Abstract
Chromosomal rearrangements resulting in gene fusions are frequently involved in carcinogenesis. Here, we describe a semiautomatic procedure for identifying fusion gene transcripts by using publicly available mRNA and EST databases. With this procedure, we have identified 96 transcript sequences that are derived from 60 known fusion genes. Also, 47 or more additional sequences appear to be derived from 20 or more previously unknown putative fusion genes. We have experimentally verified the presence of a previously unknown IRA1/RGS17 fusion in the breast cancer cell line MCF7. The fusion gene encodes the full-length RGS17 protein, a regulator of G protein-coupled signaling, under the control of the IRA1 gene promoter. This study demonstrates that databases of ESTs can be used to discover fusion genes resulting from structural rearrangement of chromosomes.
Chromosome aberrations are common characteristics of most human cancer cells (1, 2). Translocation of part of a gene to a new locus can produce altered gene expression that perturbs normal regulatory pathways and can initiate or stimulate neoplastic cell growth and cancer progression. A well known example is the translocation of the v-abl Abelson murine leukemia viral oncogene homolog 1 (ABL1) protooncogene from chromosome 9 to the breakpoint cluster region (BCR) gene locus on chromosome 22. The translocation generates the BCR/ABL1 fusion protein, which is responsible for ≈90% of the cases of chronic myelogenous leukemia (3). The Mitelman Database of Chromosome Aberrations in Cancer lists >45,000 cases of chromosomal aberrations (1). Many were uncovered by cytogenetic banding experiments. It is difficult to tell from these experiments whether the translocation creates a fusion of two genes and, if it does, to find the fusion gene. However, if a fusion gene is expressed, part or the entire transcript should be present as an entry in the mRNA and/or EST databases. Such transcripts can be identified because they are made up of portions of transcripts from two genes and map to two different locations in the human genome sequence.
These chimeric transcripts can be distinguished from artificial chimeras, which are created by accidental ligation of different cDNAs during the cloning procedure, by examining the sequence at the fusion point. The fusion point in the chimeras from true fusion genes will usually coincide with a canonical exon boundary because the genes are likely to break in an intron because introns are generally much longer than exons. In contrast, the fusion point for an artificial chimera will usually be within an exon of each gene because the fusion occurs between two cDNAs. To test this hypothesis, we have developed a semiautomated procedure for a massive identification of human fusion transcripts by using publicly available sequence databases. A procedure that uses this principle to identify heterologous, spliced mRNAs has been reported (4). Here, we report a comprehensive identification of both mRNAs and ESTs fusion genes and the experimental detection of the predicted nuclear receptor corepressor/HDAC3 complex/regulator of G protein signaling (IRA1/RGS17) fusion gene in MCF7 breast cancer cells.
Materials and Methods
Data Sets. We used the publicly available mRNA- and EST-togenome alignment data (“all_mrna,” 141,300 alignments from 128,007 mRNAs, and “all_est,” 4,957,003 alignments from 4,642,477 ESTs) from the University of California, Santa Cruz Human Genome Browser database (http://genome.ucsc.edu; October 27, 2003, release, Version hg16). These alignments were produced by blat by using mRNA and EST databases that were filtered to remove vector sequences and the human genome assembly National Center for Biotechnology Information Build 34, July 2003 freeze.
Initial Identification of Chimeric Sequences. Chimeras were collected from the databases described above by selecting the sequences that contained two parts, that were each at least 100 bp long, and that aligned to different chromosomes or, if to the same chromosome, in opposite orientations. This latter condition was imposed in order not to select sequences that are produced by transcription over a long sequence that spans two or more normally independent genes (5). To accommodate small errors in alignment that occur at the edges of the alignment blocks, we allowed gaps or overlaps of up to 10 bp between the two parts of the transcript sequence. This procedure identified 1,061 chimeric mRNAs and 9,855 chimeric ESTs. The fusion point was noted for each sequence.
Selection of Possible Fusion Genes. To determine whether the fusion point corresponded to a pair of known splice sites, we first collected a list of canonical exon-intron boundary sites that occur in “all_mrna” and “all_est” tables from the University of California, Santa Cruz Human Genome Browser database. Only the positions of the canonically spliced introns that obey the GT/AG rule were considered. The splice donor and acceptor sites, supported by at least one mRNA or two ESTs, were recorded and used as the known splice sites. We then selected from the chimeric mRNAs and ESTs only those whose fusion point was located within 5 bp of a known splice site. This procedure reduced the list to 132 mRNA and 255 EST sequences. It will miss some fusion transcripts if the sequence database does not contain sequences from both of the two individual (nonfused) genes, but it ensures that the fusion point is located at or near a known splice site of two active, individual genes. Next, we prepared an artificially fused genomic DNA sequence for each fusion transcript candidate by joining two genomic sequences, one from the aligned region of each gene. Each fusion transcript candidate was then aligned to the corresponding artificially fused genomic sequence by using the SIM4 program (6) and the alignment around the fusion point was manually inspected. Only those that aligned precisely, without a gap or overlap, were retained. Finally, the transcripts that included human repetitive sequences were removed by using the repeatmasker program (http://ftp.genome.washington.edu/RM/RepeatMasker.html).
Expression of the Fusion Genes Breast Carcinoma Amplified Sequence (BCAS) BCAS4/BCAS3 and IRA1/RGS17 by RT-PCR Analysis. Messenger RNA from the breast cancer cell line MCF7 was made by using the MicroFastTrack kit (Invitrogen). cDNA was prepared by reverse transcription by using Moloney murine leukemia virus reverse transcriptase enzyme (Invitrogen) with random hexamer priming. The PCR was performed by using the following thermocycling protocol: initial denaturation at 94°C for 1 min, 35 cycles of denaturation at 94°C for 1 min, annealing at 60°C for 1 min, and elongation at 72°C for 2 min. The PCR primer pairs used were T530 (GGGAATTTCCTTGTGCCTCCA) and T531 (TGCTGGGGCCTTCATCATCT) for IRA1/RGS17 fusion, T532 (GAGCTCGCGCTCTTCCTGAC) and T533 (AGGGGCTGGCTCTCATTGGT) for BCAS4/BCAS3 fusion, and Actin-For (GCATGGGTCAGAAGGAT) and Actin-Rev (CCAATGGTGATGACCTG) for β-actin (ACTB). The PCRs were analyzed on 1.5% agarose gels.
Fluorescence In Situ Hybridization (FISH) Analysis. Metaphase slides of MCF7 were prepared as described (7). BAC clones (IRA1, clone ID RP11-126F9, and RGS17, clone ID RP11-119G6) were obtained from Invitrogen and verified by PCR. BAC DNA was resuspended in water and labeled by nick translation according to standard procedures (8). IRA1 and RGS17 were nick-translated by using digoxigenin-11-dUTP and biotin-16-dUTP (Roche Applied Science), respectively. Each product was precipitated in the presence of 50 μl of Cot-1 DNA (Invitrogen) and 1 μl of salmon sperm DNA (Sigma). The precipitate was resuspended in 5 μl of deionized Formamide (Fluka) and 5 μl of Master Mix (20% dextran sulfate and 2× SSC). The 10-μl probe mixture (5 μl of IRA1 and 5 μl of RGS17) was denatured at 80°C for 10 min, applied to the slide, and incubated overnight at 37°C.
After hybridization, biotin-16-dUTP (RGS17) was detected with Avidin-FITC (Vector Laboratories) and digoxigenin-11-dUTP (IRA1) with mouse anti-digoxin antibodies (Sigma) followed by rabbit anti-mouse tetramethylrhodamine B isothiocyanate (Sigma). Slides were counterstained with 4,6-diaminidino-2-phenylindole (Sigma) and mounted with antifade solution (Vector Laboratories).
Image acquisition was done by using a DMRXA fluorescent microscope (Leica, Deerfield, IL) equipped with three aligned optical filters for 4,6-diaminidino-2-phenylindole, FITC, and tetramethylrhodamine B isothiocyanate (Chroma Technology, Brattleboro, VT) and a Sensys charge-coupled device camera (Photometrics, Tucson, AZ). Image analysis was performed with q-fish software (Leica Microsystems Imaging Solutions, Cambridge, U.K.).
Results
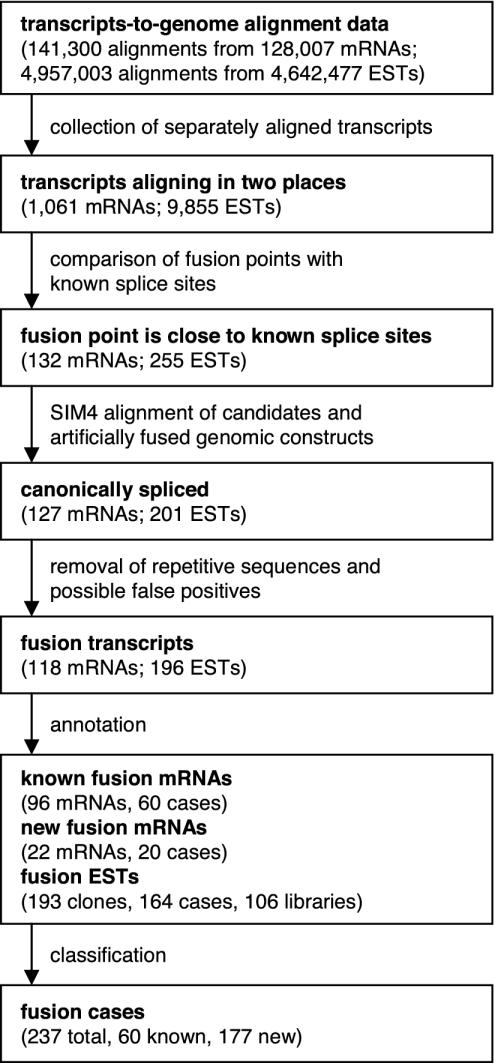
Identification of Putative Fusion Transcripts. We have developed a semiautomatic procedure to identify transcripts from possible fusion genes (Fig. 1). It involves identifying chimeric transcripts automatically using the principle outlined in the introduction, followed by a manual inspection of each candidate for a precise fit between the observed transcript sequence and the expected genomic DNA sequence at the fusion point. This procedure identified 118 mRNAs and 196 ESTs as fusion transcript sequences. These sequences are grouped into a total of 237 fusion cases (the complete list of fusion gene transcripts can be found in Table 3, which is published as supporting information on the PNAS web site).
Fig. 1.
A flow diagram showing the overall procedure for searching for the human fusion transcripts in public sequence databases.
Verification of Fusion Gene Detection. We checked GenBank annotation records of 118 mRNA sequences and found that 96 of them were reported as fusion genes, indicating that the method successfully identifies true fusion gene transcripts. These 96 known fusion mRNA sequences were derived from 60 fusion cases. Most of the remaining 22 mRNA sequences were the full insert sequences of randomly selected cDNA clones.
To verify the efficacy of the method, we examined how many of the known BCR/ABL1 fusion mRNAs deposited in the GenBank database were detected by the new procedure. We could retrieve 22 BCR/ABL1 fusion mRNAs by a text search of the GenBank mRNA database. Six of these were in our fusion mRNA list. The other 16 fusion mRNAs were missed either because they (13 mRNAs) did not have the 100-nucleotide minimum length on either side of the fusion point or because the expressed sequence did not match the expected genomic sequence precisely at the fusion point (a 3-nt deletion in two and a 55-nt insertion in one). Thus, the algorithm is conservative and will miss some genuine fusion gene transcripts. However, the long matched flanking regions and the manual inspection for exact fit ensure that only very rare accidents will produce false-positive results.
Analysis of Putative Fusion Genes. The procedure identified 177 possible fusion genes that have not been previously reported. We shall refer to these as putative fusion genes, although the connection between the observation of a chimeric transcript in the database and the actual existence of the corresponding fusion gene needs to be established by direct experiments. Most of these putative fusion genes are supported by only one transcript sequence in the databases, but 20 of these are supported by transcripts from two or more clones (Table 1). Among the partner genes involved in these newly identified putative fusion cases, 11 genes in 11 different cases are in the 148 recurrent fusion-involving genes listed in the Cancer Genome Anatomy Project Recurrent Chromosome Aberrations in Cancer Database (http://cgap.nci.nih.gov/Chromosomes/RecurrentAberrations) and/or in the 291 cancer genes recently reported by Futreal et al. (2). These are LPP, GNAS, PTEN, MLLT2, FGFR1OP, HIP1, BCL11A, NPM1, HMGA2, MSF, and PRDM16. However, their partner genes, ZNF262, ZNF288, KIAA0905, UBE2D3, TAGAP, ANGPT1, E2F5, Hs.54957, ELAC2, Hs.410998, and PCDHGC3, respectively, are not in either database.
Table 1. Number of known and putative fusion sequences and genes.
| Total | Known | New | New,≥2 clones | |
|---|---|---|---|---|
| Sequences | 314 | 96 | 218 | 47 |
| Genes | 237 | 60 | 177 | 20 |
The 237 known and putative fusion cases identified in this study involve 417 different genes. The 36 genes that participate in two or more (known or putative) fusion events detected in this study are listed in Table 2. Thirteen of these are not in either of the databases mentioned above.
Table 2. List of genes that participate in two or more fusion events.
| No. | Gene | Known status* | Cases† | 5′‡ | 3′‡ | Chromosomal band | Title |
|---|---|---|---|---|---|---|---|
| 1 | MLL | Known | 12 | 10 | 2 | 11q23.3 | Myeloid/lymphoid or mixed-lineage leukemia |
| 2 | CREBBP | Known | 4 | 2 | 2 | 16p13.3 | CREB-binding protein |
| 3 | RUNX1 | Known | 4 | 4 | 0 | 21q22.12 | Runt-related transcription factor |
| 4 | ALK | Known | 3 | 0 | 3 | 2p23.2 | Anaplastic lymphoma kinase |
| 5 | MLLT2 | Known | 3 | 2 | 1 | 4q21.3 | Myeloid/lymphoid or mixed-lineage leukemia; translocated to, 2 |
| 6 | NPM1 | Known | 3 | 2 | 1 | 5q35.1 | Nucleophosmin |
| 7 | RET | Known | 3 | 0 | 3 | 10q11.21 | Ret protooncogene |
| 8 | FUS | Known | 3 | 2 | 1 | 16p11.2 | Fusion [involved in t(12;16) in malignant liposarcoma] |
| 9 | BCR | Known | 3 | 2 | 1 | 22q11.23 | Breakpoint cluster region |
| 10 | EWSR1 | Known | 3 | 3 | 0 | 22q12.2 | Ewing sarcoma breakpoint region 1 |
| 11 | RBM15 | Known | 2 | 1 | 1 | 1p13.3 | RNA-binding motif protein 15 |
| 12 | NTRK1 | Known | 2 | 0 | 2 | 1q23.1 | Neurotrophic tyrosine kinase, receptor, type 1 |
| 13 | RPS6KC1 | New | 2 | 0 | 2 | 1q32.3 | Ribosomal protein S6 kinase, 52 kDa, polypeptide 1 |
| 14 | TFG | Known | 2 | 2 | 0 | 3q12.2 | TRK-fused gene |
| 15 | KIAA0372 | New | 2 | 1 | 1 | 5q15 | KIAA0372 |
| 16 | HINT1 | New | 2 | 0 | 2 | 5q23.3 | Histidine triad nucleotide-binding protein 1 |
| 17 | TGFBI | New | 2 | 0 | 2 | 5q31.1 | Transforming growth factor, beta-induced, 68 kDa |
| 18 | CREB3L2 | Known | 2 | 2 | 0 | 7q33 | cAMP-responsive element binding protein 3-like 2 |
| 19 | FGFR1 | Known | 2 | 1 | 1 | 8p12 | Fibroblast growth factor receptor 1 |
| 20 | MYST3 | Known | 2 | 1 | 1 | 8p11.21 | MYST histone acetyltransferase 3 |
| 21 | NR4A3 | Known | 2 | 0 | 2 | 9q22.33 | Nuclear receptor subfamily 4, group A, member 3 |
| 22 | MLLT10 | Known | 2 | 1 | 1 | 10p12.31 | Myeloid/lymphoid or mixed-lineage leukemia; translocated to, 10 |
| 23 | MYST4 | Known | 2 | 1 | 1 | 10q22.2 | MYST histone acetyltransferase 4 |
| 24 | NUP98 | Known | 2 | 2 | 0 | 11p15.4 | Nucleoporin 98 kDa |
| 25 | PICALM | Known | 2 | 0 | 2 | 11q14.2 | Phosphatidylinositol-binding clathrin assembly protein |
| 26 | MARS | New | 2 | 0 | 2 | 12q13.3 | Methionine-tRNA synthetase |
| 27 | CPM | New | 2 | 0 | 2 | 12q15 | Carboxypeptidase M |
| 28 | RARA | Known | 2 | 0 | 2 | 17q21.2 | Retinoic acid receptor, alpha |
| 29 | NBR2 | New | 2 | 2 | 0 | 17q21.31 | Neighbor of BRCA1 gene 2 |
| 30 | Hs.410998 | New | 2 | 0 | 2 | 17q23.2 | Strong similarity to protein ref:NP_006631.1 MLL septin-like fusion; septin D1 |
| 31 | OAZ1 | New | 2 | 2 | 0 | 19p13.3 | Ornithine decarboxylase antizyme 1 |
| 32 | FKBP8 | New | 2 | 0 | 2 | 19p13.11 | FK506-binding protein 8, 38 kDa |
| 33 | Hs. 102754 | New | 2 | 1 | 1 | 21q21.1 | cDNA FLJ38295 full insert sequence, clone FCBBF3012332 |
| 34 | MKL1 | Known | 2 | 1 | 1 | 22q13.1 | Megakaryoblastic leukemia 1 |
| 35 | TUBGCP6 | New | 2 | 0 | 2 | 22q13.33 | Tubulin, gamma complex-associated protein 6 |
| 36 | SHANK3 | New | 2 | 2 | 0 | 22q13.33 | SH3 and multiple ankyrin repeat domains 3 |
The genes listed in the Cancer Genome Anatomy Project Recurrent Chromosome Aberrations in Cancer Database and/or in the list of Futreal et al. (2) are considered to be known as a recurrent fusion gene.
Total number of cases in which the gene participates.
Number of cases where the gene participates as the 5′ partner (5′) or the 3′ partner (3′).
RT-PCR Detection of Predicted Fusion Gene Transcripts in MCF7 Cells.We selected two cases for experimental verification. Both were identified in the NIH_MGC_107 library prepared from an un-identified breast adenocarcinoma cell line. One is the fusion of the BCAS4 gene and the BCAS3 gene, which is observed in an mRNA and in six independent EST clones from this library (Fig. 2A). The fusion is known to be present in the MCF7 breast cancer cell line (9).
Fig. 2.
Schematic representation and RT-PCR detection of BCAS4/BCAS3 and IRA1/RGS17 fusions. The fusion genes, BCAS4/BCAS3 (A) and IRA1/RGS17 (B), are depicted. Boxes represent the exons, and broken lines are the introns. Fusion events are indicated by the arcs. Arrows indicate the transcription start sites. Exons are numbered from the 5′ to the 3′ direction as they occur in the original gene. Two BCAS4/BCAS3 fusion transcripts, BU943989 and BU957509, have an additional exon between BCAS3 gene exons 23 and 24, which is designated as 23a. Primers for the RT-PCR are indicated (T530, T531, T532, and T533). ORFs are marked with gray boxes. (C) The fusion gene transcripts for the BCAS4/BCAS3 and the IRA1/RGS17 fusions were detected in MCF7 cells. The β-actin (ACTB) was used as the positive control. The product sizes of ACTB, BCAS4/BCAS3, and IRA1/RGS17 are 600, 328, and 367 bp, respectively.
In the same library, we also observed the fusion of the IRA1 gene and the RGS17 gene. This fusion was supported by three independent clones (Fig. 2B). The 5′-UTR exon 1 of IRA1 is fused with the start codon-bearing exon 2 of RGS17, generating a fusion gene that encodes the full-length RGS17 protein under the control of IRA1 gene promoter. To test for the existence of the IRA1/RGS17 fusion in the MCF7 breast cancer cell line, a primer pair was designed that was specific for the fusion transcript. An RT-PCR product of the expected 367 bp in size was successfully detected (Fig. 2C). The PCR product was excised from the gel, cloned into a vector, and sequenced. The sequence showed 100% identity with the fusion EST sequences and with two parental genes, confirming the existence and expression of the IRA1/RGS17 fusion gene transcripts in MCF7 cells.
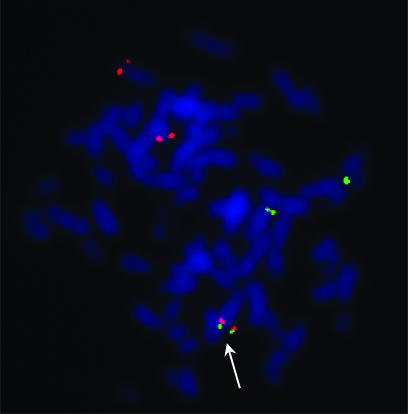
Verification of the IRA1/RGS17 Fusion in MCF7 Cells by FISH. A FISH experiment was conducted by using two BAC clones, IRA1 (3q26.32) and RGS17 (6q25.2), to determine the physical relationship of these two genes in the MCF7 cell line. The majority (16 of 22) of metaphase cells analyzed showed distinct IRA1 signals (red) present on both normal chromosomes 3 and distinct RGS17 signals (green) present on both a normal chromosome 6 and a presumed derivative chromosome 6 t(6;22) [by prior spectral karyotyping analysis of the MCF7 cell line (10)]. In addition, fusion of a red and green signal was visualized in 20/22 metaphase cells on a derivative chromosome, most likely the previously identified t(3;6)(q26;q25)del (3)(p14) (Fig. 3) (10, 11). This, together with RT-PCR detection of the fusion transcript, clearly demonstrates the presence of the IRA1/RGS17 fusion gene in MCF7 cells.
Fig. 3.
Detection of the 3;6 translocation in MCF7 cells by FISH. A representative result of the FISH experiment is presented. The IRA1 gene (red) and the RGS17 gene (green) are on the chromosomes 3 and 6, respectively. Besides two chromosomes of each chromosome 3 and 6, a 3;6 translocation was detected (white arrow).
Discussion
We have developed a semiautomatic procedure for systematic identification of fusion transcripts based on the hypothesis that a fusion transcript is created by a translocation event that fuses intronic sequences between exons from two heterologous genes on different chromosomal loci. Using this procedure, we have identified 118 mRNAs and 196 ESTs as fusion transcript sequences from publicly available databases. Among the annotated mRNA sequences, 96 were previously known as fusion transcripts. All known BCR/ABL1 fusion transcript sequences that meet the condition we used were successfully identified by the procedure. The successful rediscovery of experimentally isolated fusion mRNAs strongly confirm the validity of our reasoning and the procedure adopted. Most of newly identified fusion mRNAs were full-insert sequences of randomly selected cDNA clones from FLJ collection (12), DKFZ genes (13), or MGC clones (14), without prior biological studies on these cases. Although we cannot completely rule out the possibility of creation of a chimeric clone in the process of cDNA library construction, it is unlikely that random breakage and rejoining of two cDNAs would happen at the exact exon boundaries of two genes.
BCAS4/BCAS3 and IRA1/RGS17 Fusion in MCF7 Cells. A BCAS4/BCAS3 fusion produced by imbalanced chromosome translocation has been described in MCF7 breast cancer cells (9). Exactly the same fusion event supported by six EST clones was identified as the top candidate by our method, demonstrating the efficacy of the procedure. These ESTs were isolated from a cDNA library prepared from an anonymous breast adenocarcinoma cell line.
The second fusion gene, IRA1/RGS17, was predicted to occur in the same library and its existence was experimentally confirmed at the mRNA level by RT-PCR and at the genomic level by FISH analysis in MCF7 cells. This is a previously unreported account of a fusion involving the IRA1 gene on chromosome 3q26.32 and the RGS17 gene on 6q25.2. The IRA1 (also known as TBLR1) protein is a subunit for the nuclear receptor corepressor/HDAC3 complex that exhibits transcriptional repression (15). RGS17 (also known as RGSZ2) protein is a member of the GTPase-activating proteins that act as regulators of G protein signaling, showing enriched expression in brain (16). RGS17 protein was reported to induce dispersal of the Golgi apparatus by inactivating the G protein Gαz (17). It was also reported to preferentially inhibit G protein-coupled receptor signaling by G proteins Gi/o, Gz, and Gq (18). Components in the G protein-coupled receptor signaling pathways, including RGS proteins, are known to be involved in many cancers, including prostate cancer (19), and considered as potential therapeutic targets in cancer therapy (20).
Putative Fusion Genes in Normal Cells. Of the total 237 fusion cases, 137 are from cancer tissues, including 58 known cases, two are from noncancer diseased tissues, one is from a mixed sample, and five are from undetermined tissue sources. The remaining 92 cases are from unlabeled, presumably normal tissues. The occurrence of what appear to be genuine fusion gene transcripts from normal cells was unexpected. The normal cells include brain, placenta, eye, testis, hematopoietic cells, liver, pancreas, and others. When the two genes involved in a putative fusion are within the same chromosome, multilocus long transcription has been considered as one of the mechanisms for producing chimeric transcripts (4). But we have eliminated this possibility by selecting only inverted cases when the fusion is between two genes in the same chromosome. Another possibility is trans-splicing, in which two independently transcribed mRNA molecules are fused together, apparently with the same apparatus used for the normal cis-splicing (21). Trans-splicing has been reported to occur in higher mammals, including humans (21-23), although it is believed to occur at a very low frequency (21). Some of the newly detected chimeric sequences, in particular, those that occur only once in the database, could have been generated by this mechanism. However, even among the 20 cases that are supported by two or more clones, five are from normal tissues.
It is possible that at least some of these putative fusion transcripts are from cells that are phenotypically normal but bear a chromosomal aberration. It has been noted that there are many germ-line mutations that are silent, i.e., they do not directly cause cancer (2). A PubMed search yielded >100 articles that report phenotypically normal persons who carry a chromosomal translocation. Often, such silent translocations are discovered through the offspring who carry noticeable genetic defects (24, 25). It is also possible that silent chromosomal translocations happen as somatic mutations during normal development and differentiation processes.
EST Database as Information Source for Fusion Gene Discovery Resulting from Structural Rearrangement of Chromosomes. We demonstrated that ESTs can be used as an information source for identification of fusion genes resulting from possible chromosomal translocations or inversions. Although we have found a large number of putative fusion gene transcripts from the expressed sequence databases, this is certainly not a complete set that exists in the databases used. This lack of complete coverage is partly because we have used a conservative algorithm, which misses some candidates in favor of selecting only those with better evidence. The list will increase in size in the future as the sizes of the mRNA and EST databases increase. Combining computational prediction and experimental verification should result in a large collection of chromosomal aberrations from both cancer and phenotypically normal tissues. Such data will present an opportunity to uncover novel molecular mechanisms of tumor pathogenesis.
Supplementary Material
Acknowledgments
We thank Drs. Michael M. Gottesman and Douglas Lowy for reading the manuscript and for making valuable suggestions.
Abbreviations: ABL1, Abelson murine leukemia viral oncogene homolog 1; BCAS, breast carcinoma amplified sequence; BCR, breakpoint cluster region; FISH, fluorescence in situ hybridization; IRA1, nuclear receptor corepressor/HDAC3 complex; RGS, regulator of G protein signaling.
References
- 1.Mitelman, F., Johansson, B. & Mertens, F. (2004) Nat. Genet. 36, 331-334. [DOI] [PubMed] [Google Scholar]
- 2.Futreal, P. A., Coin, L., Marshall, M., Down, T., Hubbard, T., Wooster, R., Rahman, N. & Stratton, M. R. (2004) Nat. Rev. Cancer 4, 177-183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pane, F., Intrieri, M., Quintarelli, C., Izzo, B., Muccioli, G. C. & Salvatore, F. (2002) Oncogene 21, 8652-8667. [DOI] [PubMed] [Google Scholar]
- 4.Romani, A., Guerra, E., Trerotola, M. & Alberti, S. (2003) Nucleic Acids Res. 31, e17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Communi, D., Suarez-Huerta, N., Dussossoy, D., Savi, P. & Boeynaems, J. M. (2001) J. Biol. Chem. 276, 16561-16566. [DOI] [PubMed] [Google Scholar]
- 6.Florea, L., Hartzell, G., Zhang, Z., Rubin, G. M. & Miller, W. (1998) Genome Res. 8, 967-974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kirsch, I. R., Morton, C. C., Nakahara, K. & Leder, P. (1982) Science 216, 301-303. [DOI] [PubMed] [Google Scholar]
- 8.Sambrook, J., Fritsch, E. F. & Maniatis, T. (1989) Molecular Cloning: A Laboratory Manual (Cold Spring Harbor Lab. Press, Plainview, NY).
- 9.Barlund, M., Monni, O., Weaver, J. D., Kauraniemi, P., Sauter, G., Heiskanen, M., Kallioniemi, O. P. & Kallioniemi, A. (2002) Genes Chromosomes Cancer 35, 311-317. [DOI] [PubMed] [Google Scholar]
- 10.Roschke, A. V., Tonon, G., Gehlhaus, K. S., McTyre, N., Bussey, K. J., Lababidi, S., Scudiero, D. A., Weinstein, J. N. & Kirsch, I. R. (2003) Cancer Res. 63, 8634-8647. [PubMed] [Google Scholar]
- 11.Davidson, J. M., Gorringe, K. L., Chin, S. F., Orsetti, B., Besret, C., Courtay-Cahen, C., Roberts, I., Theillet, C., Caldas, C. & Edwards, P. A. (2000) Br. J. Cancer. 83, 1309-1317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ota, T., Suzuki, Y., Nishikawa, T., Otsuki, T., Sugiyama, T., Irie, R., Wakamatsu, A., Hayashi, K., Sato, H., Nagai, K., et al. (2004) Nat. Genet. 36, 40-45. [DOI] [PubMed] [Google Scholar]
- 13.Wiemann, S., Weil, B., Wellenreuther, R., Gassenhuber, J., Glassl, S., Ansorge, W., Bocher, M., Blocker, H., Bauersachs, S., Blum, H., et al. (2001) Genome Res. 11, 422-435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Strausberg, R. L., Feingold, E. A., Grouse, L. H., Derge, J. G., Klausner, R. D., Collins, F. S., Wagner, L., Shenmen, C. M., Schuler, G. D., Altschul, S. F., et al. (2002) Proc. Natl. Acad. Sci. USA 99, 16899-16903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yoon, H. G., Chan, D. W., Huang, Z. Q., Li, J., Fondell, J. D., Qin, J. & Wong, J. (2003) EMBO J. 22, 1336-1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Larminie, C., Murdock, P., Walhin, J. P., Duckworth, M., Blumer, K. J., Scheideler, M. A. & Garnier, M. (2004) Brain Res. Mol. Brain Res. 122, 24-34. [DOI] [PubMed] [Google Scholar]
- 17.Nagahama, M., Usui, S., Shinohara, T., Yamaguchi, T., Tani, K. & Tagaya, M. (2002) J. Cell Sci. 115, 4483-4493. [DOI] [PubMed] [Google Scholar]
- 18.Mao, H., Zhao, Q., Daigle, M., Ghahremani, M. H., Chidiac, P. & Albert, P. R. (2004) J. Biol. Chem. 279, 26314-26322. [DOI] [PubMed] [Google Scholar]
- 19.Daaka, Y. (2004) Sci. STKE 2004, re2. [DOI] [PubMed] [Google Scholar]
- 20.Liebmann, C. (2004) Curr. Pharm. Des. 10, 1937-1958. [DOI] [PubMed] [Google Scholar]
- 21.Tasic, B., Nabholz, C. E., Baldwin, K. K., Kim, Y., Rueckert, E. H., Ribich, S. A., Cramer, P., Wu, Q., Axel, R. & Maniatis, T. (2002) Mol. Cell 10, 21-33. [DOI] [PubMed] [Google Scholar]
- 22.Caudevilla, C., Serra, D., Miliar, A., Codony, C., Asins, G., Bach, M. & Hegardt, F. G. (1998) Proc. Natl. Acad. Sci. USA 95, 12185-12190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li, B. L., Li, X. L., Duan, Z. J., Lee, O., Lin, S., Ma, Z. M., Chang, C. C., Yang, X. Y., Park, J. P., Mohandas, T. K., et al. (1999) J. Biol. Chem. 274, 11060-11071. [DOI] [PubMed] [Google Scholar]
- 24.Osztovics, M. & Kiss, P. (1975) Clin. Genet. 8, 112-116. [PubMed] [Google Scholar]
- 25.Batista, D. A., Pai, G. S. & Stetten, G. (1994) Am. J. Med. Genet. 53, 255-263. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.