Abstract
Glyoxalase I forms part of the glyoxalase pathway that detoxifies reactive aldehydes such as methylglyoxal, using the spontaneously formed glutathione hemithioacetal as substrate. All known eukaryotic enzymes contain zinc as their metal cofactor, whereas the Escherichia coli glyoxalase I contains nickel. Database mining and sequence analysis identified putative glyoxalase I genes in the eukaryotic human parasites Leishmania major, Leishmania infantum, and Trypanosoma cruzi, with highest similarity to the cyanobacterial enzymes. Characterization of recombinant L. major glyoxalase I showed it to be unique among the eukaryotic enzymes in sharing the dependence of the E. coli enzyme on nickel. The parasite enzyme showed little activity with glutathione hemithioacetal substrates but was 200-fold more active with hemithioacetals formed from the unique trypanosomatid thiol trypanothione. L. major glyoxalase I also was insensitive to glutathione derivatives that are potent inhibitors of all other characterized glyoxalase I enzymes. This substrate specificity is distinct from that of the human enzyme and is reflected in the modification in the L. major sequence of a region of the human protein that interacts with the glycyl-carboxyl moiety of glutathione, a group that is conjugated to spermidine in trypanothione. This trypanothione-dependent glyoxalase I is therefore an attractive focus for additional biochemical and genetic investigation as a possible target for rational drug design.
Keywords: methylglyoxal, d-lactate, glutathione, nickel
All organisms require systems to shield them from chemical stress, such as the antioxidant enzymes that detoxify endogenous oxidants and the enzymes that metabolize exogenous toxins. However, endogenous toxins such as the reactive α-oxoaldehyde methylglyoxal (2-oxopropanal) are also by-products of metabolism (1). The main source of methylglyoxal is the nonenzymatic fragmentation of triose phosphates, but it also is formed during threonine catabolism and acetone oxidation (2). In some prokaryotes, dihydroxyacetone phosphate is enzymatically converted to methylglyoxal and inorganic phosphate by methylglyoxal synthase (EC 4.2.3.3) in response to phosphate starvation (2). Methylglyoxal reacts rapidly with both proteins and nucleic acids and thus is both toxic and mutagenic (3, 4).
The glyoxalase system is a ubiquitous detoxification pathway that protects against the cellular damage caused by methylglyoxal. This pathway comprises glyoxalase I (GLO1) (lactoylglutathione lyase, EC 4.4.1.5) and glyoxalase II (GLO2) (hydroxyacylglutathione hydrolase, EC 3.1.2.6), which act in concert to convert the spontaneously formed hemithioacetal adduct between glutathione (γ-l-glutamyl-l-cysteinylglycine) and methylglyoxal into d-lactate and free glutathione (1). Thus, glutathione acts as a cofactor in the overall reaction pathway. GLO1 is a metalloenzyme that isomerizes the glutathione hemithioacetal to S-d-lactoylglutathione through proton transfer to a metal-bound enediol intermediate (5). This metal cofactor is zinc in eukaryotes but nickel in Escherichia coli (6, 7). The difference in cofactor dependence is reflected in differences between the active sites of the human and E. coli enzymes, suggesting that the latter may be a target for antimicrobial therapy (8, 9). GLO2 acts to hydrolyze the S-d-lactoylglutathione product of GLO1 to d-lactate and glutathione (1). GLO2 is not as well characterized as GLO1 and is not seen as a target for chemotherapy, because the reaction catalyzed by GLO1 is essentially irreversible under physiological conditions; thus, inhibition of GLO2 would only cause the accumulation or excretion of the nontoxic S-d-lactoylglutathione (10). Interestingly, GLO1 inhibitors are selectively toxic to proliferating cells, possibly as a result of an inhibition of DNA synthesis produced by the accumulation of methylglyoxal (11, 12). GLO1 inhibitors have also been reported to have antimalarial (13) and antitrypanosomal (14) activities.
New antiparasitic drugs are required for the treatment of tropical diseases caused by the pathogenic trypanosomes and Leishmania. These protozoa cause Chagas' disease, sleeping sickness, and the various forms of leishmaniasis, which cause serious illness and death, particularly in many tropical regions. The currently available treatments, on the whole, are poor and suffer from serious side effects, limited efficacy, and both natural and acquired resistance (15). New drugs and drug targets are therefore urgently required (16). Such a target may be offered by the glyoxalase system of the pathogenic kinetoplastids, as a consequence of these protozoa possessing an unusual thiol metabolism. In these organisms, instead of glutathione, the major low molecular mass thiol is trypanothione [N1,N8-bis(glutathionyl)spermidine] (17). Previous investigations on Leishmania donovani showed that intact cells quantitatively converted methylglyoxal to d-lactate, suggesting the presence of an active glyoxalase system (18). However, only very low GLO1 and GLO2 activities could be detected in extracts using glutathione as a cofactor (19). These observations prompted us to investigate whether trypanothione would substitute for glutathione in the glyoxalase pathway. Here we report the cloning, expression, and characterization of a Leishmania major GLO1 that is similar to prokaryotic glyoxalases but unique in its pronounced specificity for trypanothione hemithioacetal over glutathione hemithioacetal as substrate. L. major GLO1 thus represents a member of a previously uncharacterized family of GLO1 enzymes. During our investigations into L. major GLO1, an independent study reported that GLO2 from Trypanosoma brucei was highly selective for mono- and bis-S-d-lactoyltrypanothione (20). Taken together, these observations provide compelling evidence for trypanosomatids possessing a unique trypanothione-dependent glyoxalase system.
Materials and Methods
Materials. Saccharomyces cerevisiae GLO1 was purchased from Sigma. Methylglyoxal was prepared from methylglyoxaldimethylacetal and confirmed to be free of formaldehyde, as described in ref. 21. S-p-bromobenzylglutathione was synthesized as described in ref. 22. All other reagents were standard commercial products of high purity.
Determination of Dissociation Constants. Methylglyoxal stocks were assayed by preparation of the disemicarbazone adduct (23) and with S. cerevisiae GLO1 (24), both giving identical values. The Kd values for the adducts between glutathione or trypanothione and phenylglyoxal were determined as described in ref. 24. However, the extinction coefficient of the trypanothione-phenylglyoxal adduct was determined by using excess phenylglyoxal rather than excess trypanothione, because the use of millimolar levels of trypanothione is impractically expensive. A dual-beam Shimadzu UV-2401 PC spectrophotometer was used with a blank assay lacking thiol in the reference beam. The measured extinction coefficients then were used to determine the concentration of adduct in a set of equilibrium mixtures in 100 mM (Na+) phosphate, pH 7.0. The concentrations used (expressed as [SH group]) were 100 μM phenylglyoxal plus 100 μM thiol, 100 μM phenylglyoxal plus 200 μM thiol, and 200 μM phenylglyoxal plus 100 μM thiol.
Growth and Lysis of Leishmania. The L. major Friedlin A1 clone was grown as described in ref. 25. Frozen cells were thawed on ice and resuspended in lysis buffer [75 mM (Na+) phosphate, pH 7.5/500 mM NaCl/1 mM DTT/1 mM benzamidine/3 μg·ml–1 leupeptin/250 μM 4-(2-aminoethyl)benzenesulphonyl fluoride/1 μM pepstatin A]. Cells were lysed by sonication (four 30-s bursts), with cooling to <4°C between pulses. After centrifugation (100,000 × g for 1 h), the extract was dialyzed against assay buffer plus 1 mM DTT for 2 h to remove components of <10 kDa.
Cloning and Expression of L. major GLO1. A putative L. major GLO1 gene (GenBank accession no. AY604654) was identified in the L. major genome database (www.genedb.org). This gene was amplified by PCR from genomic DNA by using the sense primer 5′-CATATGCCGTCTCGTCGTATGCTGCACACC-3′ and the antisense primer 5′-GGATCCGGACCTTACGCAGTGCCCTGCTCCTTC-3′ and fully sequenced. These primers added a5′ NdeI site and a 3′ BamHI site (underlined), allowing cloning into the expression plasmid pET15b (Novagen).
Transformed BL21(DE3)pLysS E. coli were grown in LB medium containing 100 μg·ml–1 carbenicillin and 12.5 μg·ml–1 chloramphenicol to an absorbance of 0.6 at 600 nm. Expression was induced for 4 h at 25°C with 1 mM isopropyl-β-d-galactopyranoside. Cells were lysed as described above in buffer modified by the substitution of 10 mM 2-mercaptoethanol for DTT. The supernatant after centrifugation (30,000 × g for 30 min) was applied at 2 ml·min–1 to a nickel-charged 5-ml HiTrap chelating Sepharose column (Amersham Pharmacia) in 50 mM (Na+) phosphate/200 mM NaCl, pH 7.5. GLO1 was eluted with a linear gradient of 0–500 mM imidazole in the same buffer, and the tag was removed by digestion for 4 h at 25°C with thrombin (10 units·mg–1; Pharmacia). After dialysis against 20 mM Bis-Tris, pH 6.5, the protein was applied at 2 ml·min–1 to a 5-ml HiTrap Q-Sepharose column (Amersham Pharmacia). GLO1 was eluted with a linear gradient of 0–500 mM NaCl in the same buffer.
Protein Characterization. Subunit masses were measured by matrix-assisted laser desorption ionization mass spectrometry on a Voyager-DE STR (PerSeptive Biosystems, Framingham, MA). The native Mr of GLO1 was measured by using a Superdex 200 HR 10/30 size-exclusion column (Amersham Pharmacia) in 50 mM Hepes/300 mM NaCl, pH 7.4. Unknowns and standards were applied and eluted at 0.5 ml·min–1 in the same buffer. The pKa of the protein was measured by chromatofocusing with a Mono P HR 5/20 column (Amersham Pharmacia) and a pH 7.0–4.0 gradient of Polybuffer 74.
Production and Assay of Thiols. Trypanothione disulphide (2.5 μmol; Bachem) in 0.5 ml of 400 mM Hepes buffer, pH 7.4, was reduced by the addition of a 0.5-ml suspension of Tris(2-carboxyethyl)phosphine agarose (containing 4.8 μmol of reductant; Pierce). After mixing (25°C for 30 min), the agarose was removed, HCl was added to 20 mM, and the acidified solution was stored at –20°C. Thiol concentrations were determined by titration with 5,5′-dithio-bis(2-nitrobenzoic acid) (26) by using an extinction coefficient of 14.14 mM–1·cm–1 at 412 nm for the thionitrobenzoate anion (27).
Enzyme Kinetics. Kinetic constants were determined at 27°C in 0.5-ml assays containing 100 mM (Na+) phosphate, pH 7.0. To ensure full saturation with cofactor, GLO1 was diluted with assay buffer containing 20 μM NiCl2 and 0.02% (wt/vol) BSA. The required concentration of hemithioacetal and 0.1 mM free thiol were produced by varying the thiol and aldehyde concentrations, with the concentrations quoted of trypanothione being that of the sulfydryl group. Hemithioacetal concentrations were calculated by using the published Kd values of 3 and 0.6 mM for the methylglyoxal– and phenylglyoxal–glutathione equilibria, respectively (24). Trypanothione hemithioacetal concentrations were calculated by using the Kd value of 3 mM for the glutathione–methylglyoxal equilibrium and the experimental value of 0.5 mM for the phenylglyoxal–trypanothione equilibrium. In the case of glutathione hemithioacetals, Km values were too large for accurate measurement of kinetic constants, and thus kcat/Km values were determined from the slope of plots of rate versus enzyme concentration.
Trypanothione-dependent GLO2 activity was determined at 27°C in 0.5 ml of 100 mM Mops, (pH 7.2)/150 μM trypanothione-SH/200 μM methylglyoxal (20). Lactoyltrypanothione was produced by adding 0.3 nmol of recombinant L. major GLO1, and the reaction was monitored at 240 nm. Once the reaction was complete, cell extract was added, and the rate of decrease in absorbance at 240 nm was measured. Glutathione-dependent GLO2 activity was assayed under identical conditions using 150 μM glutathione and 0.3 nmol S. cerevisiae GLO1 in place of trypanothione and L. major GLO1. Methylglyoxal reductase and methylglyoxal dehydrogenase were assayed as described in ref. 19.
Apoenzyme Production, Metal Reconstitution, and Analysis. Apoenzyme was produced by overnight dialysis at 4°C of 50 nmol of GLO1 against 1 liter of 20 mM imidazole/1 mM EDTA, pH 7.0, containing 5 g of Chelex resin and then for an additional 4 h against 2 liters of 20 mM imidazole, pH 7.0, containing 5 g of Chelex resin. Aliquots (0.1 ml) were incubated on ice with a 100-fold molar excess of metal chloride and then assayed. Chelex-treated ultrapure water and buffers were used in plastic containers at all stages in this procedure to reduce adventitious metal-ion contamination.
The zinc and nickel contents of a 0.1-μmol sample of GLO1 were determined by atomic absorption spectrophotometry. This sample was dialyzed overnight at 4°C against 2 liters of 10 mM Hepes, pH 7.0, containing 5 g of Chelex resin. The metals then were measured by using a Unicam 939 AA spectrophotometer, with conditions being according to manufacturer guidelines.
Results
Dissociation Constants and Absorbance Coefficients. The dissociation constants for the glutathione and trypanothione hemithioacetals of phenylglyoxal were determined spectrophotometrically by the change in absorbance at 280 nm after hemithioacetal formation. Because trypanothione contains two SH groups per mole, data are presented per mole of sulfydryl group for consistency with data for glutathione. With glutathione in 20-fold excess, the Δε280 nm for the phenylglyoxal-glutathione adduct was estimated to be 0.76 ± 0.02 mM–1·cm–1, which is in reasonable agreement with the value of 1.17 mM–1·cm–1 previously determined by this method (24). With phenylglyoxal in 20-fold excess, similar values of 0.83 ± 0.1 mM–1·cm–1 for the glutathione-phenylglyoxal adduct and 1.04 ± 0.04 mM–1·cm–1 for the trypanothione-phenylglyoxal adduct were obtained. These extinction coefficients then were used to determine Kd values. These values were 0.5 ± 0.04 mM for the glutathione–phenylglyoxal equilibrium and 0.5 ± 0.03 mM for the trypanothione–phenylglyoxal equilibrium, close to the previously published value of 0.6 ± 0.05 mM for the glutathione–phenylglyoxal equilibrium (24).
The values of Δε240 nm for the isomerization of the trypanothione hemithioacetals of methylglyoxal and phenylglyoxal were determined by using L. major GLO1 to convert a known quantity of trypanothione to the corresponding 2-hydroxyacyl thiol ester. For the isomerization of the trypanothione–methylglyoxal and phenylglyoxal hemithioacetals, these values were 3.0 ± 0.3 and 5.59 ± 0.06 mM–1·cm–1, respectively. Within experimental error, these are the same as the values of 2.86 and 5.69 mM–1·cm–1 found for the corresponding reactions with glutathione (28). To determine whether hemithioacetals formed by either or both sulfydryl groups in trypanothione would act as substrates, reactions containing 1 mM of the trypanothione sulfydryl group were run to completion in the presence of 3 mM methylglyoxal. The concentration of residual thiol then was determined by using 5,5′-dithio-bis(2-nitrobenzoic acid). The addition of L. major GLO1 to these assays removed 99.6 ± 0.2% of the sulfydryl groups that were measurable before addition of the enzyme, indicating that hemithioacetals formed by both sulfydryl groups of trypanothione can be isomerized by L. major GLO1.
Methylglyoxal-Catabolizing Activities in L. major. L. major extract was assayed for enzymes capable of metabolizing methylglyoxal. Only trypanothione-dependent GLO1 and NADPH-dependent methylglyoxal reductase activities could be detected, with the GLO1 activity being 10-fold higher than the methylglyoxal reductase activity (Table 1). The specific activity of methylglyoxal reductase in this extract agrees well with the previously reported level of 4.8 ± 0.7 nmol·min–1 per mg in L. donovani, as does the lack of significant GLO1 activity with glutathione hemithioacetal substrate (19). Because of the different initial absorbances and wavelengths of the various assays, the maximum amount of protein that can be added varies. Hence, the assays are most sensitive for reductase/dehydrogenases (limit of detection, <0.4 nmol·min–1 per mg), less sensitive for GLO1 (limit of detection, <5 nmol·min–1 per mg), and least sensitive for GLO2 (limit of detection, <18 nmol·min–1 per mg).
Table 1. Methylglyoxal-catabolizing activities in L. major extract.
| Enzyme and cosubstrate | Specific activity, nmol·min-1 per mg | |
|---|---|---|
| GLO1 | Trypanothione | 40 ± 7 |
| Glutathione | <5 | |
| GLO2 | Trypanothione | <2 |
| Glutathione | <2 | |
| Methylglyoxal reductase | NADPH | 4.0 ± 0.4 |
| NADH | <2 | |
| Methylglyoxal dehydrogenase | NADPH | <2 |
| NADH | <2 |
Trypanosomatid GLO1 Sequences. Putative GLO1 genes were identified in the trypanosomatid genome databases by blast searches (L. major, LmjF35.3010; Leishmania infantum, LI0762b01.p1k; and Trypanosoma cruzi, Tc00.1047053510659.240 and Tc00.1047053510743.70). These genes are highly similar to each other, with the L. infantum and T. cruzi sequences showing 97% and 72% identity, respectively, to the L. major sequence at the amino acid level. The two T. cruzi GLO1 sequences differ by only three silent nucleotide substitutions and may represent different alleles of the gene.
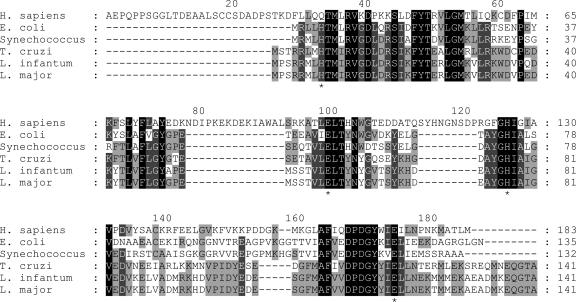
Interestingly, the putative GLO1 sequences are most similar to bacterial glyoxalases, with, for instance, the L. major GLO1 being 47% and 51% identical at the amino acid level to the glyoxalases of the proteobacteria E. coli and the cyanobacteria Synechococcus sp. WH 8102, respectively. In contrast, L. major GLO1 is only 33% identical to human GLO1, with the human glyoxalase sequence containing an N-terminal extension and two inserts that are lacking from both the bacterial and trypanosomatid enzymes (Fig. 1). The trypanosomatid GLO1 sequences are also grouped with proteobacterial and cyanobacterial GLO1 enzymes in phylogenetic analyses (data not shown).
Fig. 1.
Alignment of trypanosomatid, human, and E. coli GLO1 sequences. The L. major, L. infantum, and T. cruzi GLO1 sequences are shown aligned with the human (Swiss-Prot accession no. P78375), E. coli (Swiss-Prot accession no. P77036), and Synechococcus sp. WH 8102 (TrEMBL accession no. Q7U3T2) enzymes. The alignment is shaded according to identity: white text on a black background, complete identity; white text on a dark-gray background, >75% identity; black text on a light-gray background, >50% identity. The positions of the active-site metal-binding residues in the human and E. coli enzymes are indicated under the sequences by asterisks.
Expression and Molecular Characterization of L. major GLO1. L. major GLO1 was expressed and purified to homogeneity by metal-affinity and ion-exchange chromatography. The mass of the protein was measured by mass spectrometry before and after removal of the hexahistidine tag. Before cleavage, the mass was 18,370 ± 18 Da (predicted, 18,481 Da), with thrombin cleavage reducing this mass to 16,607 ± 17 Da (predicted, 16,599 Da). No residual hexahistidine-tagged enzyme could be detected in the final preparation by mass spectrometry or SDS/PAGE. Size-exclusion chromatography against known standards showed the protein to be a dimer in solution, with an Mr of 38,700 (predicted, 33,200). The isoelectric point was also determined to be pH 4.9 (predicted, pH 4.7).
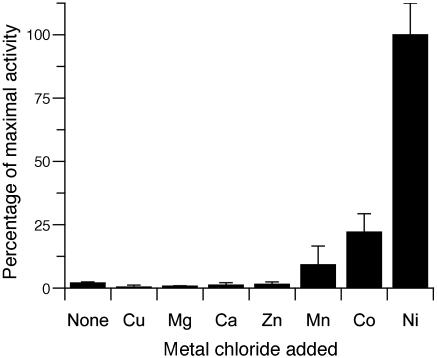
Metal Specificity of L. major GLO1. The cofactor specificity of L. major GLO1 was investigated by reconstitution of apoenzyme with metal ions (Fig. 2). Of the metals tested, only manganese, cobalt, and nickel reactivated the enzyme, with the largest recovery in activity being seen with nickel. No activity was seen with zinc, the cofactor of the human enzyme. After extensive dialysis of the recombinant protein to remove loosely bound metals, 0.9 mol of nickel and 0.1 mol of zinc per mole of GLO1 dimer were detected by atomic absorption spectrophotometry. This stoichiometry suggests that one active site in the homodimeric protein does not tightly bind metal cofactor, an observation also made with the E. coli enzyme (7).
Fig. 2.
Metal reconstitution of the L. major GLO1. Reactivation of the apo form of the L. major GLO1 by a 100-fold excess of metal ions. Assays contained 0.1 mM trypanothione hemithioacetal and 0.1 mM free trypanothione-SH. All assays were performed in triplicate, with the mean activity relative to the nickel-activated enzyme displayed.
Catalytic Activities of the L. major GLO1. With trypanothione hemithioacetal as substrate, the L. major GLO1 is a highly efficient enzyme with kcat/Km values approaching the diffusion limit (Table 2). These kinetic constants are comparable with published values for the human enzyme using glutathione hemithioacetal substrates (29). Significantly, the L. major enzyme shows a marked preference for trypanothione hemithioacetal, with a 200-fold difference in kcat/Km between this substrate and glutathione hemithioacetal. The hemithioacetal formed with methylglyoxal and the mono-thiol N1-glutathionylspermidine is also a substrate for this enzyme (data not shown), suggesting that the hemithioacetals formed by the two sulfydryl groups of trypanothione could bind to the active site independently. This would be consistent with the observation that both of these groups are substrates for the L. major GLO1. This specificity of the L. major GLO1 for glutathione amides such as trypanothione is also seen in the effect of glutathione derivatives on the activity of the enzyme. A range of S-alkyl and S-benzyl glutathione derivatives that have been characterized previously as GLO1 inhibitors were tested against the L. major enzyme. The S-methyl, S-propyl, S-butyl, S-hexyl, and S-octyl glutathione derivatives were tested at 1 mM, whereas S-p-nitrobenzyl and S-p-bromobenzylglutathione were tested at 250 μM. No inhibition was observed at these concentrations (data not shown).
Table 2. Kinetic parameters of the L. major GLO1.
| Hemithioacetal substrate | Km, μM | kcat, s-1 | kcat/Km × 107, M-1·s-1 | kcat/Km (relative) |
|---|---|---|---|---|
| Trypanothione | ||||
| Methylglyoxal | 32 ± 3 | 800 ± 30 | 2.5 | 100 |
| Phenylglyoxal | 51 ± 7 | 570 ± 40 | 1.1 | 44 |
| Glutathione | ||||
| Methylglyoxal | >1900 | ND | 0.013 | 0.5 |
| Phenylglyoxal | >80 | ND | 0.008 | 0.3 |
ND, not determined.
Discussion
Because methylglyoxal is produced from several different metabolic intermediates, almost all cells will be exposed to its toxic effects and thus require a methylglyoxal detoxification system. Methylglyoxal synthase may produce an additional toxic challenge, but the existence of this activity in trypanosomatids is controversial (18, 19). Although we identified an ORF with high similarity to prokaryotic methylglyoxal synthases in a single unassembled sequence (LM32D2b09) in the L. major genome database, this sequence is not found in other trypanosomatids. Indeed, this sequence seems to be of Pseudomonas origin, and attempts to clone this gene from L. major genomic DNA were unsuccessful.
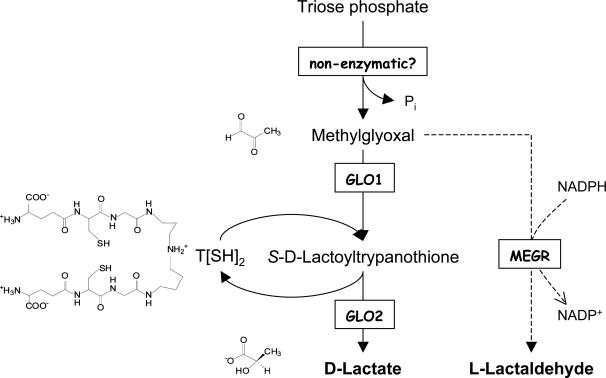
Previous studies on the metabolism of exogenous methylglyoxal by L. donovani showed that 98% was recovered as d-lactate, with only 1.2% being recovered as lactaldehyde (19). These data are entirely consistent with our observation of 10-fold higher trypanothione-dependent GLO1 than methylglyoxal reductase activity in extracts and also indicate that a GLO2-like activity must be present in these cells. The relative specific activities of methylglyoxal-catabolizing enzymes in L. major therefore suggest that this parasite has a highly active glyoxalase system that differs from other organisms in being trypanothione-dependent rather than glutathi-one-dependent (Fig. 3).
Fig. 3.
Metabolism of methylglyoxal in Leishmania spp. The major end product is d-lactate (98%) with a trace amount of l-lactaldehyde (2%) formed via methylglyoxal reductase (MEGR) (19). Enzymatic formation of methylglyoxal via methylglyoxal synthase is controversial (18).
Although the L. major genome contains a putative GLO2 (LMFLCHR12_92), which is 51% identical to T. brucei GLO2 at the amino acid level (20), we were unable to detect any GLO2 activity in cell extracts of L. major, which may be because of the insensitivity of the assay method or instability of the enzyme. T. brucei GLO2 readily loses activity because of loss of its metal cofactor (20), and thus L. major GLO2 may have lost activity during dialysis to remove trypanothione and glutathione cosubstrates. blast searches of the T. cruzi genome identified homologues of GLO1 (Tc00.1047053510659.240 and Tc00.1047053510743.70) and GLO2 (Tc00.1047053507603.230 and Tc00.1047053509429.290). The two GLO2 homologues differ by three residues at the amino acid level, with both having 66% identity to T. brucei GLO2. There are no publications on glyoxalase activity in T. cruzi, but the presence of these genes suggests that the glyoxalase pathway may be present. Unexpectedly, we were unable to identify a strong homologue of GLO1 in the almost completed T. brucei brucei genome. Although this result is consistent with an earlier report that a closely related subspecies (T. brucei gambiense) does not produce d-lactate (30), the presence of a functional GLO2 in T. brucei brucei (20) suggests that GLO1 might be present because the genome sequence for this organism is incomplete. Additional studies are required to resolve this question.
The high degree of sequence similarity and the conservation of the predicted metal-binding residues between the trypanosomatid and the E. coli GLO1 enzymes suggest that the parasite enzymes all might be nickel-dependent. However, the cofactor requirement of a GLO1 enzyme cannot be predicted from sequence alone, because the zinc-dependent Pseudomonas putida and S. cerevisiae enzymes have identical metal-binding residues as those of the E. coli enzyme (31, 32). Thus, the unambiguous demonstration of the requirement of the L. major GLO1 for nickel, but not zinc, by metal analysis and reconstitution is important in confirming the unique relationship of these eukaryotic glyoxalases to the E. coli enzyme. Importantly, the similarity in sequence to cyanobacterial enzymes suggests that the presence of these genes in the trypanosomatids may be the result of gene transfer from an ancestral endosymbiont (33). It is interesting that a rice GLO1 that is of bacterial origin and belongs to a group of plant glyoxalases with strong sequence similarity to the bacterial enzymes has been identified (34). However, these plant enzymes are twice the size of the dimeric bacterial enzymes and therefore are probably monomeric GLO1 enzymes more closely related to those of S. cerevisiae and Plasmodium sp. (35) than those of the trypanosomatids.
The high level of specificity of the L. major GLO1 for trypanothione hemithioacetals defines this enzyme as a trypanothione-dependent GLO1 (lactoyltrypanothione lyase, EC unassigned). This substrate requirement and the insensitivity of the L. major enzyme to glutathione-based inhibitors are highly significant, because this specificity contrasts with that of the human enzyme. In human and S. cerevisiae GLO1, the glutathione glycine carboxylate makes a significant contribution to the binding of substrates and transition-state analogues (36, 37). Binding occurs through an interaction of the carboxylate with main-chain atoms of a flexible loop formed from residues Pro-152–Gly-159 in the human enzyme (8). This flexible loop is also present in the E. coli structure (Pro-102–Val-109) (9), but four of these eight residues are absent from the corresponding region in the trypanosomatid sequences (Fig. 1). The lack of sequence conservation suggests that structural differences in this part of the L. major protein may confer specificity for trypanothione hemithioacetals. Another structural difference that may affect binding of hemithioacetal substrates is seen between the E. coli and human enzymes. The E. coli active site is significantly larger (9), and it is possible that it therefore may allow binding of not only the hemithioacetal with glutathione but also with glutathionyl spermidine, a metabolite that represents >80% of glutathione-containing thiols in stationary-phase E. coli cells (38).
The contrasting substrate specificities of the human and trypanosomatid enzymes suggest that GLO1 may be an attractive target for drug design. However, gene-knockout and inhibitor studies are required to determine whether this pathway plays an essential role in parasite growth, survival, or virulence.
Acknowledgments
We thank Dean Wilson for performing metal analysis, Douglas Lamont for performing mass spectrometry, and John Richardson for assistance with organic synthesis. The use of the genome sequence data from the Wellcome Trust Sanger Institute Pathogen Sequencing Unit (www.genedb.org) is gratefully acknowledged. This work was funded by the Wellcome Trust.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: GLO1, glyoxalase I; GLO2, glyoxalase II; trypanothione, N1,N8-bis(glutathionyl)spermidine.
Data deposition: The sequence reported in this paper (Leishmania major GLO1 gene) has been deposited in the GenBank database (accession no. AY604654).
References
- 1.Thornalley, P. J. (1996) Gen. Pharmacol. 27, 565–573. [DOI] [PubMed] [Google Scholar]
- 2.Cooper, R. A. (1984) Annu. Rev. Microbiol. 38, 49–68. [DOI] [PubMed] [Google Scholar]
- 3.Lo, W. C. T., Westwood, M. E., McLellan, A. C., Selwood, T. & Thornalley, P. J. (1994) J. Biol. Chem. 269, 32299–32305. [PubMed] [Google Scholar]
- 4.Marinari, U. M., Ferro, M., Sciaba, L., Finollo, R., Bassi, A. M. & Brambilla, G. (1984) Cell Biochem. Funct. 2, 243–248. [DOI] [PubMed] [Google Scholar]
- 5.Davidson, G., Clugston, S. L., Honek, J. F. & Maroney, M. J. (2001) Biochemistry 40, 4569–4582. [DOI] [PubMed] [Google Scholar]
- 6.Aronsson, A. C., Marmstal, E. & Mannervik, B. (1978) Biochem. Biophys. Res. Commun. 81, 1235–1240. [DOI] [PubMed] [Google Scholar]
- 7.Clugston, S. L., Barnard, J. F., Kinach, R., Miedema, D., Ruman, R., Daub, E. & Honek, J. F. (1998) Biochemistry 37, 8754–8763. [DOI] [PubMed] [Google Scholar]
- 8.Cameron, A. D., Ridderstrom, M., Olin, B., Kavarana, M. J., Creighton, D. J. & Mannervik, B. (1999) Biochemistry 38, 13480–13490. [DOI] [PubMed] [Google Scholar]
- 9.He, M. M., Clugston, S. L., Honek, J. F. & Matthews, B. W. (2000) Biochemistry 39, 8719–8727. [DOI] [PubMed] [Google Scholar]
- 10.Sellin, S. & Mannervik, B. (1983) J. Biol. Chem. 258, 8872–8875. [PubMed] [Google Scholar]
- 11.Ayoub, F. M., Allen, R. E. & Thornalley, P. J. (1993) Leuk. Res. 17, 397–401. [DOI] [PubMed] [Google Scholar]
- 12.Egyud, L. G. & Szent-Gyorgyi, A. (1968) Science 160, 1140. [DOI] [PubMed] [Google Scholar]
- 13.Thornalley, P. J., Strath, M. & Wilson, R. J. (1994) Biochem. Pharmacol. 47, 418–420. [DOI] [PubMed] [Google Scholar]
- 14.D'Silva, C., Daunes, S., Rock, P., Yardley, V. & Croft, S. L. (2000) J. Med. Chem. 43, 2072–2078. [DOI] [PubMed] [Google Scholar]
- 15.Fairlamb, A. H. (2003) Trends Parasitol. 19, 488–494. [DOI] [PubMed] [Google Scholar]
- 16.Croft, S. L. & Coombs, G. H. (2003) Trends Parasitol. 19, 502–508. [DOI] [PubMed] [Google Scholar]
- 17.Fairlamb, A. H., Blackburn, P., Ulrich, P., Chait, B. T. & Cerami, A. (1985) Science 227, 1485–1487. [DOI] [PubMed] [Google Scholar]
- 18.Darling, T. N. & Blum, J. J. (1988) Mol. Biochem. Parasitol. 28, 121–127. [DOI] [PubMed] [Google Scholar]
- 19.Ghoshal, K., Banerjee, A. B. & Ray, S. (1989) Mol. Biochem. Parasitol. 35, 21–29. [DOI] [PubMed] [Google Scholar]
- 20.Irsch, T. & Krauth-Siegel, R. L. (2004) J. Biol. Chem. 279, 22209–22217. [DOI] [PubMed] [Google Scholar]
- 21.Pourmotabbed, T. & Creighton, D. J. (1986) J. Biol. Chem. 261, 14240–14244. [PubMed] [Google Scholar]
- 22.Vince, R., Daluge, S. & Wadd, W. B. (1971) J. Med. Chem. 14, 402–404. [DOI] [PubMed] [Google Scholar]
- 23.Alexander, N. M. & Boyer, J. L. (1971) Anal. Biochem. 41, 29–38. [DOI] [PubMed] [Google Scholar]
- 24.Vander Jagt, D. L., Han, L. P. & Lehman, C. H. (1972) Biochemistry 11, 3735–3740. [DOI] [PubMed] [Google Scholar]
- 25.Ariyanayagam, M. R. & Fairlamb, A. H. (2001) Mol. Biochem. Parasitol. 115, 189–198. [DOI] [PubMed] [Google Scholar]
- 26.Beutler, E., Duron, O. & Kelly, B. M. (1963) J. Lab. Clin. Med. 61, 882–888. [PubMed] [Google Scholar]
- 27.Collier, B. (1973) Anal. Biochem. 56, 310–311. [DOI] [PubMed] [Google Scholar]
- 28.Vander Jagt, D. L., Daub, E., Krohn, J. A. & Han, L. P. (1975) Biochemistry 14, 3669–3675. [DOI] [PubMed] [Google Scholar]
- 29.Ridderstrom, M. & Mannervik, B. (1996) Biochem. J. 314, 463–467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Darling, T. N., Balber, A. E. & Blum, J. J. (1988) Mol. Biochem. Parasitol. 30, 253–257. [DOI] [PubMed] [Google Scholar]
- 31.Saint-Jean, A. P., Phillips, K. R., Creighton, D. J. & Stone, M. J. (1998) Biochemistry 37, 10345–10353. [DOI] [PubMed] [Google Scholar]
- 32.Frickel, E. M., Jemth, P., Widersten, M. & Mannervik, B. (2001) J. Biol. Chem. 276, 1845–1849. [DOI] [PubMed] [Google Scholar]
- 33.Hannaert, V., Saavedra, E., Duffieux, F., Szikora, J. P., Rigden, D. J., Michels, P. A. M. & Opperdoes, F. R. (2003) Proc. Natl. Acad. Sci. USA 100, 1067–1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Usui, Y., Nakase, M., Hotta, H., Urisu, A., Aoki, N., Kitajima, K. & Matsuda, T. (2001) J. Biol. Chem. 276, 11376–11381. [DOI] [PubMed] [Google Scholar]
- 35.Iozef, R., Rahlfs, S., Chang, T., Schirmer, H. & Becker, K. (2003) FEBS Lett. 554, 284–288. [DOI] [PubMed] [Google Scholar]
- 36.Hamilton, D. S. & Creighton, D. J. (1992) Biochim. Biophys. Acta 1159, 203–208. [DOI] [PubMed] [Google Scholar]
- 37.Murthy, N. S., Bakeris, T., Kavarana, M. J., Hamilton, D. S., Lan, Y. & Creighton, D. J. (1994) J. Med. Chem. 37, 2161–2166. [DOI] [PubMed] [Google Scholar]
- 38.Smith, K., Borges, A., Ariyanayagam, M. R. & Fairlamb, A. H. (1995) Biochem. J. 312, 465–469. [DOI] [PMC free article] [PubMed] [Google Scholar]