Abstract
After binding to cellular receptors and proteolytic activation, the protective antigen component of anthrax toxin forms a heptameric prepore. The prepore later undergoes pH-dependent conversion to a pore, mediating translocation of the edema and lethal factors to the cytosol. We describe structures of the prepore (3.6 Å) and a prepore:receptor complex (4.3 Å) that reveal the location of poreforming loops and an unexpected interaction of the receptor with the pore-forming domain. Lower pH is required for prepore-to-pore conversion in the presence of the receptor, indicating that this interaction regulates pH-dependent pore formation. We present an example of a receptor negatively regulating pH-dependent membrane insertion.
Many bacteria that colonize mammalian hosts have evolved mechanisms for introducing bacterial enzymes into the cytosolic compartment of host cells. These enzymes disrupt metabolism in various ways, disabling professional phagocytes and/or other cells of the host's immune system. Bacillus anthracis accomplishes this disruption by secreting a tripartite toxin–anthrax toxin, consisting of two intracellularly acting enzymes together with a multifunctional protein that delivers the enzymes to the cytosol (1). The two enzymes are: edema factor (EF, 89 kDa), an adenylate cyclase (2), and lethal factor (LF, 90 kDa), a metalloprotease specific for mitogen-activated protein kinase kinases (3, 4). The delivery component, termed protective antigen (PA, 83 kDa), is a receptor-binding protein that forms a pore in the endosomal membrane, enabling EF and LF to cross to the cytosol.
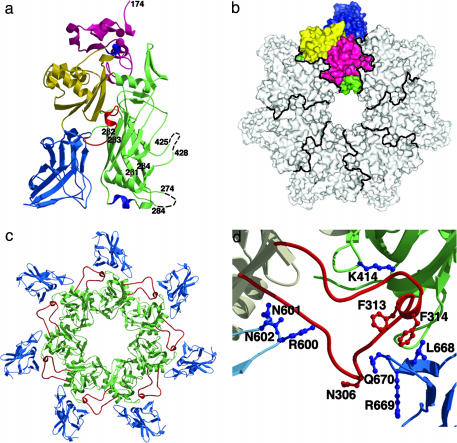
PA, EF, and LF combine at the surface of receptor-bearing cells to form a series of toxic noncovalent complexes (1). PA is a four-domain molecule that binds to either of two cell-surface receptors, capillary morphogenesis protein 2 (CMG2) or anthrax toxin receptor/tumor endothelial marker 8 (ATR/TEM8) (5–7). Proteolysis at a furin-sensitive cleavage site within domain 1 (residues 1–258) removes a 20-kDa fragment, PA20, from the N terminus (8), leaving a 63-kDa fragment, PA63, bound to the receptor. The remaining part of domain 1 (residues 168–258) forms the N terminus of PA63 and functions in oligomerization and in binding EF and LF (Fig. 1a). In the absence of PA20, PA63 self-associates to form a ring-shaped heptamer (9), termed the prepore, the structure of which was previously determined at 4.5 Å (5). Domain 2 (residues 259–487) has a β-barrel core structure and lines the lumen of the heptamer (Fig. 1 a and b). There is a large amphipathic loop between strands 2β2 and 2β3 (residues 302–323) that is disordered in the crystal structure of monomeric PA and is believed to insert into the membrane as a hairpin to generate a 14-stranded β-barrel pore (10) (Fig. 1a). For this loop to reach and span the membrane, the 2β2 and 2β3 strands are predicted to peel away from the domain 2 core and, together with 2β1, 2β4, and the amphipathic hairpin, form an extended β-barrel involving residues 285–340 (5, 10, 11). Domain 3 (residues 488–595) is located on the outside of the heptamer and contains several key residues for oligomerization (12) (Fig. 1b). Domain 4 (residues 596–735) has limited contact with the rest of PA and has been implicated by blocking antibodies, proteolysis, and mutagenesis to function in host–cell receptor binding (13–16) (Fig. 1a).
Fig. 1.
The (PA63)7 prepore structure. (a) A single monomer from the 3.6-Å (PA63)7 structure. Domains 1, 2, 3, and 4 are colored in pink, green, yellow and blue, respectively. The previously unresolved 303–322 (red), 343–350 (blue) and 512–515 (blue) loops are visible whereas the 275–283 and 426–427 loops remain unstructured (black dotted lines). (b) An aerial view (domain 1′ is at the top, closest to the viewer) of the PA63 heptamer with one monomer colored as in a. Domain 2 lines the prepore lumen whereas domains 3 and 4 are located on the outside of the heptamer ring. (c) Domains 2 (green) and 4 (blue) from the 3.6-Å (PA63)7 structure, as viewed from the bottom. The domain 2 insertion loop (red) projects out to bind the neighboring monomer in a groove between domains 2 and 4. (d) The domain 2 insertion loop contacts domain 4 from its own monomer (residues 600–602) and domains 2 (residue 414) and 4 (residues 668–670) from the neighboring monomer. N306 was mutated to cysteine and labeled with pyrene for the experiments described in Fig. 3b. Residues F313 and F314 are predicted to form the tip of the membrane inserted β-hairpin (10).
Both of the known PA receptors, ATR/TEM8 and CMG2, are type 1 membrane proteins with an extracellular von Willebrand factor A (VWA) domain (6, 7). The VWA domains have 60% amino acid identity and, as in the case of the α-integrin inserted (I) domains, include a metal ion-dependent adhesion site (MIDAS) motif that is thought to mediate their interaction with ligand (6, 7). The crystal structure of the CMG2 VWA domain alone revealed a Mg2+ ion bound in the MIDAS along with a carboxylate ligand mimetic (17). Mutagenesis studies suggest that the carboxylate sidechain of PA D683 completes the MIDAS metal coordination in ATR/TEM8 (18). Significant additional contacts are anticipated, because the affinity of the ATR/TEM8 VWA domain for PA is tighter than the μM-mM affinities observed between integrin I domains and their physiological ligands (H. Scobie, personal communication, and ref. 19). Given their sequence homology, we anticipate that ATR/TEM8 and CMG2 will interact with PA similarly. The interaction between PA and the CMG2 VWA domain (Kd = 170 pM) is much tighter than that with ATR/TEM8 (H. Scobie, personal communication, and ref. 20), however, indicating the likelihood of some differences in contacts.
In this article, we describe a 3.6-Å structure of the heptameric PA prepore alone and a 4.3-Å structure of the prepore complexed with the VWA domain of CMG2. The prepore structure reveals the location of the seven 2β2–2β3 loops from domain 2 that are implicated in formation of a transmembrane β-barrel. The structure of the complex shows that CMG2 has contacts not only with domain 4 of PA, as expected, but also with domain 2. The notion that receptor-domain 2 contacts could regulate the pH at which pore formation occurs is supported by experiments showing a major shift in the prepore-to-pore transition toward lower pHs in the presence of CMG2. These findings demonstrate that a receptor can negatively regulate the pH-dependent insertion of a protein into a membrane.
Materials and Methods
Preparation of Proteins. We prepared the PA heptameric prepore (PA63)7 and CMG2 VWA domain (residues 38–218) proteins as described (20). The PA heptameric prepore:CMG2 VWA domain complex, (PA63)7(CMG2)7, was prepared by incubating the purified proteins for 15 min at room temperature using a >10-fold molar excess of CMG2. The complex was then isolated by gel filtration on a Sephacryl-300 column (Pharmacia) and concentrated for crystallization.
Crystallization and Data Collection. Crystals of (PA63)7 were grown by hanging drop vapor diffusion at room temperature by mixing 2 μl of protein solution [8.6 mg/ml in 320 mM NaCl, 40 mM β-octyl glucoside, 20 mM Tris (pH 8.0)] with 2 μl of reservoir solution (50 mM Hepes, pH 8.25) and 0.4 μl of 100 mM spermine. Crystals grew in space group P1 with cell dimensions a = 149.6 Å, b = 167.0 Å, c = 168.2 Å, α = 77.6°, β = 75.7°, γ = 76.0°, and two heptamers per asymmetric unit. Glycerol was added for cryoprotection, and the data were collected at 100 K on beamline 7-1 of the Stanford Synchrotron Radiation Laboratory (SSRL).
Crystals of (PA63)7(CMG2)7 grew at 4°C when 2 μl of protein [12 mg/ml in 20 mM Tris (pH 8.5), 200 mM NaCl, 1 mM MgCl2) were mixed with 2 μl of reservoir solution [6% polyethylene glycol (PEG) 8000/10 mM NaCl/50 mM Tris, pH 8.5). The crystals were in space group P1 (a = 157.3 Å, b = 158.0 Å, c = 212.5 Å, α = 69.7°, β = 69.4°, γ = 65.6°), with two heptameric complexes per cell. Ethylene glycol was used for cryoprotection, and data were collected at beamline F-1 of the Cornell High Energy Synchrotron Source (CHESS). Both datasets were processed with HKL2000 (21) and sharpened with a B factor of –50.
Structure Determination. The positions of the two PA63 heptamers in the (PA63)7 data were determined by molecular replacement using the 4.5-Å heptamer structure (5) as a search model. Rigid body refinement of the 14 monomer subunits was then performed at 5 Å, and the noncrystallographic symmetry (ncs) operators were calculated and used as strict constraints in energy minimization. Repeating this cycle at 4.5 Å, 4.0 Å, and finally 3.6 Å allowed for convergence of the ncs operators and stable refinement. Positional refinement, simulated annealing with torsion angle dynamics, and grouped B-factor refinement were carried out in cns (22) with strict 14-fold ncs restraints. Iterative cycles of refinement and rebuilding in o (23) resulted in a 3.6-Å structure with an overall R factor of 31.4% (Rfree = 31.7%, Table 1).
Table 1. Crystallographic statistics.
| (PA63)7 | (PA63)7 (CMG2)7 | |
|---|---|---|
| Wavelength, Å | 1.0800 | 0.9186 |
| Resolution, Å | 30-3.6 | 20-4.3 |
| Unique reflections | 163,249 | 120,016 |
| Redundancy | 2.0 (1.7) | 2.7 (2.5) |
| Completeness, % | 93.4 (68.8) | 97.3 (98.1) |
| I/σ(I) | 10.6 (1.6) | 6.8 (1.8) |
| Rsym, % | 4.8 (29.1) | 16.5 (44.9) |
| Rcryst/Rfree | 31.4/31.7 | 32.2/33.0 |
| rms deviation bond length, Å | 0.005 | 0.004 |
| rms deviation bond angle, o | 1.1 | 0.9 |
Numbers in parentheses refer to the values for the outer shell.
One heptamer from the refined 3.6-Å structure of (PA63)7 was used as the molecular replacement search model for the (PA63)7(CMG2)7 data. The positions of the two PA heptamers were identified, improved by rigid body refinement, and provided sufficient phase information to manually dock the refined 1.5-Å structure of the CMG2 VWA domain (PDB = 1SHU) into an fo-fc map. Rigid body, positional, and grouped B-factor refinement was performed in two cycles, before and after one round of minor rebuilding in o, by using cns and strict 14-fold noncrystallographic symmetry restraints. The final model has an R factor of 32.2% (Rfree = 33.0%, Table 1) and contains residues 174–274, 285–425, 428–735, and two calcium ions from PA and residues 38–218 (numbered as 1038–1218) and one magnesium ion from CMG2 in 14 copies.
pH-Dependent Conversion of Prepore to SDS-Resistant State. Five microliters of 200-μM CMG2 was added to six aliquots of 40-μl 4.5-μM (PA63)7 prepore and left for 15 min to allow complete binding at room temperature. Six identical (PA63)7 aliquots contained no CMG2. Individual aliquots were incubated for 1 h with 40 μl of the following buffers: 1 M Bistris ([bis(2-hydroxyethyl)amino]tris(hydroxymethyl)methane), pH 5.5; 1 M Bistris, pH 6.0; 1 M Bistris, pH 6.5; 1 M Hepes, pH 7.0; 1 M Hepes, pH 7.5; and 1 M Hepes, pH 8.0. Samples were exposed to 1.25% SDS for 20 min, separated on a 4–20% Tris-Glycine gel in SDS running buffer, and visualized with Coomassie.
Pyrene Fluorescence. Mutant N306C PA83 was expressed and purified with 0.5-mM Tris-(2-carboxyethyl)phosphine present during the final step of purification. Pyrene modification was carried out as described (24) with modifications. Briefly, a 10-fold molar excess of N-(1-pyrene)maleimide (PM) in N,N-dimethylformamide was added to N306C monomer in 20 mM Tris·HCl (pH 7.5) and reacted 15 h at 4°C. Labeled protein was then separated from free dye by elution over a G-50 Sephadex column and proteolytically activated to form heptamer. Mixing experiments with CMG2 were done by incubating 1 μM N306-PM heptamer with a 10-fold molar excess of CMG2 for 10 min in 20 mM Tris·HCl (pH 8.5) with 2% n-dodecyl-β-d-maltopyranoside, followed by addition of 0.02 equivalents of either 1 M Tris, pH 8; 1 M Hepes, pH 7; 1 M Bistris, pH 6; or 1 M acetate, pH 4.9. The resulting pH for each sample was 8.3, 7.0, 6.2, and 4.9, respectively. After a 30-min incubation, samples were analyzed in an ISS (Champagne, IL) fluorimeter in a 1 × 0.5-cm quartz cuvette using an excitation wavelength of 341 nm. Emission was measured from 360 to 600 nm and normalized against the maximal fluorescence at 384 nm.
Results and Discussion
The 3.6-Å resolution structure of the PA heptamer prepore was determined by molecular replacement using the 4.5-Å prepore model (5). The final model contains residues 174–274, 285–425, 428–735, and two calcium ions (in 14 copies, because there were two heptamers per asymmetric unit). The missing loops, 275–283 and 425–427, are within the lumen of the prepore and are probably mobile, because they are predicted to undergo structural changes in the formation of pore (11, 25) (Figs. 1a and 2b). The improved resolution of the data, along with 14-fold averaging, allows for visualization of some functionally important regions of structure that were unresolved in the previous PA monomer and heptamer structures (5) (Fig. 1a). The 343–350 domain 2 helix interacts unexpectedly with receptor (see below), and the 512–515 domain 3 loop makes contacts with a neighboring monomer to stabilize the heptamer, a result anticipated by mutagenesis (12). Most notably, the new prepore structure contains electron density for the 303–322 membrane insertion loop. This loop projects out, away from the monomer, and packs between domains 2 and 4 of the neighboring monomer (Fig. 1 c and d). These contacts are not likely to contribute in a major way to prepore stability because a 300–320 deletion mutant is still able to form heptamers (24). However, the contacts the membrane insertion loop makes with domain 4 of its own monomer (600–602) and domains 2 and 4 of the neighboring monomer (414 and 668–670) probably affect the loop's conformation and motility (Fig. 1d).
Fig. 2.
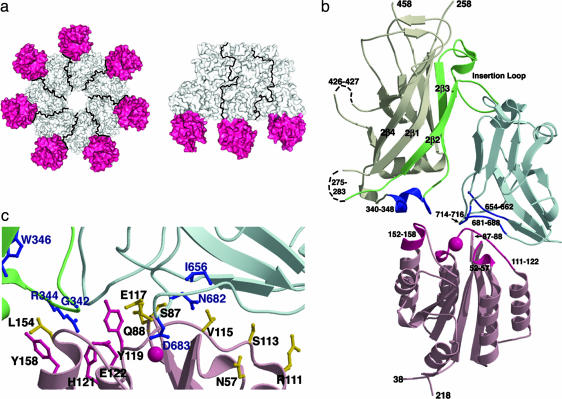
The (PA63)7(CMG2)7 structure. (a) In these bottom and side views, the CMG2 VWA domains are depicted in pink. Only three PA:CMG2 monomers are shown in the side view for clarity. (b) The CMG2 VWA domain (pink) binds both PA domain 2 (white, green, and dark blue) and PA domain 4 (light and dark blue). Direct contacts within the interface are depicted in dark pink (CMG2) and dark blue (PA). The PA insertion loop and the contiguous 2β2 and 2β3 β-strands (green) are predicted to peel away from the domain 2 β-barrel core to form a pore. The CMG2 VWA domain bound to the PA 340–348 loop is likely to impede this rearrangement. (c) This close-up of the PA:CMG2 interface is colored as in b. Previous PA mutagenesis studies suggest the importance of residues G342, W346, I656, N682, and D683 (blue) in intoxication, although the G342C and W346C mutants may reflect structural instability (16). The domain 2 R344 (blue) is buried within the PA:CMG2 interface and may form a salt bridge with CMG2 E122 (pink). Residues Y119, H121, E122, and Y158 of CMG2 (pink) are strictly conserved in ATR/TEM8 and cluster at the PA domain 2 interface, suggesting that ATR/TEM8 binding will also block PA insertion loop rearrangement. Other CMG2 residues at the interface with PA (yellow) are not conserved in ATR/TEM8.
The structure of the PA heptamer:CMG2 VWA domain complex was determined at 4.3 Å and reveals the CMG2 MIDAS occupied by PA D683 (Fig. 2c). In addition, there are significant contacts between four CMG2 loops (52–57, 87–88, 111–122, and 152–158) and four PA loops (340–348, 654–662, 681–688, and 714–716), burying 2000 Å2 of total accessible surface area (Fig. 2b). The protein–protein interface is significantly larger than that observed in other MIDAS I domain–ligand interactions [the α2 integrin-collagen (26) and αL integrin-intercellular adhesion molecule-1 (ICAM-1) (27) interactions both bury ≈1,250 Å2], and this result may explain the tighter CMG2:PA binding affinity. The CMG2:PA interaction also differs from the α2 integrin-collagen (26) and αL integrin-ICAM-1 (27) interactions in that the CMG2 MIDAS is not located in the center of the binding site (Fig. 2b). Extensive site-directed mutagenesis in PA corroborates the importance of some of the domain 4 interface residues (I656, N682, and D683) in receptor binding and toxicity (15, 16). Although a similar analysis has not yet been performed for CMG2, four of the CMG2 residues in the CMG2-PA interface (Y119, H121, E122, and Y158) are strictly conserved in ATR/TEM8 and probably result in a common interaction with PA (Fig. 2c). Many of the other CMG2 residues that constitute the PA:CMG2 interface (N57, S87, Q88, R111, S113, V115, E117, and L154) are not conserved in ATR/TEM8 and may account for their differences in affinity (Fig. 2c).
Whereas the majority of the PA:receptor interaction is mediated through PA domain 4, CMG2 has significant contact with the 340–348 loop of PA domain 2 (Fig. 2 b and c). This loop is flexible as evidenced by its disorder in the monomeric PA83 structure and is predicted to move upon pore formation to accommodate the extended β-barrel composed of residues 285–340 (5, 11). The interaction of the 340–348 PA loop with CMG2 may impede this rearrangement, and we hypothesized that this result could contribute to a lowered pH threshold for prepore to pore conversion.
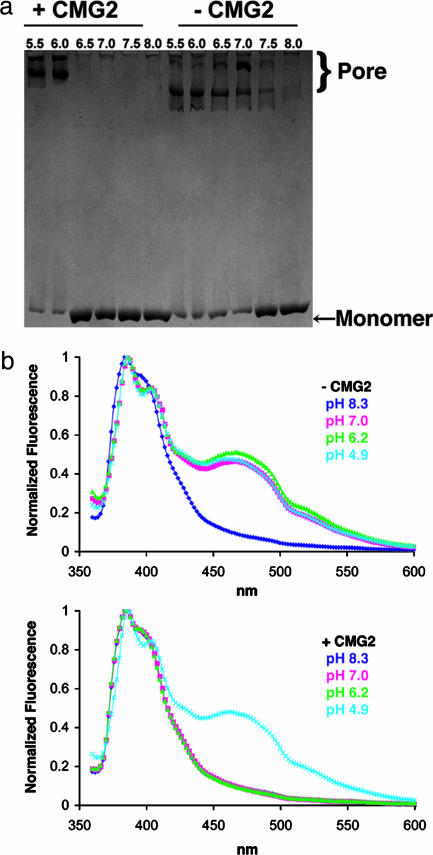
Conversion of the prepore to pore in solution was found to require a significantly lower pH in the presence of receptor (Fig. 3). In one set of experiments, SDS resistance was used as a reporter for pore formation (24). At pH 7.5 or higher, the heptamer dissociated in SDS to the PA63 monomer, but, at pH 7 or below, it converted to an SDS-resistant form that is correlated with pore formation (Fig. 3a). In the presence of CMG2 (or ATR/TEM8, not shown), this conversion to an SDS-resistant state occurred only after the pH had been lowered to between 6 and 6.5 (Fig. 3a). A similar shift in the pH threshold was observed in a fluorescence assay in which the PA insertion loop was labeled at N306C with pyrene and samples were exposed to various pHs in the presence or absence of CMG2. The pyrene labels are too far apart in the prepore structure to interact, but their proximity within the β-barrel of the pore leads to excimer formation and fluorescence (24). This event occurred between pH 7 and 8.3 in the absence of receptor but required a pH between 4.9 and 6.2 in its presence (Fig. 3b). Both techniques show that the pH required for pore formation drops at least a full unit when receptor is added. The results are consistent with previous studies showing a requirement for lower pH on receptor-bearing cells than in solution (24) and indicate that receptor binding acts as a barrier to pore formation. By binding the 340–348 loop of domain 2, CMG2 is likely to block the unfolding of the 2β3 strand (328–335) adjacent to the insertion loop (Fig. 2b). Further, CMG2 binding tethers PA domain 2 to domain 4 and could therefore affect the motility of the insertion loop from the neighboring monomer, which is bound between these two domains (Fig. 1c). The molecular basis by which pH triggers these rearrangements remains unknown. Given a pH profile consistent with the titration of histidines, however, it is notable that there are four histidines located in the 285–340 stretch of domain 2 (H299, H304, H310, and H336) and an additional histidine from CMG2 (H121) located at the domain 2 interface.
Fig. 3.
The pH-dependent conversion of (PA63)7 prepore to pore. (a) Formation of an SDS-resistant state correlates with the formation of pore in solution (24). In the absence of CMG2, this conversion takes place between pH 7 and 7.5. In the presence of CMG2 (and ATR/TEM8, not shown), a pH between 6 and 6.5 is required. The slower pore migration in the presence of CMG2 may indicate that CMG2 remains bound, although the differences between the multiple pore bands are not yet understood. (b) Excimer formation by pyrene-labeled N306C (PA63)7 occurs at a pH between 7.0 and 8.3 (Upper) but requires a pH between 4.9 and 6.2 in the presence of the CMG2 VWA domain (Lower).
Receptor binding seems to block the rearrangement of the PA insertion loop until the pH is well below 7 and therefore confines toxin pore formation to the endosomal membrane and not the plasma membrane. This restriction eliminates the possibility of premature pore formation before full assembly with the catalytic factors. Further, it ensures exposure of EF and LF to endosomal pH, thought to be critical in the unfolding required for their translocation (B. A. Krantz, personal communication, and ref. 28).
Many bacterial and viral pathogens exploit host cell-surface proteins as receptors and vehicles for cellular internalization. For those pathogens that reach endosomal compartments, many use acidic pH to trigger membrane lysis, translocation, or fusion. In some cases (for example in avian sarcoma and leukosis virus and the paromyxovirus SV5 SER isolate), it is thought that the conformational rearrangements required for fusion are triggered by both a receptor-priming event and acidic pH (29, 30). The results reported here, indicating that the anthrax toxin receptor, CMG2, acts as a barrier to pore formation, provide a twist in understanding the tightly orchestrated roles of receptor and low pH in microbial pathogenesis.
Acknowledgments
We thank Jonah Rainey, Heather Scobie, and John Young for their critical review of the manuscript and Lauren Greene and Ruth Pimental for help with protein expression and purification. For their assistance with x-ray crystallography, we thank Piotr Sliz and Ben Spiller, and we thank the staffs of the Cornell High Energy Synchrotron Source beamline F1 and the Stanford Synchrotron Radiation Laboratory beamline 7-1 for access and support. This work was supported by National Institutes of Health Grants AI022021, AI048489, and AI056013.
Abbreviations: EF, edema factor; LF, lethal factor; PA, protective antigen; CMG2, capillary morphogenesis protein 2; ATR/TEM8, anthrax toxin receptor/tumor endothelial marker 8; VWA, von Willebrand factor A; MIDAS, metal ion-dependent adhesion site; (PA63)7, PA heptameric prepore; (PA63)7(CMG2)7, PA heptameric prepore:CMG2 VWA domain complex.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID codes 1TZO and 1TZN).
Note. After completing the work for this manuscript, we learned of a related report, detailing the structure of the PA83 monomer bound to the CMG2 VWA domain (31). The structure, which was determined at 2.5 Å, should be helpful in dissecting the contributions of individual amino acids to this high affinity, pH-dependent binding interaction.
References
- 1.Collier, R. J. & Young, J. A. (2003) Annu. Rev. Cell Dev. Biol. 19, 45–70. [DOI] [PubMed] [Google Scholar]
- 2.Leppla, S. H. (1982) Proc. Natl. Acad. Sci. USA 79, 3162–3166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Duesbery, N. S., Webb, C. P., Leppla, S. H., Gordon, V. M., Klimpel, K. R., Copeland, T. D., Ahn, N. G., Oskarsson, M. K., Fukasawa, K., Paull, K. D., et al. (1998) Science 280, 734–737. [DOI] [PubMed] [Google Scholar]
- 4.Vitale, G., Pellizzari, R., Recchi, C., Napolitani, G., Mock, M. & Montecucco, C. (1998) Biochem. Biophys. Res. Commun. 248, 706–711. [DOI] [PubMed] [Google Scholar]
- 5.Petosa, C., Collier, R. J., Klimpel, K. R., Leppla, S. H. & Liddington, R. C. (1997) Nature 385, 833–838. [DOI] [PubMed] [Google Scholar]
- 6.Bradley, K. A., Mogridge, J., Mourez, M., Collier, R. J. & Young, J. A. (2001) Nature 414, 225–229. [DOI] [PubMed] [Google Scholar]
- 7.Scobie, H. M., Rainey, G. J., Bradley, K. A. & Young, J. A. (2003) Proc. Natl. Acad. Sci. USA 100, 5170–5174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Molloy, S. S., Bresnahan, P. A., Leppla, S. H., Klimpel, K. R. & Thomas, G. (1992) J. Biol. Chem. 267, 16396–16402. [PubMed] [Google Scholar]
- 9.Milne, J. C., Furlong, D., Hanna, P. C., Wall, J. S. & Collier, R. J. (1994) J. Biol. Chem. 269, 20607–20612. [PubMed] [Google Scholar]
- 10.Benson, E. L., Huynh, P. D., Finkelstein, A. & Collier, R. J. (1998) Biochemistry 37, 3941–3948. [DOI] [PubMed] [Google Scholar]
- 11.Nassi, S., Collier, R. J. & Finkelstein, A. (2002) Biochemistry 41, 1445–1450. [DOI] [PubMed] [Google Scholar]
- 12.Mogridge, J., Mourez, M. & Collier, R. J. (2001) J. Bacteriol. 183, 2111–2116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brossier, F., Sirard, J. C., Guidi-Rontani, C., Duflot, E. & Mock, M. (1999) Infect. Immun. 67, 964–967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Varughese, M., Teixeira, A. V., Liu, S. & Leppla, S. H. (1999) Infect. Immun. 67, 1860–1865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rosovitz, M. J., Schuck, P., Varughese, M., Chopra, A. P., Mehra, V., Singh, Y., McGinnis, L. M. & Leppla, S. H. (2003) J. Biol. Chem. 278, 30936–30944. [DOI] [PubMed] [Google Scholar]
- 16.Mourez, M., Yan, M., Lacy, D. B., Dillon, L., Bentsen, L., Marpoe, A., Maurin, C., Hotze, E., Wigelsworth, D., Pimental, R.-A., et al. (2003) Proc. Natl. Acad. Sci. USA 100, 13803–13808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lacy, D. B., Wigelsworth, D. J., Scobie, H. M., Young, J. A. & Collier, R. J. (2004) Proc. Natl. Acad. Sci. USA 101, 6367–6372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bradley, K. A., Mogridge, J., Rainey, G. J., Batty, S. & Young, J. A. (2003) J. Biol. Chem. 278, 49342–49347. [DOI] [PubMed] [Google Scholar]
- 19.Shimaoka, M., Takagi, J. & Springer, T. A. (2002) Annu. Rev. Biophys. Biomol. Struct. 31, 485–516. [DOI] [PubMed] [Google Scholar]
- 20.Wigelsworth, D. J., Krantz, B. A., Christensen, K. A., Lacy, D. B., Juris, S. J. & Collier, R. J. (2004) J. Biol. Chem. 279, 23349–23356. [DOI] [PubMed] [Google Scholar]
- 21.Otwinowski, Z. & Minor, W. (1997) Methods Enzymol. 276, 307–326. [DOI] [PubMed] [Google Scholar]
- 22.Brunger, A. T., Adams, P. D., Clore, G. M., DeLano, W. L., Gros, P., Grosse-Kunstleve, R. W., Jiang, J. S., Kuszewski, J., Nilges, M., Pannu, N. S., et al. (1998) Acta. Crystallogr. D 54, 905–921. [DOI] [PubMed] [Google Scholar]
- 23.Jones, T. A., Zou, J. Y., Cowan, S. W. & Kjeldgaard. (1991) Acta. Crystallogr. A 47, 110–119. [DOI] [PubMed] [Google Scholar]
- 24.Miller, C. J., Elliott, J. L. & Collier, R. J. (1999) Biochemistry 38, 10432–10441. [DOI] [PubMed] [Google Scholar]
- 25.Sellman, B. R., Nassi, S. & Collier, R. J. (2001) J. Biol. Chem. 276, 8371–8376. [DOI] [PubMed] [Google Scholar]
- 26.Emsley, J., Knight, C. G., Farndale, R. W., Barnes, M. J. & Liddington, R. C. (2000) Cell 101, 47–56. [DOI] [PubMed] [Google Scholar]
- 27.Shimaoka, M., Xiao, T., Liu, J. H., Yang, Y., Dong, Y., Jun, C. D., McCormack, A., Zhang, R., Joachimiak, A., Takagi, J., et al. (2003) Cell 112, 99–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wesche, J., Elliott, J. L., Falnes, P. O., Olsnes, S. & Collier, R. J. (1998) Biochemistry 37, 15737–15746. [DOI] [PubMed] [Google Scholar]
- 29.Mothes, W., Boerger, A. L., Narayan, S., Cunningham, J. M. & Young, J. A. (2000) Cell 103, 679–689. [DOI] [PubMed] [Google Scholar]
- 30.Seth, S., Vincent, A. & Compans, R. W. (2003) J. Virol. 77, 6520–6527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Santelli, E., Bankston, L. A., Leppla, S. H. & Liddington, R. C. (July 4, 2004) Nature, 10.1038/nature02763. [DOI] [PubMed]