does not absolutely require the threonine at position 182. These data were confirmed with functional studies of 7E3-induced inhibition of adhesion to immobilized fibrinogen.
Puzon-McLaughlin et al. (7) also reported that swapping the murine W129–N133 sequence into β3Hu resulted in partial loss of 7E3 binding. We did not observe a loss of 7E3 binding when the two individual substitutions in this region (W129S and N133T) were made, suggesting that both substitutions are required to affect 7E3 binding.
Because the loss of 7E3 binding upon altering one or more amino acids in β3 provides only presumptive evidence for a direct role of the amino acid in 7E3 binding, we sought more convincing evidence by attempting to re-engineer β3M so that 7E3 would bind to it when complexed with human αIIb. We demonstrated that insertion of the human C177–C184 sequence into β3M supported partial binding of 7E3 as judged by flow cytometry, but the binding appeared to be of low affinity because 7E3 did not immunoprecipitate the receptor, and 7E3 only partially inhibited the adhesion to immobilized fibrinogen of cells expressing this construct. When the W129S substitution was added to β3M containing the human C177–C184 loop, however, 7E3 not only reacted with the receptor by flow cytometry and immunoprecipitation, but also eliminated cell adhesion. Thus, substitutions in both regions of β3M are required for 7E3 binding to cells coexpressing human αIIb.
Analysis of species differences in the ability of 7E3 to inhibit platelet αIIbβ3 is also consistent with the importance of both human C177–C184 and W129 in 7E3 binding (19–21). As shown in Table 4, a decreasing order of binding affinity of 7E3 for human (5), dog (22), and rat (23) platelets is observed, whereas 7E3 does not bind at all to platelets from pig (B.S.C., unpublished observation) and mouse. Dog β3 shares the same 177–184 sequence as human, but has the W129S substitution found in β3M. Rat β3 has the same W129S and N133T substitutions as mouse, but it does not have the T182N substitution found to be so important in the loss of 7E3 binding to β3M; instead it has a conservative T182S substitution. Pig β3 is particularly instructive because the pig and human 177–184 sequences are identical, but pig has a W129E substitution that introduces a charged amino acid in place of a hydrophobic residue.
Table 4. 7E3 binding in different species and amino acid sequences of Site1 and Site2 on β3.
| Species | 7E3 activity | 129-133 | 177-184 |
|---|---|---|---|
| Human | +++++ | 129WSIQN133 | 177CYDMKTTC184 |
| Dog | +++ | 129S----133 | 177--------184 |
| Rat | + | 129S---T133 | 177--T--ST-184 |
| Pig | - | 129E----133 | 177--------184 |
| Mouse | - | 129S---T133 | 177--N--NA-184 |
Although W129 is not continuous with C177–C184 in the primary sequence of β3Hu, the crystal structure of αVβ3 shows these regions to be contiguous in the folded protein (18). These data provide very strong complementary support for the biochemical data indicating that both regions participate in 7E3 binding. Moreover, this region is very close to the β3 metal ion-dependent adhesion site (MIDAS), which participates in ligand binding. Thus, although we cannot exclude the possibility of allosteric effects, we propose that the 7E3 epitope is on or near the ≈15-Å ridge that extends between the C177–C184 loop and W129 (Fig. 5 A and B). This proposed location will, however, require direct confirmation by crystallographic analysis. This region contains the conserved residues Y122 and K125. Y122 may also be important to the 7E3 epitope because Y122 appears to form hydrogen bonds with both K125 and T182. Thus, we found that the K125Q substitution, which alters the hydrogen-bonding pattern that links the C177–C184 loop to α-helix 1, decreased 7E3 binding by ≈50%, strengthening the hypothesis that α-helix 1, which contains W129, is important in maintaining the 7E3 epitope.
The proximity of the proposed 7E3 epitope to the MIDAS provides an explanation for the ability of 7E3 to inhibit ligand binding and suggests that 7E3 inhibits ligand binding by steric hindrance, an allosteric effect on the RGD binding region, or both effects. Our previous studies demonstrating that RGD peptides in solution do not inhibit 7E3 binding to αIIbβ3 (24), but that 7E3 inhibits the binding to αIIbβ3 of RGD peptides immobilized on beads (24), are also consistent with 7E3 operating through steric hindrance, allosteric changes in the RGD binding region, or both effects. The data in this study also provide an explanation for our previous observation that 7E3 blocks the binding of antibody LM609 to αVβ3 because previous studies by Puzon-McLaughlin et al. (7) demonstrated that LM609 did not bind to human αVβ3 when the region immediately adjacent to the C177–C184 loop was swapped with the murine sequence.
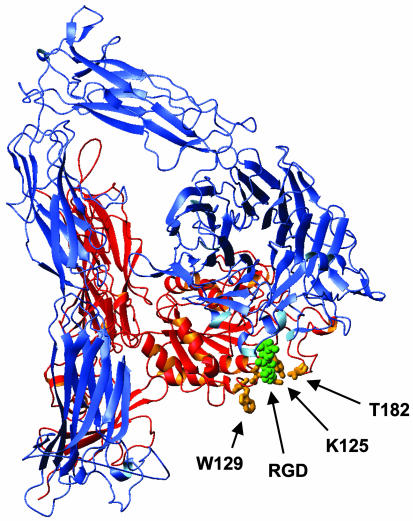
Based on the crystallographic data of Xiong et al. (18) and our NMR and electron microscopic data (10, 25) we previously proposed that the αVβ3 receptor assumes variably bent conformations. Thus, if αIIbβ3 is similarly bent, the 7E3 epitope may be only partially accessible on the most highly bent forms of the receptor as a result of steric hindrance from the legs of αIIb and/or β3. If the legs of the receptor enter the plasma membrane with a perpendicular orientation, the plasma membrane may also limit access of 7E3 to its epitope (Fig. 7) (25); a model based on electron cryomicroscopy, however, suggests that the legs do not enter the membrane with a perpendicular orientation, (26) and thus this issue remains open.
Fig. 7.
Crystal structure of entire extracellular domain of αVβ3 with amino acids Y122, K125, W129, and K181, which are implicated in the 7E3 epitope, shown in orange. The RGD peptide is shown in green. Figure was prepared with mol mol (32).
We previously observed that 7E3 IgG binds more slowly to unactivated platelets than the smaller 7E3 Fab fragment and that multimers of 7E3 (dimers, trimers, and tetramers) bind more slowly than 7E3 IgG (5). Moreover, we observed that the on rate of 7E3 IgG increases by >2-fold with platelet activation, but the on rate of 7E3 Fab increases much less with activation (9). These observations are consistent with our model in which the extent of receptor bending is variable and there is a dynamic equilibrium between the different bent conformations (10, 25). They are also consistent with our data that activation results in the receptor assuming a less bent or perhaps fully extended conformation (10). These data do not, however, exclude the possibility that additional changes occur in the head and tail regions of the molecule (2, 27–30). Our previous studies with RGD peptides tethered to small beads by variably sized glycine tethers also are consistent with this working model because beads containing peptides with long, but not short, tethers could bind to both unactivated and activated platelets, whereas beads with peptides with short tethers bound much better to activated platelets (24).
To test the effect of locking the receptor in a bent position on 7E3 binding, we analyzed 7E3 IgG binding to cells expressing αIIb-R320C β3-R563C, in which a disulfide bond constrains the receptor in a bent conformation, and found that the binding was reduced as compared to 7E3 IgG binding to cells expressing normal αIIbβ3. Consistent with the reduction in binding being a result of the disulfide bond locking the receptor in a bent position, mild reduction of the receptor with DTT markedly reduced the difference in 7E3 IgG binding to mutant and native αIIbβ3. Further support for a size-dependent access mechanism comes from our findings that the binding of the much smaller 7E3 Fab to cells expressing αIIb-R320C β3-R563C or native αIIbβ3 was nearly equivalent and that DTT treatment only minimally increased 7E3 Fab binding to the mutant receptor.
In conclusion, we provide data on 7E3 binding and propose a location for the 7E3 epitope in the 3D structures of αIIbβ3 and αVβ3. The strategic localization of this region relative to the β3 ligand binding site provides a structural explanation for the inhibitory effects of 7E3 on ligand binding. Moreover, combining the data on the size and the activation-dependent nature of 7E3 binding to platelets with structural data on αVβ3 provides insights into the molecular dynamics of the receptor under basal conditions and the conformational changes it undergoes with activation.
Acknowledgments
This work was supported in part by General Clinical Research Center Grant M01-RR00102 from the National Center for Research Resources at the National Institutes of Health and National Heart, Lung, and Blood Institute Grant HL 19278 (to B.S.C); the American Heart Association Heritage Affiliate, Ilma F. Kern Foundation in honor of John Halperin, M.D. (D.L.F); The Charles Slaughter Foundation (D.L.F); and funds from Stony Brook University.
This contribution is part of the special series of Inaugural Articles by members of the National Academy of Sciences elected on April 29, 2003.
Abbreviations: RGD, arginine-glycine-aspartic acid; CHO, Chinese hamster ovary; β3Hu, human β3; β3M, mouse β3; MFI, mean fluorescent intensity; MIDAS, metal ion-dependent adhesion site.
See accompanying Biography on page 13111.
References
- 1.Humphries, M. J. (2000) Biochem. Soc. Trans. 28, 311–339. [PubMed] [Google Scholar]
- 2.Hynes, R. (2002) Cell 110, 673–687. [DOI] [PubMed] [Google Scholar]
- 3.Coller, B. S. (1985) J. Clin. Invest. 76, 101–108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Charo, I. F., Bekeart, L. S. & Phillips, D. R. (1987) J. Biol. Chem. 262, 9935–9938. [PubMed] [Google Scholar]
- 5.Coller, B. S., Peerschke, E. I., Seligsohn, U., Scudder, L. E., Nurden, A. T. & Rosa, J. P. (1986) J. Lab. Clin. Med. 107, 384–392. [PubMed] [Google Scholar]
- 6.Reginelli, J. P. & Topol, E. J. (2000) Am. J. Med. 109, 252–254. [DOI] [PubMed] [Google Scholar]
- 7.Puzon-McLaughlin, W., Kamata, T. & Takada, Y. (2000) J. Biol. Chem. 275, 7795–7802. [DOI] [PubMed] [Google Scholar]
- 8.Takagi, J., DeBottis, D. P., Erickson, H. P. & Springer, T. A. (2002) Biochemistry 41, 4339–4347. [DOI] [PubMed] [Google Scholar]
- 9.Coller, B. S. (1986) J. Cell Biol. 103, 451–456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Takagi, J., Petre, B., Walz, T. & Springer, T. (2002) Cell 110, 599–611. [DOI] [PubMed] [Google Scholar]
- 11.Takagi, J. & Springer, T. A. (2002) Immunol. Rev. 186, 141–163. [DOI] [PubMed] [Google Scholar]
- 12.Newman, P. J., Allen, R. W., Kahn, R. A. & Kunicki, T. J. (1985) Blood 65, 227–232. [PubMed] [Google Scholar]
- 13.Grimaldi, C. M., Chen, F., Scudder, L. E., Coller, B. S. & French, D. L. (1996) Blood 88, 1666–1675. [PubMed] [Google Scholar]
- 14.Chen, C. A. & Okayama, H. (1988) BioTechniques 6, 632–638. [PubMed] [Google Scholar]
- 15.Suehiro, K., Smith, J. W. & Plow, E. F. (1996) J. Biol. Chem. 271, 10365–10371. [DOI] [PubMed] [Google Scholar]
- 16.Mitchell, W. B., Li, J. H., Singh, F., Michelson, A. D., Bussel, J., Coller, B. S. & French, D. L. (2003) Blood 101, 2268–2276. [DOI] [PubMed] [Google Scholar]
- 17.MacKerell, A. D., Brooks, C. L., Nilsson, L., Roux, B., Won, Y. & Karplus, M. (1998) in The Encyclopedia of Computational Chemistry, eds. Schleyer, P. V. R., Schreiner, P. R., Allinger, N. L., Clark, T., Gasteiger, J., Kollman, P. & Schaefer, H. F., III (Wiley, New York), pp. 271–277.
- 18.Xiong, J. P., Stehle, T., Diefenbach, B., Zhang, R., Dunker, R., Scott, D. L., Joachimiak, A., Goodman, S. L. & Arnaout, M. A. (2001) Science 294, 339–345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cieutat, A. M., Rosa, J. P., Letourneur, F., Poncz, M. & Rifat, S. (1993) Biochem. Biophys. Res. Commun. 193, 771–778. [DOI] [PubMed] [Google Scholar]
- 20.Lipscomb, D. L., Bourne, C. & Boudreaux, M. K. (1999) J. Lab. Clin. Med. 134, 313–321. [DOI] [PubMed] [Google Scholar]
- 21.Sanz, L. M., Jimenez-Marin, A., Yerle, M., Llanes, D., Barbancho, M. J. & Pavon, J. J. (2002) Anim. Genet. 33, 239–240. [DOI] [PubMed] [Google Scholar]
- 22.Gold, H. K., Coller, B. S., Yasuda, T., Saito, T., Fallon, J. T., Guerrero, J. L., Leinbach, R. C., Ziskind, A. A. & Collen, D. (1988) Circulation 77, 670–677. [DOI] [PubMed] [Google Scholar]
- 23.Sassoli, P. M., Emmell, E. L., Tam, S. H., Trikha, M., Zhou, Z., Jordan, R. E. & Nakada, M. T. (2001) Thromb. Haemostasis 85, 896–902. [PubMed] [Google Scholar]
- 24.Beer, J. H., Springer, K. T. & Coller, B. S. (1992) Blood 79, 117–128. [PubMed] [Google Scholar]
- 25.Beglova, N., Blacklow, S. C., Takagi, J. & Springer, T. A. (2002) Nat. Struct. Biol. 9, 282–287. [DOI] [PubMed] [Google Scholar]
- 26.Adair, B. D. & Yeager, M. (2002) Proc. Natl. Acad. Sci. USA 99, 14059–14064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Humphries, M. J. (2002) Arthritis Res. 4, Suppl. 3, S69–S78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Arnaout, M. A. (2002) Immunol. Rev. 186, 125–140. [DOI] [PubMed] [Google Scholar]
- 29.Liddington, R. C. & Ginsberg, M. H. (2002) J. Cell Biol. 158, 833–839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Luo, B. H., Springer, T. A. & Takagi, J. (2003) Proc. Natl. Acad. Sci. USA 100, 2403–2408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xiong, J. P., Stehle, T., Zhang, R., Joachimiak, A., Frech, M., Goodman, S. L. & Arnaout, M. A. (2002) Science 296, 151–155. [DOI] [PubMed] [Google Scholar]
- 32.Koradi, R., Billeter, M. & Wuthrich, K. (1996) J. Mol. Graphics 14, 51–32. [DOI] [PubMed] [Google Scholar]