Abstract
Flavin-containing Baeyer–Villiger monooxygenases employ NADPH and molecular oxygen to catalyze the insertion of an oxygen atom into a carbon–carbon bond of a carbonylic substrate. These enzymes can potentially be exploited in a variety of biocatalytic applications given the wide use of Baeyer–Villiger reactions in synthetic organic chemistry. The catalytic activity of these enzymes involves the formation of two crucial intermediates: a flavin peroxide generated by the reaction of the reduced flavin with molecular oxygen and the “Criegee” intermediate resulting from the attack of the flavin peroxide onto the substrate that is being oxygenated. The crystal structure of phenylacetone monooxygenase, a Baeyer–Villiger monooxygenase from the thermophilic bacterium Thermobifida fusca, exhibits a two-domain architecture resembling that of the disulfide oxidoreductases. The active site is located in a cleft at the domain interface. An arginine residue lays above the flavin ring in a position suited to stabilize the negatively charged flavin-peroxide and Criegee intermediates. This amino acid residue is predicted to exist in two positions; the “IN” position found in the crystal structure and an “OUT” position that allows NADPH to approach the flavin to reduce the cofactor. Domain rotations are proposed to bring about the conformational changes involved in catalysis. The structural studies highlight the functional complexity of this class of flavoenzymes, which coordinate the binding of three substrates (molecular oxygen, NADPH, and phenylacetone) in proximity of the flavin cofactor with formation of two distinct catalytic intermediates.
Keywords: flavoenzyme, mechanism of catalysis, biocatalysis, Baeyer–Villiger reaction, crystallography
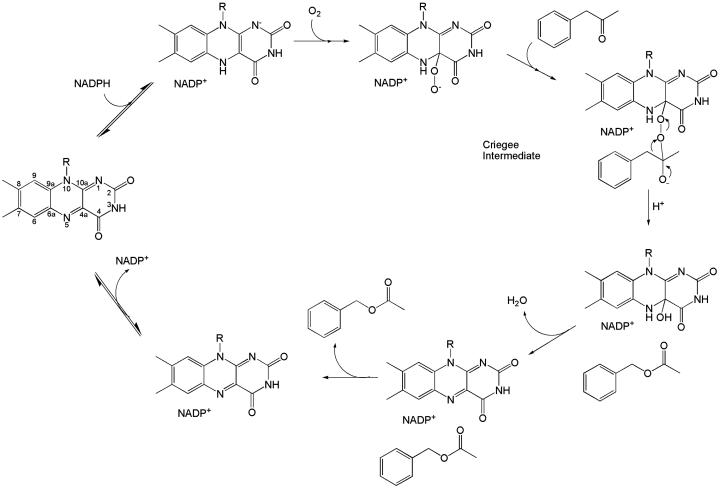
At the end of the 19th century, Baeyer and Villiger (1) discovered that cyclic ketones react with oxidants, such as peroxymonosulfuric acid, to yield lactones. The mechanism of the Baeyer–Villiger reaction involves a nucleophilic attack of a peroxide to the ketone reagent to generate the so-called “Criegee” intermediate (Fig. 1), which is followed by an intramolecular rearrangement that leads to the migration of an alkyl group to an oxygen atom, generating the lactone product. Baeyer–Villiger reactions are of enormous value in synthetic organic chemistry, and the number of their applications is countless.
Fig. 1.
Schematic representation of the overall catalytic reaction of the Baeyer–Villiger monooxygenases with reference to PAMO (mainly based on the kinetic analysis by Sheng et al. (7) of cyclohexanone monooxygenase). The atomic numbering of the flavin ring is shown on the left (corresponding to the initial step of the reaction).
Several microorganisms produce enzymes capable to catalyze Baeyer–Villiger reactions. These proteins are extensively studied for their exploitation in biocatalytic applications (2, 3). This interest follows the problems related to the toxicity and instability of the oxidizing reactants that are currently being used in chemical processes. In addition, enzymatic reactions exhibit a superior degree of enantio- and regioselectivity. Baeyer–Villiger monooxygenases are classified depending on the nature of their flavin cofactor (4). The type I enzymes are the most extensively investigated (5). They are FAD-dependent proteins that use NADPH and molecular oxygen to insert an oxygen atom into their substrate. Kinetic and spectroscopic data have shown that the mechanism underlying the enzyme reaction is conceptually identical to that of the equivalent nonenzymatic processes (6). The key feature is the reaction of the NADPH-reduced flavin with molecular oxygen to generate a stable flavin-peroxide intermediate, which attacks the carbonyl carbon of the substrate (Fig. 1). Rearrangement of the resulting Criegee intermediate yields the oxygenated product and a hydroxy-flavin adduct, which must be hydrolyzed to regenerate the oxidized flavin. Oxidized NADP+ remains tightly bound throughout the catalytic process and does not dissociate before decay of the flavin-peroxide intermediate (Fig. 1) (7).
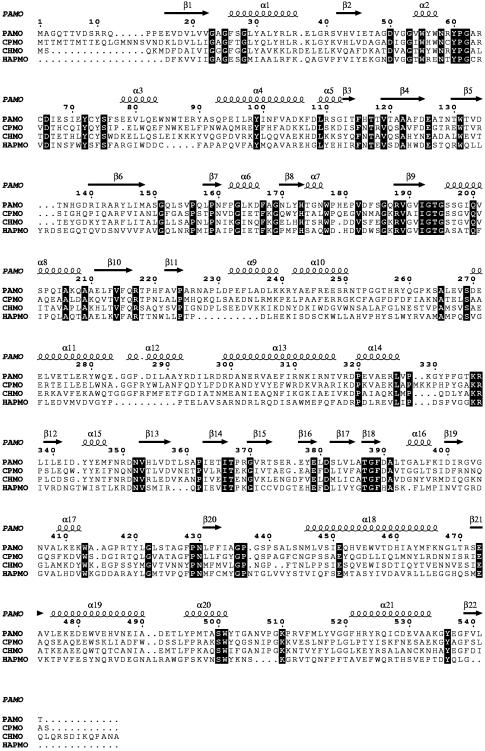
Here, we describe the three-dimensional structure of a Baeyer–Villiger monooxygenase. We have expressed and purified several monooxygenases (2, 8), and the only protein that so far produced crystals suited for x-ray analysis is phenylacetone monooxygenase (PAMO) from the moderate thermophilic bacterium Thermobifida fusca. PAMO is a monomeric 62-kDa enzyme that catalyses the conversion of phenylacetone to phenylacetate (Fig. 1) and displays high homology to other well characterized type I Baeyer–Villiger monooxygenases (Fig. 2).
Fig. 2.
Sequence alignment of representative Baeyer–Villiger monooxygenases. PAMO is from T. fusca (8); CPMO is cyclopentanone monooxygenase from Comamonas testosteroni (34); CHMO is cyclohexanone monooxygenase from Acinetobacter calcoaceticus (NCIMB catalog no. 9871) (35); HAPMO is 4-hydroxyacetophenone monooxygenase from Pseudomonas fluorescens (36). The sequence alignment was calculated with clustalw (37). The secondary structure elements present in the PAMO three-dimensional structure are numbered sequentially. The first 140 amino acids of 4-hydroxyacetophenone monooxygenase are omitted because they form an additional domain normally not found in Baeyer–Villiger monooxygenases. Strictly conserved residues are shown in filled boxes.
Materials and Methods
Protein Expression, Purification, and Crystallization. The pamo gene from genomic DNA of T. fusca was cloned in a pBAD (Invitrogen) expression vector. Transformed Escherichia coli TOP10 cells were grown at 37°C in Terrific Broth medium supplemented with 100 μg/ml ampicillin and 0.1% (wt/vol) arabinose. To obtain the selenomethionine-substituted PAMO, transformed BL21(DE3)RP cells were grown in a minimal M9 medium (64 g/liter dibasic sodium phosphate/15 g/liter monobasic potassium phosphate/2.5 g/liter sodium chloride/5 g/liter ammonium chloride) supplemented with Lys, Thr, Phe, Leu, Ile, Val, Pro, and l-selenomethionine (100 mg of amino acid per liter)/2 mM magnesium sulfate/0.1 mM calcium chloride/1 mM thiamine/1% (vol/vol) glycerol/100 μg/ml ampicillin.
E. coli cells were harvested by centrifugation and resuspended in 50 mM sodium phosphate buffer/5 mM 2-mercaptoethanol/35 μg/ml DNase/1 mM PMSF, pH 7.0. The cell extract was centrifuged and then incubated for 1 h at 50°C. We added 1.5 M ammonium sulfate to the supernatant, which was then loaded onto a butyl-Sepharose column (16/10) preequilibrated with 50 mM sodium phosphate/1.5 M ammonium sulfate, pH 7.0. PAMO was eluted with a descending gradient of 1.5–0.7 M ammonium sulfate and dialyzed against 50 mM sodium phosphate buffer, pH 7.0. The dialyzed sample was loaded onto a preparative Superdex 200 (16/60) column preequilibrated with the same buffer. Pooled fractions were loaded onto a Mono Q HR 5/5 column and eluted with a linear ascending gradient of 0–1 M NaCl. During the purification procedures, the protein lost the FAD cofactor. Therefore, an excess of FAD (5 mM) was added to the final protein sample by incubation at 50°C for 1 h. The purification of selenomethionine-substituted PAMO was performed with the same protocol used for native protein, except for the addition of 5 mM DTT.
Crystals of PAMO were grown at 293 K by the vapor diffusion method. Hanging drops were formed by mixing equal volumes of 18 mg of protein per ml in 5 mM FAD and 50 mM sodium phosphate, pH 7.0, and of a well solution consisting of 1.5 M ammonium sulfate and 0.5 M lithium chloride.
Structure Determination. PAMO crystals belong to space group I222. The asymmetric unit contains a PAMO monomer with a solvent content of 63%. Diffraction data were measured at 100 K on Beamlines BM14 and ID14-EH1 of the European Synchrotron Radiation Facility (Grenoble, France). The data sets were processed (Table 1) by using mosflm (9) and the ccp4 suite of programs (10).
Table 1. Data collection statistics.
| Native | Seleno-methionine substituted | |
|---|---|---|
| No. of crystals | 1 | 2 |
| a/b/c, Å | 86.47/115.65/165.25 | 86.32/117.01/161.98 |
| Resolution, Å | 1.7 | 2.8 |
| Rsym*† | 0.10 (0.39) | 0.16 (0.38) |
| Completeness,† % | 96.2 (80.3) | 99.9 (99.9) |
| I/σ(I)† | 8.5 (3.1) | 16.9 (3.9) |
| Unique reflections | 94,458 | 20,527 |
| Redundancy | 3.7 | 13.1 |
| Mean FOM‡ (centric/acentric) | — | 0.084/0.25 |
| Phasing power | — | 0.73 |
—, Not applicable.
Rsym = ΣhΣi|I(hkl) - 〈I(hkl)〉|/ΣhΣi/(hkl).
Data for the highest resolution shell are given in parentheses.
FOM, overall figure of merit calculated by the program sharp (12).
The structure of PAMO was solved by using the single-wavelength anomalous dispersion method. The six selenium sites were found by using shelxd (11). Phasing with the program sharp (12) produced an electron density map of good quality. The program warp (13) was used to automatically build 470 of 542 residues. The subsequent refinement was carried out by using the native 1.7-Å-resolution data with refmac (14). Model building was done with the program o (15). The refined model has good stereochemical parameters (Table 2). The N-terminal nine amino acids are disordered and not visible in the electron density map.
Table 2. Refinement statistics.
| Resolution range, Å | 15.0-1.7 |
| Rfactor (Rfree)* | 0.212 (0.243) |
| No. of non-hydrogen protein atoms | 4,323 |
| No. of water molecules | 536 |
| No. of FAD atoms | 53 |
| Average B-factor, Å2 | |
| Protein atoms | 26.6 |
| FAD | 19.7 |
| Water molecules | 39.3 |
| rmsd bond length,† Å | 0.012 |
| rmsd bond angle,†o | 1.25 |
| Ramachandran plot‡ | |
| Most favorable region, % | 92.4 |
| Disallowed regions, % | 0.00 |
Model analysis was carried out with dali (16), procheck (17), o (15), and programs of the ccp4 package (10). Pictures were generated with molscript (18), bobscript (19), raster3d (20), and esprit (21).
Results
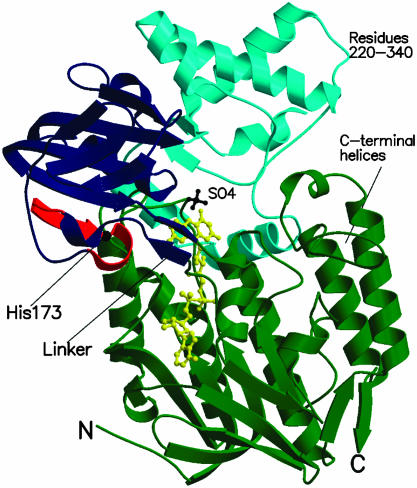
Overall Structure. PAMO consists of two domains (Fig. 3): the FAD-binding domain (residues 10–158 and 390–542) and the NADP-binding domain (residues 159–389). Both domains exhibit the typical dinucleotide-binding fold (22). A search of the Protein Data Bank (23) with the program dali (16) showed that PAMO has the highest structural similarity with the flavoprotein class of the disulfide oxidoreductases (24). The closest structural homologues of PAMO are NADH peroxidase (rms deviation of 3.8 Å for 278 Cα atoms with 18% sequence identity) (25) and human glutathione reductase (rms deviation of 4.1 Å for 276 Cα atoms with 14% sequence identity) (26). Comparison with the disulfide oxidoreductases shows that the basic dinucleotide-binding scaffold is conserved, although with some differences. The FAD-binding domain of PAMO contains three C-terminal α-helices (Figs. 2 and 3) that are not present in the corresponding domain of the disulfide oxidoreductases. In addition, the NADP-binding domain of PAMO exhibits a large insertion of 120 aa (residues 220–340) just after the βαβ unit, which is at the heart of the NADP-binding site. The inserted residues form a α-helical subdomain, which interacts with the FAD-binding domain, forming part of the active site (Fig. 3). The subdomain residues display rather poor sequence conservation (Fig. 2). A dali (16) search shows that there are no other three-dimensional protein structures that have an inserted subdomain of this topology, which seems, therefore, characteristic of the Baeyer–Villiger monooxygenase class of enzymes (27, 28).
Fig. 3.
Ribbon diagram of the PAMO monomer. The FAD-binding domain (residues 10–158 and 390–542) is shown in green, and the NADP-binding domain (159–389) is shown in blue. Residues 220–340, which form a subdomain inserted into the canonical NADP-binding domain topology, are depicted in cyan. The C-terminal helical extension (residues 475–542) found in the dinucleotide-binding fold of the FAD domain is labeled. The fingerprint residues (167–177), which characterize the Baeyer–Villiger monooxygenases (27), are outlined in red. His-173 is a strictly conserved residue of the fingerprint motif that has been shown to be crucial for catalysis. The N-terminal and C-terminal residues are labeled by N and C, respectively. The FAD cofactor and bound sulfate ion are shown in yellow and black ball-and-stick representations, respectively.
The FAD-Binding Site. The recombinant PAMO is purified mostly in the apo form because the cofactor is lost during purification. Incubation with FAD fully restores the enzymatic activity, and the crystalline protein binds the cofactor with full occupancy (see Fig. 5). The cofactor exhibits the same binding mode found in flavoenzyme structures with similar topology (Fig. 3). The Ade ring is partly solvent-accessible, whereas the remaining part of the cofactor is fully embedded by the protein, establishing a number of van der Waals and H-bond interactions.
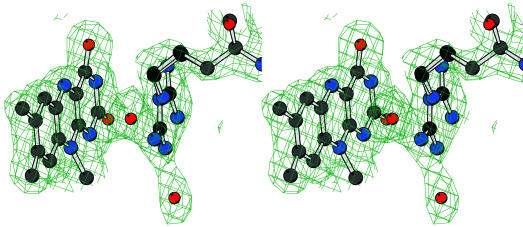
Fig. 5.
Stereo view of the final 2Fo – Fc electron density map for the flavin and Arg-337 in approximately the same orientation as in Fig. 3. The contour level is 1 σ. Atom colors are as in Fig. 4.
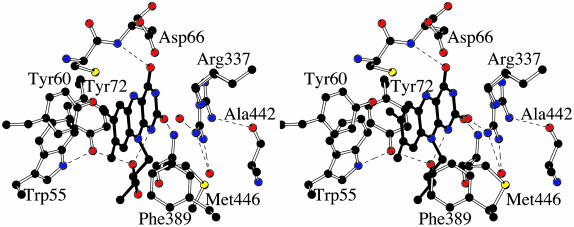
The flavin ring binds at the bottom of the cleft between the two domains of the protein (Fig. 3). The O2 and O4 atoms of the flavin ring (the flavin atomic numbering is shown in Fig. 1) are engaged in H-bonds with the backbone N atoms of Met-446 and Asp-66 (Fig. 4). The N1 atom establishes an intramolecular H-bond with the 3′-OH group of the ribityl chain. No H-bonds involve the N5 atom, which is at 3.5 and 3.7 Å from the N and OD2 atoms of Asp-66, respectively. On the si side, the flavin makes extensive van der Waals contacts with a cluster of conserved aromatic amino acids (Trp-55, Tyr-60, and Tyr-72) (Fig. 4). Conversely, the side chain of an Arg residue (Arg-337) lays on the re side of the flavin ring (Figs. 4 and 5). Arg-337 exhibits a rather broad electron density. We have observed this feature in all density maps calculated with the many data sets that were measured in the course of the crystallographic analysis. Based on these findings, Arg-337 has been modeled in a double conformation (Figs. 4 and 5). This interpretation is corroborated by the usage of the automated side-chain docking algorithm of the program warp (13), which identifies the two alternate conformations as equally compatible with the electron density map. The disorder is confined to the side-chain atoms because the backbone atoms of Arg-337 and of its neighboring residues are in well defined electron density and display B-factors that do not differ from the average protein B-factor (Table 2).
Fig. 4.
Stereo view of the flavin-binding site. The orientation is approximately the same as that of Fig. 3. The flavin has a planar conformation although its N10 atom exhibits a considerable degree of pyramidalization, which positions the C1 atom of the ribityl chain out of the flavin plane. Carbons are shown in black, oxygens are shown in red, and nitrogens are shown in blue. Ordered water molecules are shown as red spheres. H-bond interactions are outlined by the dashed lines.
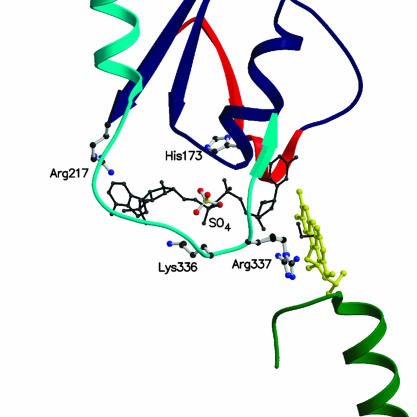
The NADP-Binding Site. We have performed extensive cocrystallization and soaking experiments with NADP+, NADPH, and their analogs. However, none of them was successful in that the resulting crystals did not contain a bound ligand molecule as gathered from the inspection of the electron density map. The protein region forming the NADP-binding site can be inferred from the comparison with the structures of the NAD(P)(H) complexes of flavoenzymes with similar folding topology. We have exploited the structures of human glutathione reductase (complex with NADPH) (29), NADH peroxidase (complex with NADH) (25) and E. coli thioredoxin reductase (complex with a NADP+ analogue) (30). Based on the similarity with these enzymes, NADP(H) is predicted to bind in an extended conformation on the surface of the NADP-binding domain (Fig. 6). The Ade-ribose moiety binds at the C terminus of the central β-sheet of the domain, whereas the nicotinamide ring points toward the re side of the flavin ring at the domain interface. A key feature of the proposed NADP binding mode is that the Ade ring binds in a region involved in crystal-packing interactions. In particular, residues 506–513 of a symmetry-related molecule form a long protruding loop that occupies the Ade site. Such a steric hindrance caused by crystal packing provides an explanation for the failure of all soaking experiments.
Fig. 6.
Close-up view of the NADP-binding site at the domain interface. The FAD cofactor is shown in yellow. Ribbons are colored as in Fig. 3. The picture shows the position of residues that have been shown to be crucial for catalysis and/or NADPH binding. A NADPH molecule is shown modeled in the conformation observed in topologically similar enzymes (black). The image was generated by superimposing the NADP-binding domain of human glutathione reductase in complex with NADPH (29) to the corresponding domain of PAMO. The resulting position of the NADPH molecule bound to glutathione reductase is depicted. As a reference, a model for the peroxide adduct is shown in black. The adduct has been modeled based on the three-dimensional structure of 4a,5-epoxyethano-3-methyl-4a,5-dihydrolumiflavin (38).
Several observations support the proposed binding site of NADP(H). The ligand pyrophosphate group is expected to bind at the N terminus of the α-helix α8 (Fig. 2) on the first βαβ unit of the NADP-binding domain. In the crystal structure, a sulfate ion (Figs. 3 and 6) is present in this position, which, therefore, appears to be able to bind a negatively charged group. In 4-hydroxyacetophenone monooxygenase (Fig. 2), replacement of the residue corresponding to Arg-217 of PAMO with an Ala side chain greatly reduces the affinity for NADPH (28). Consistent with this finding, the three-dimensional structure reveals that Arg-217 is an integral part of the binding site for the NADP Ade moiety. The side chain of Lys-336 is close to the predicted location of the 2′-phosphate of NADP, and its amino group appears to be properly positioned to establish a direct or water-mediated H-bond with the NADP phosphate (Fig. 6). A Lys→Ala mutation targeting the corresponding residue in 4-hydroxyacetophenone monooxygenase causes a drastic reduction in the affinity for NADPH and effectively switches the enzyme specificity toward NADH (28). Conservation of this lysine among type I Baeyer–Villiger monooxygenases (Fig. 2) agrees with the strict NADPH specificity observed for members of this enzyme family (2).
Analysis of the stereospecificity in 4-hydroxyacetophenone monooxygenase (unpublished data) shows that the reaction involves the transfer of the pro-R hydrogen from the nicotinamide to the flavin. This stereospecificity is opposite to that of the disulfide oxidoreductases (31), implying that, compared with the binding mode observed in these enzymes, the nicotinamide ring bound to PAMO and similar monooxygenases has a flipped orientation with respect to the flavin. A detailed analysis of the NADPH-binding mode must await the crystal structure determination of a protein complex.
The Central Role of Arg-337. Arg-337 is a strictly conserved amino acid among Baeyer–Villiger monooxygenases (Fig. 2), and the crystal structure now shows that it is part of the active site, located on the re side of the flavin. In 4-hydroxyacetophenone monooxygenase, mutation of the corresponding Arg residue to Ala makes the enzyme completely inactive (28). The mutation does not affect binding of NADPH nor flavin reduction by NADPH (Fig. 1), but it completely knocks out the ability to catalyze the insertion of the oxygen atom into the substrate. This finding suggested that this Arg residue stabilizes the negatively charged flavin-peroxide intermediate (28). The crystal structure fully supports this conclusion by showing that Arg-337 is in front of flavin C4a atom, the locus where the peroxide adduct is formed (Fig. 1). In both conformations found in the crystal structure, the Arg-337 side chain would appear to be able to directly interact with the intermediate, possibly forming direct H-bonds (Fig. 6).
The conformation and position of Arg-337 suggests an even more complex role in catalysis than simply intermediate stabilization. In the crystal structure, Arg-337 is not fixed in a single conformation, but it adopts two alternate conformations (Fig. 5). This feature underlies an inherent flexibility that can be functionally relevant. Reduction of the flavin by NADPH involves the direct transfer of a hydride anion from the nicotinamide ring to the N5 atom of the cofactor. As indicated by the three-dimensional structures of many flavoenzymes (24), hydride transfer is brought about by positioning the nicotinamide adjacent to the flavin so that the two rings overlap each other. In the case of PAMO, this process must necessarily involve a movement of the Arg-337 side chain, which must shift away from the position observed in the crystal structure to allow the binding of nicotinamide with the catalytically competent stereochemistry. Arg-337 is predicted to exist in two positions: (i) an “IN” position, corresponding to that found in the crystal structure and involved in the stabilization of the flavin-peroxide intermediate during catalysis and (ii) an “OUT” position, which provides the space for the nicotinamide ring to perform cofactor reduction. Support for drastic changes in the flavin environment occurring during catalysis is provided by analysis of cyclohexanone monooxygenase, which exhibits extraordinarily large spectral changes upon NADP+ binding (7).
Conformational Changes in Catalysis. The two-domain architecture of PAMO suggests that the catalytically relevant conformational changes might be brought about partly by a movement of the NADP-binding domain. The intriguing feature of this idea is that a domain rotation would couple the “moving IN/OUT” of the NADP nicotinamide ring with the moving OUT/IN of the Arg-337 side chain, which also belongs to the NADP-binding domain (Fig. 6). The comparison of the PAMO structure with those of NAD(P)H complexes of disulfide oxidoreductases (25, 29, 30) shows that the NADP-binding domain of PAMO adopts a generally more open conformation with respect to that of the topologically similar proteins. Domain movements are not unprecedented in proteins with this domain topology; for instance, a 67° rotation of the NADPH domain has been shown to occur during catalysis of thioredoxin reductase (30). The domain linker of PAMO comprises two antiparallel β-strands (residues 158–160 and 387–389) (Fig. 3), which are highly conserved in amino acid sequence among Baeyer–Villiger monooxygenases (Fig. 2). This “double-hinge” β-sheet construction is similar to that of the domain linker of thioredoxin reductase, and it has been found at the hinge points in several proteins that exhibit domain motions (32).
The amino acid sequences of the Baeyer–Villiger monooxygenases are characterized by the conserved sequence motif FXGXXXHXXXW (27), which in PAMO corresponds to residues 167–177 (Figs. 2, 3, and 6). Most remarkably, the three-dimensional structure shows that the fingerprint amino acids belong to the linker segment that connects the FAD-binding domain to the NADP-binding domain and are not directly part of the active site. Mutagenesis studies have shown that the strictly conserved His residue belonging to the fingerprint sequence (His-173 in PAMO) is crucial for catalysis and FAD binding. In cyclohexanone monooxygenase (33), the replacement of this His residue with Gln causes a 10-fold reduction in activity, whereas in 4-hydroxyacetophenone monooxygenase (27), the His→Ala mutation makes the enzyme inactive. In the crystal structure, His-173 is fully solvent exposed being part of the domain linker region at 16 Å from the flavin and at >10 Å from the NADP-binding site (Figs. 3 and 6). Rather than being directly involved in substrate binding and catalysis, His-173 and the fingerprint motif residues are likely to have a critical role by taking part in the domain rotations and conformational changes that occur during the catalytic cycle.
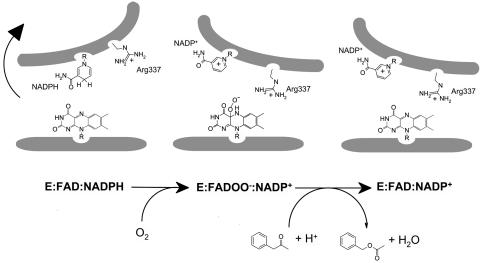
A Model for Catalysis. The crystallographic analysis provides a structural framework for the mechanism of the catalytic reaction proposed on the bases of the kinetic data (7, 28, 33) measured on several Baeyer–Villiger monooxygenases (Figs. 1 and 7).
Fig. 7.
Schematic representation of the main structural features and conformational changes occurring in catalytic steps of the overall reaction of Baeyer–Villiger monooxygenases. E, enzyme.
The reaction starts with the binding of NADPH, which is in the proper position to perform flavin reduction. Simultaneously, Arg-337 adopts the OUT position.
The reduced enzyme reacts with oxygen to form the flavinperoxide intermediate. This step requires a conformational change, possibly involving a domain rotation coupled to a conformational change of Arg-337 and neighboring residues, which would bring the Arg-337 side chain in proximity of the flavin (IN position) to stabilize the negatively charged peroxide intermediate.
The substrate binds and is oxygenated by the flavin. Arg-337 might be involved in the stabilization of the tetrahedral Criegee intermediate formed by the attack of the flavin-OO– group onto the substrate. As suggested by the kinetic analysis of cyclohexanone monooxygenase (7), NADP+ remains bound to the NADP-binding domain, which is expected to be in an open conformation similar to that present in the crystal structure. NADP+ bound to this domain conformation does not block accessibility to the active site (Fig. 6).
The enzyme ternary complex (E:NADP+:product) dissociates, and the products are released.
This initial model for enzyme catalysis needs to be validated and understood in more detail by future studies.
Acknowledgments
We thank Dr. M. Walsh for outstanding support during data collection and Dr. R. van den Heuvel for MS analysis. This work was supported by Ministero dell'Istruzione, dell'Universitá, e della Ricerca Grants COFIN03, FIRB, and Legge 449/97 and by the Centro Interuniversitario Biotecnologie.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviation: PAMO, phenylacetone monooxygenase.
Data deposition: The atomic coordinates have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 1W4X).
References
- 1.Baeyer, A. & Villiger, V. (1899) Chem. Ber. 32, 3625–3633. [Google Scholar]
- 2.Kamerbeek, N. M., Janssen, D. B., Van Berkel, W. J. H. & Fraaije, M. W. (2003) Adv. Synth. Catal. 345, 667–678. [Google Scholar]
- 3.Alphand, V., Carrea, G., Wohlgemuth, R., Furstoss, R. & Woodley, J. M. (2003) Trends Biotechnol. 21, 318–323. [DOI] [PubMed] [Google Scholar]
- 4.Willetts, A. (1997) Trends Biotechnol. 15, 55–62. [DOI] [PubMed] [Google Scholar]
- 5.Mihovilovic, M. D., Muller, B. & Stanetty, P. (2002) Eur. J. Org. Chem. 22, 3711–3730. [Google Scholar]
- 6.Kelly, D. R., Wan, P. W. H. & Tang, J. (1998) in Biotechnology, eds. Rehm, H.-J. & Reed, G. (Wiley, New York), Vol. 8a, pp. 536–587. [Google Scholar]
- 7.Sheng, D., Ballou, D. P. & Massey, V. (2001) Biochemistry 40, 11156–11167. [DOI] [PubMed] [Google Scholar]
- 8.Fraaije, M. W., Kamerbeek, N. M., Heidekamp, A. J., Fortin, R. & Janssen, D. B. (2004) J. Biol. Chem. 279, 3354–3360. [DOI] [PubMed] [Google Scholar]
- 9.Leslie, A. G. (1999) Acta Crystallogr. D 55, 1696–1702. [DOI] [PubMed] [Google Scholar]
- 10.Computational Project Number 4 (1994) Acta Crystallogr. D 50, 760–776.15299374 [Google Scholar]
- 11.Schneider, T. R. & Sheldrick, G. M. (2002) Acta Crystallogr. D 58, 1772–1779. [DOI] [PubMed] [Google Scholar]
- 12.Bricogne, G., Vonrhein, C., Flensburg, C., Schiltz, M. & Paciorek, W. (2003) Acta Crystallogr. D 59, 2023–2030. [DOI] [PubMed] [Google Scholar]
- 13.Perrakis, A., Morris, R. J. & Lamzin, V. S. (1999) Nat. Struct. Biol. 6, 458–463. [DOI] [PubMed] [Google Scholar]
- 14.Murshudov, G. N., Vagin, A. A. & Dodson, E. J. (1997) Acta Crystallogr. D 53, 240–255. [DOI] [PubMed] [Google Scholar]
- 15.Jones, T. A., Zou, J. Y., Cowan, S. W. & Kjeldgaard, M. (1991) Acta Crystallogr. A 47, 110–119. [DOI] [PubMed] [Google Scholar]
- 16.Holm, L. & Sander, C. (1993) J. Mol. Biol. 233, 123–138. [DOI] [PubMed] [Google Scholar]
- 17.Laskowski, R. A., MacArthur, M. W., Moss, D. S. & Thornton, J. M. (1993) J. Appl. Crystallogr. 26, 283–291. [Google Scholar]
- 18.Kraulis, P. J. (1991) J. Appl. Crystallogr. 24, 946–950. [Google Scholar]
- 19.Esnouf, R. M. (1999) Acta Crystallogr. D 55, 938–940. [DOI] [PubMed] [Google Scholar]
- 20.Merritt, E. A. & Bacon, D. J. (1997) Methods Enzymol. 277, 505–524. [DOI] [PubMed] [Google Scholar]
- 21.Gouet, P., Courcelle, E., Stuart, D. I. & Metoz, F. (1999) Bioinformatics 15, 305–308. [DOI] [PubMed] [Google Scholar]
- 22.Wierenga, R. K., Drenth, J. & Schulz, G. E. (1983) J. Mol. Biol. 167, 725–739. [DOI] [PubMed] [Google Scholar]
- 23.Berman, H. M., Battistuz, T., Bhat, T. N., Bluhm, W. F., Bourne, P. E., Burkhardt, K., Feng, Z., Gilliland, G. L., Iype, L., Jain, S., et al. (2002) Acta Crystallogr. D 58, 89–907. [DOI] [PubMed] [Google Scholar]
- 24.Pai, E. F. (1991) Curr. Opin. Struct. Biol. 1, 796–803. [Google Scholar]
- 25.Stehle, T., Claiborne, A. & Schulz, G. E. (1993) Eur. J. Biochem. 211, 221–226. [DOI] [PubMed] [Google Scholar]
- 26.Karplus, P. A. & Schulz, G. E. (1987) J. Mol. Biol. 195, 701–729. [DOI] [PubMed] [Google Scholar]
- 27.Fraaije, M. W., Kamerbeek, N. M., van Berkel, W. J. H. & Janssen, D.B. (2002) FEBS Lett. 518, 43–47. [DOI] [PubMed] [Google Scholar]
- 28.Kamerbeek, N. M., Fraaije, M. W. & Janssen, D. B. (2004) Eur. J. Biochem. 271, 1–10. [Google Scholar]
- 29.Karplus, P. A. & Schulz, G. E. (1989) J. Mol. Biol. 210, 163–180. [DOI] [PubMed] [Google Scholar]
- 30.Lennon, B. W., Williams, C. H., Jr., & Ludwig, M. L. (2000) Science 289, 1190–1194. [DOI] [PubMed] [Google Scholar]
- 31.Manstein, D. J., Massey, V., Ghisla, S. & Pai, E. F. (1988) Biochemistry 27, 2300–2305. [DOI] [PubMed] [Google Scholar]
- 32.Hayward, S. (1999) Proteins Struct. Funct. Genet. 36, 425–435.10450084 [Google Scholar]
- 33.Cheesman, M. J., Byron Kneller, M. & Rettie, A. E. (2003) Chem. Biol. Interact. 146, 157–164. [DOI] [PubMed] [Google Scholar]
- 34.Iwaki, H., Hasegawa, Y., Wang, S., Kayser, M. M. & Lau, P. C. (2002) Appl. Environ. Microbiol. 68, 5671–5684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen, Y. C., Peoples, O. P. & Walsh, C. T. (1988) J. Bacteriol. 170, 781–789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kamerbeek, N. M., Moonen, M. J., Van Der Ven, J. G., Van Berkel, W. J. H., Fraaije, M. W. & Janssen, D. B. (2001) Eur. J. Biochem. 268, 2547–2557. [DOI] [PubMed] [Google Scholar]
- 37.Pearson, W. R. & Lipman, D. J. (1988) Proc. Natl. Acad. Sci. USA 85, 2444–2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bolognesi, M., Ghisla, S. & Incoccia, L. (1978) Acta Crystallogr. B 34, 821–828. [Google Scholar]