Abstract
The emergence of antimicrobial resistance severely threatens our ability to treat bacterial infections. While acquired resistance has received considerable attention, relatively little is known of intrinsic resistance that allows bacteria to naturally withstand antimicrobials. Gene products that confer intrinsic resistance to antimicrobial agents may be explored for alternative antimicrobial therapies, by potentiating the efficacy of existing antimicrobials. In this study, we identified the intrinsic resistome to a broad spectrum of antimicrobials in the human pathogen, Staphylococcus aureus. We screened the Nebraska Transposon Mutant Library of 1920 single-gene inactivations in S. aureus strain JE2, for increased susceptibility to the anti-staphylococcal antimicrobials (ciprofloxacin, oxacillin, linezolid, fosfomycin, daptomycin, mupirocin, vancomycin, and gentamicin). Sixty-eight mutants were confirmed by E-test to display at least twofold increased susceptibility to one or more antimicrobial agents. The majority of the identified genes have not previously been associated with antimicrobial susceptibility in S. aureus. For example, inactivation of genes encoding for subunits of the ATP synthase, atpA, atpB, atpG and atpH, reduced the minimum inhibitory concentration (MIC) of gentamicin 16-fold. To elucidate the potential of the screen, we examined treatment efficacy in the Galleria mellonella infection model. Gentamicin efficacy was significantly improved, when treating larvae infected with the atpA mutant compared to wild type cells with gentamicin at a clinically relevant concentration. Our results demonstrate that many gene products contribute to the intrinsic antimicrobial resistance of S. aureus. Knowledge of these intrinsic resistance determinants provides alternative targets for compounds that may potentiate the efficacy of existing antimicrobial agents against this important pathogen.
Keywords: Staphylococcus aureus, antimicrobial agents, intrinsic resistance, potentiator targets, Galleria mellonella
Introduction
Antibiotic resistant bacteria are a growing global issue that reduces our ability to cure bacterial infections (Gould, 2009; Davies and Davies, 2010). The limited success in identifying promising new antimicrobial entities led to a search for new approaches to counter the increasing problems of antibiotic resistance (Butler et al., 2013). One approach suggested is to re-sensitize resistant bacteria to an antimicrobial agent by potentiating the efficacy of an antimicrobial with a helper-drug. The helper-drug can target gene products that by any mechanism aid bacteria to resist higher concentrations of an antimicrobial (Pieren and Tigges, 2012). Besides the ability of bacteria to acquire antimicrobial resistance via horizontal gene transfer or spontaneous mutations, they can also be intrinsically resistant to antimicrobials (Cox and Wright, 2013). Intrinsic resistance to antimicrobials has traditionally been attributed to reduced permeability of the cell envelope, presence of inactivating enzymes or efflux pumps that can extrude the antimicrobial agents (Cox and Wright, 2013). Clinical use of potentiators have been applied successfully to the antimicrobial class of β-lactams, where β-lactamase inhibitors can significantly enhance the efficacy of β-lactams (Drawz and Bonomo, 2010). An analogous approach has been pursued by limiting the active efflux of antimicrobial agents by efflux pump inhibitors (Lomovskaya and Bostian, 2006), which have been shown to potentiate the efficacy of, e.g., levofloxacin in Pseudomonas aeruginosa (Renau et al., 1999) and norfloxacin in Staphylococcus aureus (Stermitz et al., 2000). However, efflux pumps inhibitors have not yet been approved for treatment of human infections due to tolerability issues (Fernebro, 2011).
It has recently been established from genome-wide studies of intrinsic resistance determinants in the Gram-negative bacteria Escherichia coli (Tamae et al., 2008; Liu et al., 2010), Acinetobacter baylyi (Gomez and Neyfakh, 2006) and Pseudomonas aeruginosa (Fajardo et al., 2008; Dötsch et al., 2009; Alvarez-Ortega et al., 2010; Gallagher et al., 2011; Krahn et al., 2012) that large and complex networks of both established and yet uncharacterized gene products contribute to reduce the inhibitory activity of antimicrobial agents. Equivalent comprehensive genome-wide studies of intrinsic resistance determinants in Gram-positive bacteria have not been performed, except for a single study that determined the intrinsic resistance of S. aureus to vancomycin, nisin and daptomycin (Blake and O’Neill, 2013). Staphylococcus aureus is an opportunistic pathogen with the capability to cause a wide range of diseases, ranging from systemic to skin infections (Lowy, 1998). The ability to treat S. aureus infections has been greatly hampered by the ability of this pathogen to develop resistance to antimicrobials (Sakoulas and Moellering, 2008; Chambers and DeLeo, 2009), which necessitates an understanding of determinants that contribute to the reduced susceptibility of S. aureus to antimicrobial agents.
In the present study, we identified genetic determinants contributing to the intrinsic resistance of S. aureus to eight different antimicrobials (ciprofloxacin, oxacillin, linezolid, fosfomycin, daptomycin, mupirocin, vancomycin, and gentamicin). We employed the Nebraska Transposon Mutant Library of 1920 single-gene inactivations in S. aureus JE2 (Fey et al., 2013) to screen for mutants that were unable to grow at sub-inhibitory concentrations of the antimicrobials. We identified multiple genes not previously recognized as modulators of antibacterial sensitivity, thus providing novel targets for the development of antibacterial sensitizer compounds.
Materials and Methods
Bacterial Strains, Growth Conditions and Chemicals
The strains used in the study include S. aureus JE2 (plasmid-cured derivative of USA300 LAC) and all derivative strains within the Nebraska Transposon Mutant Library (NTML), consisting of 1920 unique transposon mutants with inactivation of non-essential genes (Fey et al., 2013). The bursa aurealis transposon used to create the collection contains the resistance cassette ermB conferring resistance to erythromycin (Fey et al., 2013). All bacterial strains were cultured at 37°C in tryptic soy broth (TSB) or on tryptic soy agar (TSA), with antimicrobial agents added as indicated. Antimicrobial agents used in the study were erythromycin (Sigma), ciprofloxacin (Sigma), oxacillin (Sigma), linezolid (Sigma), fosfomycin (Sigma), daptomycin (Santa Cruz Biotechnology), mupirocin (Sigma), vancomycin (Sigma) and gentamicin (Sigma). Transduction of atpA::ΦNΣ into wild type (WT) JE2 was performed with bacteriophage φ11, by selecting for transductants on erythromycin plates (5 μg/ml) (Fey et al., 2013).
Screening for Increased Antimicrobial Susceptibility
To screen for increased susceptibility among the NTML strains, we first determined the minimum inhibitory concentration (MIC) of WT strain JE2 to ciprofloxacin, oxacillin, linezolid, fosfomycin, daptomycin, mupirocin, vancomycin, and gentamicin. MIC was determined using a twofold microbroth dilution assay according to Clinical and Laboratory Standards Institute (CLSI) guidelines (CLSI, 2009), except that cation-adjusted Mueller Hinton broth was substituted with TSB.
The NTML is stored in glycerol at -80°C in 20 96-well microtiter plates. Material from the frozen stock was transferred directly with a Deutz 96 cryoreplicator (Duetz et al., 2000) from the 96-wells microtiter plates onto TSA plates supplemented with 5 μg/ml erythromycin (to prevent the growth of contaminants during the screen, as all the strains in the NTML are resistant to erythromycin; Fey et al., 2013) and 0.5x MIC of the respective antimicrobial agent. The plates were incubated at 37°C for 24 h and visually inspected for lack of growth of individual mutants.
Determination of MIC of Hypersusceptible Mutants
The MIC was determined for all strains displaying growth deficiency in the hypersusceptibility screen to the respective antimicrobial agent and WT strain JE2 using E-tests (bioMérieux) performed on TSA plates. No erythromycin was supplemented to the plates during MIC determination with E-tests. The MIC was determined upon incubation at 37°C for 22 h and interpreted according to the guidelines of the manufacturer.
Treatment of Galleria mellonella With Gentamicin
To investigate if increased antimicrobial susceptibility could be detected in vivo, we employed the Galleria mellonella infection model (Desbois and Coote, 2011; Ramarao et al., 2012). Healthy 5th-instar wax moth larvae weighting approximately 250 mg were randomly picked from a batch purchased at a local pet store and divided into six groups with 20 larvae in each. Virulence of S. aureus WT and the atpA mutant were compared by injecting 20 larvae with 10 μl (containing 1 × 107 CFU) with a Hamilton syringe into the hemocoel through the lowest left proleg. Survival of G. mellonella was monitored for 120 h. For treatment efficacy of gentamicin against S. aureus, 20 larvae for each group were injected with 1 × 107 CFU of WT or the atpA mutant. One hour post infection, the larvae were injected with 10 μl gentamicin (1 mg/kg bodyweight) in the lowest right proleg. Gentamicin therapy was repeated every 24 h for a total of 72 h and survival of G. mellonella was monitored for 120 h. A control group for toxicity of gentamicin (1 mg/kg bodyweight) as well as for non-treated (inoculated with phosphate-buffered saline) were included. The data was analyzed in GraphPad Prism 4 (GraphPad Software Inc.) using the Kaplan–Meier method and statistical difference determined using log rank test.
Chromosomal Reconstruction of the atpA Mutant
Chromosomal reconstruction of the atpA mutant was achieved by use of the temperature-sensitive shuttle vector pBASE6 (Geiger et al., 2012). A chromosomal region encompassing atpA was PCR amplified from WT S. aureus JE2 chromosomal DNA using primer pair: 5′-ATATGAGCTCGAAGAGTTAGATAAGATTGTCAAACTAG-3′/5′-GATACAAGATCTGATGGTTTGTATTGCTACTTGC-3′ and cloned into pBASE6 via SacI/BglII. This plasmid was purified from E. coli IM08B (Monk et al., 2015) and transformed directly into JE2 atpA::ΦNΣ (NE592) at 30°C followed by chromosomal integration by plating on TSA (10 μg/ml chloramphenicol) at 44°C overnight. Plasmid cross-out was performed by passage at 30°C followed by plating on TSA (500 ng/ml anhydrotetracycline) and successful allelic exchange of the transposon insertion with the intact atpA gene was selected for by replica plating of colonies and screening for sensitivity toward erythromycin and chloramphenicol. Reconstruction of the atpA locus was verified by PCR amplification using primers 5′-CAAGTATGCTAAAGCATTATTTGACGTGTC-3′/5′-CGTAATTTCTGCTTGTCTCGCTCTG-3′ positioned outside the chromosomal region used for homologous recombination.
Results and Discussion
Screening for Hyper-Susceptibility to Antimicrobial Agents
The complete NTML of 1920 single-gene inactivations (Fey et al., 2013) was screened for mutants displaying increased susceptibility to eight antimicrobial agents with different mechanism of action, by inspecting for lack of growth on agar plates supplemented with 0.5x MIC of the respective antimicrobials. The antimicrobials tested were ciprofloxacin, oxacillin, linezolid, fosfomycin, daptomycin, mupirocin, vancomycin, and gentamicin (Table 1). For the mutants identified in the initial screen as being unable to grow at 0.5x MIC, the MIC was determined using E-tests. A total of 68 mutants were confirmed to display twofold or greater increase in susceptibility to one or more antimicrobial agents compared to the WT strain S. aureus JE2 (Figure 1). Eight of these strains displayed increased sensitivity to multiple agents.
Table 1.
Antimicrobial agents used in the screening for intrinsic resistance determinants, as well as the primary target and process affected by the respective agents.
| Antimicrobial | Primary target | Process affected | JE2 MIC (μg/ml) |
|---|---|---|---|
| Ciprofloxacin | DNA gyrase | DNA replication | 32 |
| Oxacillin | Transpeptidase | Cell wall synthesis | 0.5 |
| Linezolid | 50S rRNA subunit | Protein synthesis | 3 |
| Fosfomycin | MurA | Cell wall synthesis | 0.75 |
| Daptomycin | Cytoplasmic membrane | Membrane permeability | 0.19 |
| Mupirocin | Isoleucyl t-RNA | Protein synthesis | 0.125 |
| Vancomycin | NAM/NAG peptides | Cell wall synthesis | 1.5 |
| Gentamicin | 30S rRNA subunit | Protein synthesis | 1.5 |
The MIC (μg/ml) of wild type strain JE2 is displayed for each agent.
FIGURE 1.
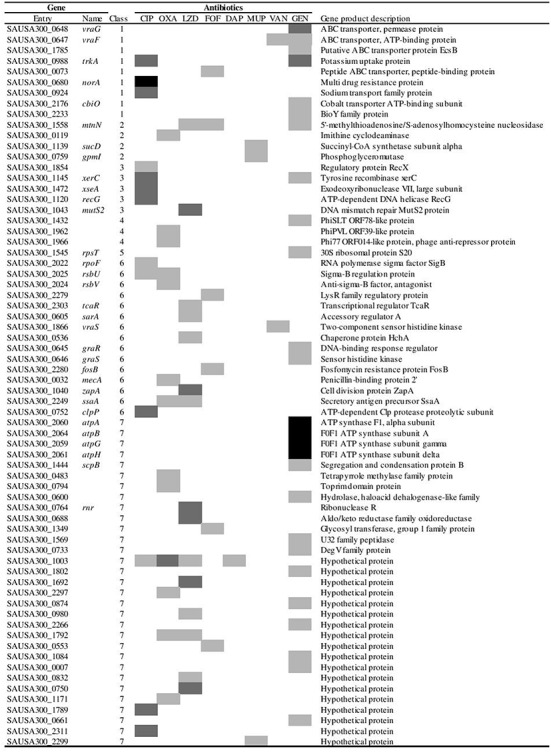
Gene inactivations affecting susceptibility to antimicrobial agents to one or more of the eight antimicrobial agents tested. The effect on susceptibility is displayed using a gray scale indicating fold reductions in sensitivity:  ,
,  and
and  . The genes are divided into the categories: (1) Membrane transporters, (2) Metabolism, (3) Replication and DNA repair, (4) Pro-phage encoded genes, (5) Protein synthesis, (6) Transcriptional regulators, chaperones, cell wall and membrane stress mechanisms and (7) Unassigned and hypothetical genes. CIP (Ciprofloxacin), OXA (Oxacillin), LZD (Linezolid), FOF (Fosfomycin), DAP (Daptomycin), MUP (Mupirocin), VAN (Vancomycin) and GEN (Gentamicin).
. The genes are divided into the categories: (1) Membrane transporters, (2) Metabolism, (3) Replication and DNA repair, (4) Pro-phage encoded genes, (5) Protein synthesis, (6) Transcriptional regulators, chaperones, cell wall and membrane stress mechanisms and (7) Unassigned and hypothetical genes. CIP (Ciprofloxacin), OXA (Oxacillin), LZD (Linezolid), FOF (Fosfomycin), DAP (Daptomycin), MUP (Mupirocin), VAN (Vancomycin) and GEN (Gentamicin).
Inactivation of Acquired Resistance Genes Increase Susceptibility to Oxacillin and Fosfomycin
The S. aureus JE2 strain carries the acquired resistance genes mecA and fosB on the chromosome (Fey et al., 2013), which confer resistance to β-lactams (Zapun et al., 2008) and fosfomycin (Thompson et al., 2014), respectively. As a verification of the screen, we indeed identified the mecA and fosB mutants as more susceptible to oxacillin and fosfomycin, respectively (Figure 1). In our experimental setup the MIC values of oxacillin and fosfomycin for WT were within the susceptibility range according to CLSI (CLSI, 2009), even though they possess mecA and fosB. Choice of medium and NaCl concentration may affect the absolute MIC value for oxacillin (Huang et al., 1993) and discrepancies in absolute fosfomycin MIC between E-test and liquid assays has been reported (Díez-Aguilar et al., 2013; Mihailescu et al., 2014). Additionally, S. aureus JE2 is resistant to ciprofloxacin due to mutations in the essential topoisomerase genes gyrA and grlA (Sreedharan et al., 1990; Ferrero et al., 1995; Diep et al., 2006; Highlander et al., 2007; Kennedy et al., 2008). The lowering in MIC for these three antimicrobials is potentially related to the effect of the resistance genes or resistance mutations and may not necessarily classify as intrinsic resistance determinants. S. aureus JE2 is susceptible toward the remaining antimicrobials and the inactivations of genes conferring increased susceptibility are considered intrinsic resistance determinants.
Gentamicin
Gentamicin belongs to the bactericidal antibiotic class of aminoglycosides, which interferes with protein synthesis through binding to the 16S rRNA of the 30S small ribosomal unit causing mistranslation of proteins (Shakil et al., 2008). Aminoglycosides are polycationic compounds, which is an important feature for their cell interactions and uptake (Shakil et al., 2008). Twenty six mutants displayed increased sensitivity to gentamicin (Figure 1). Inactivation of the gene rpsT encoding 30S ribosomal protein S20 was found to render the cell more susceptible to gentamicin suggesting that alteration of the ribosome assembly can influence gentamicin sensitivity through some currently unknown mechanism.
Six mutants with annotated membrane transport proteins exhibited increased sensitivity to gentamicin, with the most profound effect on sensitivity observed for vraG and trkA, displaying sixfold reductions in MIC. Gentamicin uptake into the cell is known to be dependent on membrane potential, where hyperpolarization of the membrane leads to increased uptake, while depolarization reduces the uptake of gentamicin (Taber et al., 1987). Hyperpolarization and accordingly increased gentamicin uptake is suspected for the trkA mutant, as a trkA deletion mutant in Mycobacterium smegmatis displays hyperpolarization and increased susceptibility to aminoglycosides (Castañeda-García et al., 2011). Lack of the TrkA transporter also potentiates the efficacy of aminoglycosides in Pseudomonas aeruginosa (Lee et al., 2009; Gallagher et al., 2011). Inactivation of vraG has previously been shown to increase the susceptibility toward gentamicin, vancomycin and cationic antimicrobial peptides in S. aureus, potentially by alteration of the cell surface charge (Meehl et al., 2007; Yang et al., 2012). The greatest sensitization for gentamicin, being a 16-fold reduction in MIC, was observed upon inactivation of genes encoding for subunits of the ATP synthase, namely atpA, atpB, atpG and atpH. Transduction of atpA into WT JE2, confirmed that transposon inactivation of atpA increases sensitivity of S. aureus toward gentamicin. Inactivation of atpA also potentiated efficacy of gentamicin in E. coli, which was hypothesized to occur via increased uptake of aminoglycosides due to elevated membrane potentials arising from altered respiration (Lobritz et al., 2015). While we were unable to complement the atpA-inactivated mutant with a functional atpA gene on a plasmid, we successfully performed allelic exchange of the transposon insertion with the intact atpA gene, generating a strain displaying gentamicin sensitivity as WT (1.5 μg/ml).
Besides the numerous established genes modulating gentamicin sensitivity, the NTML screen identified several novel genes of unknown function contributing to the intrinsic sensitivity to gentamicin (Figure 1).
Ciprofloxacin
Ciprofloxacin is a fluoroquinolone interfering with bacterial replication via inhibition of the enzymes DNA gyrase and topoisomerase IV (Blondeau, 2004). DNA gyrase, composed of two subunits GyrA and GyrB, is essential for replication initiation as the enzyme introduces negative supercoils ahead of the replication fork (Drlica and Zhao, 1997), while topoisomerase IV, composed of GrlA and GrlB, is responsible for decatenation of the replicated daughter chromosomes (Drlica and Zhao, 1997). Interaction of ciprofloxacin with these enzymes leads to a halt in replication and ultimately to DNA strands breakage (Drlica and Zhao, 1997; Blondeau, 2004). S. aureus JE2 is resistant to ciprofloxacin, due to amino acid changes in the DNA gyrase GyrA (Ser84Leu) and topoisomerase IV GrlA (Ser80Tyr), encoded by gyrA and grlA respectively (Sreedharan et al., 1990; Ferrero et al., 1995).
Elucidation of the intrinsic resistome in E. coli revealed that multiple gene products related to DNA replication and repair aid this bacterium to survive DNA damage generated by fluoroquinolone treatment (Tamae et al., 2008; Liu et al., 2010). Four of the 13 identified mutants in our screen with increased sensitivity to ciprofloxacin also display inactivation of genes related to DNA replication and repair, namely recG, xerC, recX and xseA (Figure 1). The RecG protein is a DNA helicase and strains with deletion of recG display reduced recombination and DNA repair (Al-Deib et al., 1996). Identification of recG corroborates previous work in S. aureus on increased susceptibility to quinolones in a recG mutant (Niga et al., 1997). A mutant of xseA, encoding the large subunit of exonuclease VII, was also observed in E. coli to be hypersensitive to ciprofloxacin (Tamae et al., 2008).
Three mutants with inactivated membrane transport systems, norA, trkA and SAUSA300_0924, are at least fourfold more sensitive to ciprofloxacin than the WT strain. This might be due to decreased efflux of this agent, as ciprofloxacin has been observed to being subjected to cellular efflux by pump activity of multiple efflux systems, e.g., NorA (Yamada et al., 1997; Muñoz-Bellido et al., 1999). Moreover, it was confirmed that inactivation of the stress response sigma factor σB encoded by rpoF and the σB activator RsbU potentiated the effect of ciprofloxacin, which is in agreement with previous observations in S. aureus (Riordan et al., 2006). We also identified the clpP gene encoding the proteolytic subunit of the ClpXP two-component protease to be more susceptible to ciprofloxacin, corroborating a study in P. aeruginosa (Fernández et al., 2012). A possible link between clpP and ciprofloxacin susceptibility is that clpP has been shown to interfere with activation of the SOS DNA-damage response (Cohn et al., 2011). Additionally, inactivation of several hypothetical genes (SAUSA300_1789 and SAUSA300_2311) also conferred a fourfold or greater sensitization to ciprofloxacin relative to the WT strain.
Linezolid
Linezolid belongs to the antimicrobial class of oxazolidinones (Moellering, 2003). Linezolid interacts with the A-site of the 50S ribosomal subunit and inhibits formation of the initiation complex, thus inhibiting protein synthesis (Wilson, 2014). A total of 15 mutants were more susceptible to linezolid (Figure 1), with six mutants displaying at least a fourfold increased sensitivity. None of the identified genes have to our knowledge previously been associated with increased linezolid susceptibility. Interestingly, these genes exert widely different functions, such as zapA encoding the divisome protein ZapA, which stabilizes Z-ring formation (Adams and Errington, 2009), mutS2 encoding MutS2 that in Helicobacter pylori function as an inhibitor of recombination (Pinto et al., 2005) and the ribonuclease R.
Mupirocin
The target of mupirocin is the isoleucyl-tRNA synthetase (IleRS), thus inhibiting aminoacylation of isoleucine to the cognate tRNA and thereby prevents protein synthesis (Yanagisawa et al., 1994; Pope et al., 1998). Only three mutants exhibited increased sensitivity toward mupirocin (Figure 1). None of the mutants, sucD, gpmI and SAUSA300_2299, have previously been associated with altered susceptibility to mupirocin. The gene SAUSA300_2299 has been annotated as a putative multidrug resistance transporter and could be involved in active efflux of mupirocin from the cytoplasm.
Oxacillin
Oxacillin is a β-lactam interfering with cell wall biosynthesis. The bactericidal activity of oxacillin derives from its binding to penicillin-binding proteins (PBPs), thus preventing cross-linking of peptidoglycan units of the cell wall, which eventually causes cell lysis (Kotra and Mobashery, 1998). In our study, we identified 13 genes displaying increased susceptibility to oxacillin (Figure 1). S. aureus JE2 contains the mecA gene encoding the alternative penicillin binding protein 2a (PBP2a) that enables cross-linking of peptidoglycan units in the presence of β-lactams (Zapun et al., 2008). Indeed inactivation of mecA rendered the strain more susceptible to oxacillin. A functional σB response is important for oxacillin susceptibility (Singh et al., 2003; Schulthess et al., 2009) and RsbU together with RsbV are activators of this stress response (Palma and Cheung, 2001), both of these determinants were confirmed as targets for oxacillin sensitizers in our screen. In addition to the previously established contributors to oxacillin intrinsic resistance, several genes of unknown function were identified (SAUSA300_1792 and SAUSA300_2297) that upon inactivation increased susceptibility to oxacillin equally well as mecA inactivation. It was also observed that inactivation of the unknown gene SAUSA300_1003 displayed even greater sensitivity to oxacillin than the mecA mutant. Moreover, this SAUSA300_1003 mutant displayed increased sensitivity to ciprofloxacin, linezolid and daptomycin and thereby it could be of interest to investigate how this gene can potentiate the efficacy of multiple antimicrobial agents.
Fosfomycin
Fosfomycin is a phosphoenolpyruvate analog that inhibits the first enzymatic step in peptidoglycan biosynthesis, by binding to the enzyme MurA and thereby prevent the formation of N-acetylmuramic acid, an essential precursor of the peptidoglycan cell wall (Michalopoulos et al., 2011; Karageorgopoulos et al., 2012). S. aureus JE2 harbors the fosfomycin resistance gene (fosB) that chemically inactivates fosfomycin (Rigsby et al., 2005; Castañeda-García et al., 2013) and in agreement, the fosB mutant showed increased sensitivity to fosfomycin. Moreover, inactivation of the gene SAUSA300_2279 encoding a regulatory protein of the LysR family also increased susceptibility to fosfomycin. This gene is located directly upstream of fosB and could potentially be a positive regulator of fosB or the transposon insertion in SAUSA300_2279 could prevent proper transcription of fosB. Four other genes also affected fosfomycin susceptibility, with a hypothetical (SAUSA300_0553) and a putative peptide transporter (SAUSA300_0073) being most prominent with threefold greater sensitivity to fosfomycin, which equal the potentiating effect of the fosB mutant.
Vancomycin
Vancomycin is a glycopeptide antibiotic readily used in the treatment of methicillin-resistant S. aureus (Liu et al., 2011). The bactericidal activity of vancomycin is derived from the inhibition of cell wall biosynthesis. Vancomycin binds to the D-Ala-D-Ala residues of the pentapeptides on the peptidoglycan precursors N-acetylmuramic acid and N-acetylglucosamine and thus prevents peptide cross-linking of the polysaccharide backbone in peptidoglycan biosynthesis (Reynolds, 1989; Boger, 2001). Inactivation of two genes increased the susceptibility to vancomycin, with a maximum two-fold reduction in MIC (Figure 1). We confirmed the previously established vancomycin intrinsic resistance determinants vraS (Kuroda et al., 2003; Gardete et al., 2012) and vraF of the VraFG ABC transporter system (Meehl et al., 2007). Although our screen identified recognized vancomycin intrinsic resistance determinants, we did not observe mutants of the dlt operon as seen previously in a corresponding vancomycin hypersusceptibility screen (Blake and O’Neill, 2013). No strains in the NTML exist with inactivation of any of the four genes in the dlt operon, which is involved in adding positively charged D-alanine to teichoic acids (Peschel et al., 1999).
Daptomycin
Daptomycin is a cyclic lipopeptide displaying bactericidal activity against many Gram-positive organisms (Steenbergen et al., 2005). The bactericidal activity of daptomycin is attributed to the insertion of the lipophilic tail into the bacterial cell membrane, leading to rapid membrane depolarization and potassium ion efflux, with downstream arrest of DNA, RNA and protein synthesis (Steenbergen et al., 2005).
Only a single mutant, SAUSA300_1003, displayed increased susceptibility to daptomycin. The gene SAUSA300_1003 encodes a protein of unknown function and this mutant is also more susceptible to several of the other antimicrobial agents tested.
Potentiating the Effect of Gentamicin in the Galleria mellonella Infection Model
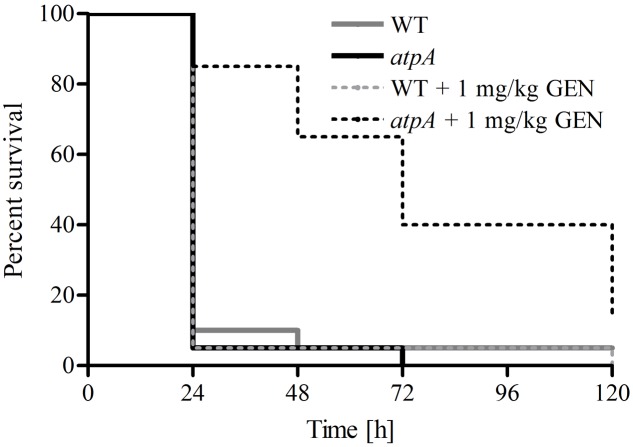
To verify the potential of enhancing the efficacy of antimicrobial agents by targeting a non-essential gene product in S. aureus, we employed the G. mellonella infection model (Ramarao et al., 2012). We tested the efficacy of gentamicin at a clinically relevant dose (1 mg/kg bodyweight) to promote survival of G. mellonella infected with a lethal dose (107 cells/larvae) of S. aureus JE2 (WT) and the atpA mutant (Figure 2). Gentamicin significantly prolonged survival of G. mellonella infected with the atpA mutant compared to the treatment of WT (P < 0.0001). None of G. mellonella larvae in the group only receiving gentamicin treatment died during the experiment (Data not shown).
FIGURE 2.
Effect of gentamicin treatment of G. mellonella larvae inoculated with 1 × 107 CFU of S. aureus JE2 and the atpA mutant. Gentamicin (1 mg/kg bodyweight) were administered 1 h post infection with follow-up treatments at 24, 48, and 72 h. Prolonged survival of G. mellonella inoculated with atpA due to gentamicin treatment was observed. All larvae survived treatment with only gentamicin (1 mg/kg bodyweight).
Conclusion
In this work, we have identified genetic determinants that increase the sensitivity of S. aureus to eight antimicrobial agents. The many modulators of antimicrobial susceptibility uncovered here for S. aureus JE2 comprise both previously established determinants in addition to numerous novel genes. To our knowledge this study provides the first whole genome overview of intrinsic resistance genes of S. aureus to agents with different modes of action. With the results obtained, it will be feasible to select particular genes for further investigation as targets for antimicrobial potentiators. The mechanisms by which the majority of these determinants contribute to modulate antibiotic susceptibility remain unknown and further work is required to establish this.
It is important to note that S. aureus JE2 carries resistance determinants to oxacillin and fosfomycin as well as mutations conferring resistance to ciprofloxacin and the genes identified for these agents cannot per se be classified as intrinsic resistance genes, unless gene inactivation is examined in a susceptible strain. However, the gene inactivations identified here to reduce the MIC of these agents could potentially be explored for re-sensitizing resistant S. aureus isolates.
Staphylococcus aureus JE2 was not phenotypically resistant to the remaining antibiotics tested (linezolid, vancomycin, gentamicin, daptomycin and mupirocin). However, an antimicrobial agent such as gentamicin has limited use against S. aureus due to the risk of toxicity at the required clinical concentrations (Rayner and Munckhof, 2005). Therefore, based on our results co-administration of an inhibitor targeting the ATP synthase could potentially lower the concentration of gentamicin needed for treatment of S. aureus and thus increase the usefulness of this drug. In the G. mellonella infection model we observed significantly increased survival of larvae infected with atpA compared to WT, when treated with gentamicin at similar concentrations used for human therapy (Rayner and Munckhof, 2005).
Improved activity of vancomycin could also be of clinical significance, even for strains that are already susceptible to this agent. There is an inverted association between MIC and treatment efficiency of vancomycin against methicillin-resistant S. aureus infections, even with MIC values well within the susceptibility range (Sakoulas et al., 2004; Lodise et al., 2008). A further reduction of the vancomycin MIC with a helper-drug could thus increase the probability of vancomycin treatment success.
Taken together, our observations provide a framework for understanding the contribution of chromosomal determinants that affect the susceptibility to antimicrobial agents in S. aureus and provide novel targets for further exploration in development of small molecules to be used as antimicrobial potentiators.
Author Contributions
MV and HI conceived and designed the study. Experiments were performed by MV, BL, MB, and JH. All authors contributed in analysis of data and drafting the manuscript. All authors read and approved the final manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The Danish Research Council for Independent Research, Technology and Production nr. 12-127417.
References
- Adams D. W., Errington J. (2009). Bacterial cell division: assembly, maintenance and disassembly of the Z ring. Nat. Rev. Microbiol. 7 642–653. 10.1038/nrmicro2198 [DOI] [PubMed] [Google Scholar]
- Al-Deib A. A., Mahdi A. A., Lloyd R. G. (1996). Modulation of recombination and DNA repair by the RecG and PriA helicases of Escherichia coli K-12. J. Bacteriol. 178 6782–6789. 10.1128/jb.178.23.6782-6789.1996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alvarez-Ortega C., Wiegand I., Olivares J., Hancock R. E., Martínez J. L. (2010). Genetic determinants involved in the susceptibility of Pseudomonas aeruginosa to β-lactam antibiotics. Antimicrob. Agents Chemother. 54 4159–4167. 10.1128/AAC.00257-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blake K. L., O’Neill A. J. (2013). Transposon library screening for identification of genetic loci participating in intrinsic susceptibility and acquired resistance to antistaphylococcal agents. J. Antimicrob. Chemother. 68 12–16. 10.1093/jac/dks373 [DOI] [PubMed] [Google Scholar]
- Blondeau J. M. (2004). Fluoroquinolones: mechanism of action, classification, and development of resistance. Surv. Ophthalmol. 49 S73–S78. 10.1016/j.survophthal.2004.01.005 [DOI] [PubMed] [Google Scholar]
- Boger D. L. (2001). Vancomycin, teicoplanin, and ramoplanin: synthetic and mechanistic studies†. Med. Res. Rev. 21 356–381. 10.1002/med.1014 [DOI] [PubMed] [Google Scholar]
- Butler M. S., Blaskovich M. A., Cooper M. A. (2013). Antibiotics in the clinical pipeline in 2013. J. Antibiot. 66 571–591. 10.1038/ja.2013.86 [DOI] [PubMed] [Google Scholar]
- Castañeda-García A., Blázquez J., Rodríguez-Rojas A. (2013). Molecular mechanisms and clinical impact of acquired and intrinsic fosfomycin resistance. Antibiotics 2 217–236. 10.3390/antibiotics2020217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castañeda-García A., Do T. T., Blázquez J. (2011). The K+ uptake regulator TrkA controls membrane potential, pH homeostasis and multidrug susceptibility in Mycobacterium smegmatis. J. Antimicrob. Chemother. 66 1489–1498. 10.1093/jac/dkr165 [DOI] [PubMed] [Google Scholar]
- Chambers H. F., DeLeo F. R. (2009). Waves of resistance: Staphylococcus aureus in the antibiotic era. Nat. Rev. Microbiol. 7 629–641. 10.1038/nrmicro2200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- CLSI (2009). Performance Standards for Antimicrobial Disk Susceptibility Tests: Approved Standard, 10th Edn. Wayne, PA: Clinical and Laboratory Standards Institute. [Google Scholar]
- Cohn M. T., Kjelgaard P., Frees D., Penadés J. R., Ingmer H. (2011). Clp-dependent proteolysis of the LexA N-terminal domain in Staphylococcus aureus. Microbiology 157 677–684. 10.1099/mic.0.043794-0 [DOI] [PubMed] [Google Scholar]
- Cox G., Wright G. D. (2013). Intrinsic antibiotic resistance: mechanisms, origins, challenges and solutions. Int. J. Med. Microbiol. 303 287–292. 10.1016/j.ijmm.2013.02.009 [DOI] [PubMed] [Google Scholar]
- Davies J., Davies D. (2010). Origins and evolution of antibiotic resistance. Microbiol. Mol. Biol. Rev. 74 417–433. 10.1128/MMBR.00016-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desbois A. P., Coote P. J. (2011). Wax moth larva (Galleria mellonella): an in vivo model for assessing the efficacy of antistaphylococcal agents. J. Antimicrob. Chemother. 66 1785–1790. 10.1093/jac/dkr198 [DOI] [PubMed] [Google Scholar]
- Diep B. A., Gill S. R., Chang R. F., Phan T. H., Chen J. H., Davidson M. G., et al. (2006). Complete genome sequence of USA300, an epidemic clone of community-acquired meticillin-resistant Staphylococcus aureus. Lancet 367 731–739. 10.1016/S0140-6736(06)68231-7 [DOI] [PubMed] [Google Scholar]
- Díez-Aguilar M., Morosini M.-I., del Campo R., García-Castillo M., Zamora J., Cantón R. (2013). In vitro activity of fosfomycin against a collection of clinical Pseudomonas aeruginosa isolates from 16 Spanish hospitals: establishing the validity of standard broth microdilution as susceptibility testing method. Antimicrob. Agents Chemother. 57 5701–5703. 10.1128/AAC.00589-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dötsch A., Becker T., Pommerenke C., Magnowska Z., Jänsch L., Häussler S. (2009). Genomewide identification of genetic determinants of antimicrobial drug resistance in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 53 2522–2531. 10.1128/AAC.00035-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drawz S. M., Bonomo R. A. (2010). Three decades of β-lactamase inhibitors. Clin. Microbiol. Rev. 23 160–201. 10.1128/CMR.00037-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drlica K., Zhao X. (1997). DNA gyrase, topoisomerase IV, and the 4-quinolones. Microbiol. Mol. Biol. Rev. 61 377–392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duetz W. A., Rüedi L., Hermann R., O’Connor K., Büchs J., Witholt B. (2000). Methods for intense aeration, growth, storage, and replication of bacterial strains in microtiter plates. Appl. Environ. Microbiol. 66 2641–2646. 10.1128/AEM.66.6.2641-2646.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fajardo A., Martínez-Martín N., Mercadillo M., Galán J. C., Ghysels B., Matthijs S., et al. (2008). The neglected intrinsic resistome of bacterial pathogens. PLoS ONE 3:e1619 10.1371/journal.pone.0001619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernández L., Breidenstein E. B., Song D., Hancock R. E. (2012). Role of intracellular proteases in the antibiotic resistance, motility, and biofilm formation of Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 56 1128–1132. 10.1128/AAC.05336-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernebro J. (2011). Fighting bacterial infections—future treatment options. Drug Resist. Updat. 14 125–139. 10.1016/j.drup.2011.02.001 [DOI] [PubMed] [Google Scholar]
- Ferrero L., Cameron B., Crouzet J. (1995). Analysis of gyrA and grlA mutations in stepwise-selected ciprofloxacin-resistant mutants of Staphylococcus aureus. Antimicrob. Agents Chemother. 39 1554–1558. 10.1128/AAC.39.7.1554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fey P. D., Endres J. L., Yajjala V. K., Widhelm T. J., Boissy R. J., Bose J. L., et al. (2013). A genetic resource for rapid and comprehensive phenotype screening of nonessential Staphylococcus aureus genes. MBio 4:e537-12 10.1128/mBio.00537-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallagher L. A., Shendure J., Manoil C. (2011). Genome-scale identification of resistance functions in Pseudomonas aeruginosa using Tn-seq. MBio 2:e315-10 10.1128/mBio.00315-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardete S., Kim C., Hartmann B. M., Mwangi M., Roux C. M., Dunman P. M., et al. (2012). Genetic pathway in acquisition and loss of vancomycin resistance in a methicillin resistant Staphylococcus aureus (MRSA) strain of clonal type USA300. PLoS Pathog. 8:e1002505 10.1371/journal.ppat.1002505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geiger T., Francois P., Liebeke M., Fraunholz M., Goerke C., Krismer B., et al. (2012). The stringent response of Staphylococcus aureus and its impact on survival after phagocytosis through the induction of intracellular PSMs expression. PLoS Pathog. 8:e1003016 10.1371/journal.ppat.1003016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez M. J., Neyfakh A. A. (2006). Genes involved in intrinsic antibiotic resistance of Acinetobacter baylyi. Antimicrob. Agents Chemother. 50 3562–3567. 10.1128/AAC.00579-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gould I. (2009). Antibiotic resistance: the perfect storm. Int. J. Antimicrob. Agents 34 S2–S5. 10.1016/S0924-8579(09)70549-7 [DOI] [PubMed] [Google Scholar]
- Highlander S. K., Hultén K. G., Qin X., Jiang H., Yerrapragada S., Mason E. O., et al. (2007). Subtle genetic changes enhance virulence of methicillin resistant and sensitive Staphylococcus aureus. BMC Microbiol. 7:99 10.1186/1471-2180-7-99 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang M., Gay T., Baker C., Banerjee S., Tenover F. (1993). Two percent sodium chloride is required for susceptibility testing of staphylococci with oxacillin when using agar-based dilution methods. J. Clin. Microbiol. 31 2683–2688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karageorgopoulos D. E., Wang R., Yu X.-H., Falagas M. E. (2012). Fosfomycin: evaluation of the published evidence on the emergence of antimicrobial resistance in Gram-negative pathogens. J. Antimicrob. Chemother. 67 255–268. 10.1093/jac/dkr466 [DOI] [PubMed] [Google Scholar]
- Kennedy A. D., Otto M., Braughton K. R., Whitney A. R., Chen L., Mathema B., et al. (2008). Epidemic community-associated methicillin-resistant Staphylococcus aureus: recent clonal expansion and diversification. Proc. Natl. Acad. Sci. U.S.A. 105 1327–1332. 10.1073/pnas.0710217105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotra L., Mobashery S. (1998). β-Lactam antibiotics, β-lactamases and bacterial resistance. Bull. Inst. Pasteur 96 139–150. 10.1016/S0020-2452(98)80009-2 [DOI] [Google Scholar]
- Krahn T., Gilmour C., Tilak J., Fraud S., Kerr N., Lau C. H.-F., et al. (2012). Determinants of intrinsic aminoglycoside resistance in Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 56 5591–5602. 10.1128/AAC.01446-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuroda M., Kuroda H., Oshima T., Takeuchi F., Mori H., Hiramatsu K. (2003). Two-component system VraSR positively modulates the regulation of cell-wall biosynthesis pathway in Staphylococcus aureus. Mol. Microbiol. 49 807–821. 10.1046/j.1365-2958.2003.03599.x [DOI] [PubMed] [Google Scholar]
- Lee S., Hinz A., Bauerle E., Angermeyer A., Juhaszova K., Kaneko Y., et al. (2009). Targeting a bacterial stress response to enhance antibiotic action. Proc. Natl. Acad. Sci. U.S.A. 106 14570–14575. 10.1073/pnas.0903619106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu A., Tran L., Becket E., Lee K., Chinn L., Park E., et al. (2010). Antibiotic sensitivity profiles determined with an Escherichia coli gene knockout collection: generating an antibiotic bar code. Antimicrob. Agents Chemother. 54 1393–1403. 10.1128/AAC.00906-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C., Bayer A., Cosgrove S. E., Daum R. S., Fridkin S. K., Gorwitz R. J., et al. (2011). Clinical practice guidelines by the Infectious Diseases Society of America for the treatment of methicillin-resistant Staphylococcus aureus infections in adults and children. Clin. Infect. Dis. 52 e18–e55. 10.1093/cid/ciq146 [DOI] [PubMed] [Google Scholar]
- Lobritz M. A., Belenky P., Porter C. B., Gutierrez A., Yang J. H., Schwarz E. G., et al. (2015). Antibiotic efficacy is linked to bacterial cellular respiration. Proc. Natl. Acad. Sci. U.S.A. 112 8173–8180. 10.1073/pnas.1509743112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lodise T., Graves J., Evans A., Graffunder E., Helmecke M., Lomaestro B., et al. (2008). Relationship between vancomycin MIC and failure among patients with methicillin-resistant Staphylococcus aureus bacteremia treated with vancomycin. Antimicrob. Agents Chemother. 52 3315–3320. 10.1128/AAC.00113-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lomovskaya O., Bostian K. A. (2006). Practical applications and feasibility of efflux pump inhibitors in the clinic—a vision for applied use. Biochem. Pharmacol. 71 910–918. 10.1016/j.bcp.2005.12.008 [DOI] [PubMed] [Google Scholar]
- Lowy F. D. (1998). Staphylococcus aureus infections. N. Engl. J. Med. 339 520–532. 10.1056/NEJM199808203390806 [DOI] [PubMed] [Google Scholar]
- Meehl M., Herbert S., Götz F., Cheung A. (2007). Interaction of the GraRS two-component system with the VraFG ABC transporter to support vancomycin-intermediate resistance in Staphylococcus aureus. Antimicrob. Agents Chemother. 51 2679–2689. 10.1128/AAC.00209-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michalopoulos A. S., Livaditis I. G., Gougoutas V. (2011). The revival of fosfomycin. Int. J. Infect. Dis. 15 e732–e739. 10.1016/j.ijid.2011.07.007 [DOI] [PubMed] [Google Scholar]
- Mihailescu R., Tafin U. F., Corvec S., Oliva A., Betrisey B., Borens O., et al. (2014). High activity of fosfomycin and rifampin against methicillin-resistant Staphylococcus aureus biofilm in vitro and in an experimental foreign-body infection model. Antimicrob. Agents Chemother. 58 2547–2553. 10.1128/AAC.02420-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moellering R. C. (2003). Linezolid: the first oxazolidinone antimicrobial. Ann. Intern. Med. 138 135–142. 10.7326/0003-4819-138-2-200301210-00015 [DOI] [PubMed] [Google Scholar]
- Monk I. R., Tree J. J., Howden B. P., Stinear T. P., Foster T. J. (2015). Complete bypass of restriction systems for major Staphylococcus aureus lineages. MBio 6:e00308-15 10.1128/mBio.00308-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muñoz-Bellido J. L., Manzanares M. A., Andrés J. M., Zufiaurre M. G., Ortiz G., Hernández M. S., et al. (1999). Efflux pump-mediated quinolone resistance inStaphylococcus aureus strains wild type for gyrA, gyrB, grlA, and norA. Antimicrob. Agents Chemother. 43 354–356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niga T., Yoshida H., Hattori H., Nakamura S., Ito H. (1997). Cloning and sequencing of a novel gene (recG) that affects the quinolone susceptibility of Staphylococcus aureus. Antimicrob. Agents Chemother. 41 1770–1774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palma M., Cheung A. L. (2001). ςB activity in Staphylococcus aureus is controlled by RsbU and an additional factor (s) during bacterial growth. Infect. Immun. 69 7858–7865. 10.1128/IAI.69.12.7858-7865.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peschel A., Otto M., Jack R. W., Kalbacher H., Jung G., Götz F. (1999). Inactivation of the dlt operon in Staphylococcus aureus confers sensitivity to defensins, protegrins, and other antimicrobial peptides. J. Biol. Chem. 274 8405–8410. [DOI] [PubMed] [Google Scholar]
- Pieren M., Tigges M. (2012). Adjuvant strategies for potentiation of antibiotics to overcome antimicrobial resistance. Curr. Opin. Pharmacol. 12 551–555. 10.1016/j.coph.2012.07.005 [DOI] [PubMed] [Google Scholar]
- Pinto A. V., Mathieu A., Marsin S., Veaute X., Ielpi L., Labigne A., et al. (2005). Suppression of homologous and homeologous recombination by the bacterial MutS2 protein. Mol. Cell. 17 113–120. 10.1016/j.molcel.2004.11.035 [DOI] [PubMed] [Google Scholar]
- Pope A. J., Moore K. J., McVey M., Mensah L., Benson N., Osbourne N., et al. (1998). Characterization of isoleucyl-tRNA synthetase from Staphylococcus aureus II. Mechanism of inhibition by reaction intermediate and pseudomonic acid analogues studied using transient and steady-state kinetics. J. Biol. Chem. 273 31691–31701. 10.1074/jbc.273.48.31691 [DOI] [PubMed] [Google Scholar]
- Ramarao N., Nielsen-Leroux C., Lereclus D. (2012). The insect Galleria mellonella as a powerful infection model to investigate bacterial pathogenesis. J. Vis. Exp. 70:e4392 10.3791/4392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rayner C., Munckhof W. (2005). Antibiotics currently used in the treatment of infections caused by Staphylococcus aureus. Intern. Med. J. 35 S3–S16. 10.1111/j.1444-0903.2005.00976.x [DOI] [PubMed] [Google Scholar]
- Renau T. E., Léger R., Flamme E. M., Sangalang J., She M. W., Yen R., et al. (1999). Inhibitors of Efflux Pumps in ∗Pseudomonas aeruginosa potentiate the activity of the fluoroquinolone antibacterial levofloxacin. J. Med. Chem. 42 4928–4931. 10.1021/jm9904598 [DOI] [PubMed] [Google Scholar]
- Reynolds P. E. (1989). Structure, biochemistry and mechanism of action of glycopeptide antibiotics. Eur. J. Clin. Microbiol. Infect. Dis. 8 943–950. 10.1007/BF01967563 [DOI] [PubMed] [Google Scholar]
- Rigsby R. E., Fillgrove K. L., Beihoffer L. A., Armstrong R. N. (2005). Fosfomycin resistance proteins: a nexus of glutathione transferases and epoxide hydrolases in a metalloenzyme superfamily. Methods Enzymol. 401 367–379. 10.1016/S0076-6879(05)01023-2 [DOI] [PubMed] [Google Scholar]
- Riordan J. T., O’Leary J. O., Gustafson J. E. (2006). Contributions of sigB and sarA to distinct multiple antimicrobial resistance mechanisms of Staphylococcus aureus. Int. J. Antimicrob. Agents 28 54–61. 10.1016/j.ijantimicag.2006.01.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakoulas G., Moellering R. C. (2008). Increasing antibiotic resistance among methicillin-resistant Staphylococcus aureus strains. Clin. Infect. Dis. 46 S360–S367. 10.1086/533592 [DOI] [PubMed] [Google Scholar]
- Sakoulas G., Moise-Broder P. A., Schentag J., Forrest A., Moellering R. C., Eliopoulos G. M. (2004). Relationship of MIC and bactericidal activity to efficacy of vancomycin for treatment of methicillin-resistant Staphylococcus aureus bacteremia. J. Clin. Microbiol. 42 2398–2402. 10.1128/JCM.42.6.2398-2402.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schulthess B., Meier S., Homerova D., Goerke C., Wolz C., Kormanec J., et al. (2009). Functional characterization of the σB-dependent yabJ-spoVG operon in Staphylococcus aureus: role in methicillin and glycopeptide resistance. Antimicrob. Agents Chemother. 53 1832–1839. 10.1128/AAC.01255-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shakil S., Khan R., Zarrilli R., Khan A. U. (2008). Aminoglycosides versus bacteria–a description of the action, resistance mechanism, and nosocomial battleground. J. Biomed. Sci. 15 5–14. 10.1007/s11373-007-9194-y [DOI] [PubMed] [Google Scholar]
- Singh V. K., Schmidt J. L., Jayaswal R., Wilkinson B. J. (2003). Impact of sigB mutation on Staphylococcus aureus oxacillin and vancomycin resistance varies with parental background and method of assessment. Int. J. Antimicrob. Agents 21 256–261. 10.1016/S0924-8579(02)00359-X [DOI] [PubMed] [Google Scholar]
- Sreedharan S., Oram M., Jensen B., Peterson L., Fisher L. (1990). DNA gyrase gyrA mutations in ciprofloxacin-resistant strains of Staphylococcus aureus: close similarity with quinolone resistance mutations in Escherichia coli. J. Bacteriol. 172 7260–7262. 10.1128/jb.172.12.7260-7262.1990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steenbergen J. N., Alder J., Thorne G. M., Tally F. P. (2005). Daptomycin: a lipopeptide antibiotic for the treatment of serious Gram-positive infections. J. Antimicrob. Chemother. 55 283–288. 10.1093/jac/dkh546 [DOI] [PubMed] [Google Scholar]
- Stermitz F. R., Lorenz P., Tawara J. N., Zenewicz L. A., Lewis K. (2000). Synergy in a medicinal plant: antimicrobial action of berberine potentiated by 5’-methoxyhydnocarpin, a multidrug pump inhibitor. Proc. Natl. Acad. Sci. U.S.A. 97 1433–1437. 10.1073/pnas.030540597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taber H. W., Mueller J., Miller P., Arrow A. (1987). Bacterial uptake of aminoglycoside antibiotics. Microbiol. Rev. 51 439–457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamae C., Liu A., Kim K., Sitz D., Hong J., Becket E., et al. (2008). Determination of antibiotic hypersensitivity among 4,000 single-gene-knockout mutants of Escherichia coli. J. Bacteriol. 190 5981–5988. 10.1128/JB.01982-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson M. K., Keithly M. E., Goodman M. C., Hammer N. D., Cook P. D., Jagessar K. L., et al. (2014). Structure and function of the genomically encoded fosfomycin resistance enzyme, FosB, from Staphylococcus aureus. Biochemistry 53 755–765. 10.1021/bi4015852 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson D. N. (2014). Ribosome-targeting antibiotics and mechanisms of bacterial resistance. Nat. Rev. Microbiol. 12 35–48. 10.1038/nrmicro3155 [DOI] [PubMed] [Google Scholar]
- Yamada H., Kurose-Hamada S., Fukuda Y., Mitsuyama J., Takahata M., Minami S., et al. (1997). Quinolone susceptibility of norA-disrupted Staphylococcus aureus. Antimicrob. Agents Chemother. 41 2308–2309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanagisawa T., Lee J. T., Wu H. C., Kawakami M. (1994). Relationship of protein structure of isoleucyl-tRNA synthetase with pseudomonic acid resistance of Escherichia coli. A proposed mode of action of pseudomonic acid as an inhibitor of isoleucyl-tRNA synthetase. J. Biol. Chem. 269 24304–24309. [PubMed] [Google Scholar]
- Yang S.-J., Bayer A. S., Mishra N. N., Meehl M., Ledala N., Yeaman M. R., et al. (2012). The Staphylococcus aureus two-component regulatory system, GraRS, senses and confers resistance to selected cationic antimicrobial peptides. Infect. Immun. 80 74–81. 10.1128/IAI.05669-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zapun A., Contreras-Martel C., Vernet T. (2008). Penicillin-binding proteins and β-lactam resistance. FEMS Microbiol. Rev. 32 361–385. 10.1111/j.1574-6976.2007.00095.x [DOI] [PubMed] [Google Scholar]