Abstract
We have determined the crystal structure of the GDP complex of the YjeQ protein from Thermotoga maritima (TmYjeQ), a member of the YjeQ GTPase subfamaily. TmYjeQ, a homologue of Escherichia coli YjeQ, which is known to bind to the ribosome, is composed of three domains: an N-terminal oligonucleotide/oligosaccharide-binding fold domain, a central GTPase domain, and a C-terminal zinc-finger domain. The crystal structure of TmYjeQ reveals two interesting domains: a circularly permutated GTPase domain and an unusual zinc-finger domain. The binding mode of GDP in the GTPase domain of TmYjeQ is similar to those of GDP or GTP analogs in ras proteins, a prototype GTPase. The N-terminal oligonucleotide/oligosaccharide-binding fold domain, together with the GTPase domain, forms the extended RNA-binding site. The C-terminal domain has an unusual zinc-finger motif composed of Cys-250, Cys-255, Cys-263, and His-257, with a remote structural similarity to a portion of a DNA-repair protein, rad51 fragment. The overall structural features of TmYjeQ make it a good candidate for an RNA-binding protein, which is consistent with the biochemical data of the YjeQ subfamily in binding to the ribosome.
Keywords: ribosome binding, oligonucleotide/oligosaccharide-binding fold, zinc-finger motif
The GTPase superclass can be divided into two large classes: TRAFAC (named after translation factors), including enzymes involved in translation (initiation, elongation, and release factors), signal transduction (in particular, the extended ras-like family), and SIMIBI (after signal recognition particle, MinD, and BioD), consisting of signal recognition particle GTPases and metabolic enzymes (1). The YlqF/YawG family belonging to the TRAFAC superclass consists of five distinct subfamilies, typified by the proteins YlqF, YqeH (both from Bacillus subtilis), YjeQ (from Escherichia coli), MJ1464 (from Methanococcus jannaschii), and YawG (from Schizosaccharomyces pombe; ref. 1). All of these protein sequence show a circular permutation of the GTPase signature motifs.
Members of the YjeQ subfamily are broadly conserved in bacteria. YjeQ from E. coli and B. subtilis has been shown to be indispensable for cell growth (2). Proteins of the YjeQ subfamily contain all sequence motifs typical of the vast class of P-loop-containing GTPases (3), but show a circular permutation in sequence (Fig. 1). All of the YjeQ subfamily proteins display a unique domain architecture composed of a predicted N-terminal oligonucleotide/oligosaccharide binding (OB-fold) RNA-binding domain, a central permuted GTPase module, and a C-terminal cysteine cluster forming a zinc-finger motif (4). This domain architecture suggests a possible role for YjeQ as a regulator of translation. In vitro, recombinant E. coli YjeQ protein interacted strongly with the 30S ribosomal subunit, with high affinity in the presence of the nonhydrolyzable GTP analog 5′-[β,γ-imido]triphosphate. Likewise, association with the 30S subunit resulted in a 160-fold stimulation of E. coli YjeQ GTPase activity, which reached a maximum with stoichiometric amounts of ribosomes. It is also noteworthy that stoichiometric amounts of E. coli YjeQ in a cell is ≈1 per 200 ribosomes, implying a critical but narrow role in a subset of translating ribosomes (5).
Fig. 1.
Sequence comparison between TmYjeQ and its homologues. Bs, B. subtilis; Ec, E. coli; Pp, Pseudomonas putida; Mpe, Mycoplasma penetrans HF-2; Mp, Mycoplasma pneumoniae. Domains are represented as by the following colors: the OB-fold domain (yellow), the GTPase domain (light blue), and the zinc-finger domain (pink). G1 and G2, representing the 310-helices, are green. The G-1 to G-5 region of characteristic GTPase loops is purple. Green characters represent the conserved residues present in more than five species. –, gaps; *, all conserved residues. Colons represent homologous residues having hydrophobic side chains, and periods represent homologue residues having polar side chains. The whole-sequence identities of Bs, Ec, Pp, Mpe, and Mp against TmYjeQ are 38.3%, 34.2%, 33.2%, 32.5%, and 26.4%, respectively. The whole-sequence identities against the OB-fold domain are 21.2%, 16.7%, 22.7%, 19.7%, and 18.2%, respectively. The whole-sequence identities against the GTPase domain are 43.1%, 38.9%, 38.3%, 37.7%, and 29.9%, respectively. The whole-sequence identities against the zinc-finger domain are 43.5%, 40.3%, 30.6%, 32.3%, and 25.8%, respectively.
To elucidate the molecular function of the YjeQ subfamily, we have determined the three-dimensional structure of YjeQ from Thermotoga maritima (TmYjeQ; gi 4981923) that shows 34% sequence identity with E. coli YjeQ (Fig. 1) and discussed its structural characteristics in relationship to its function.
Materials and Methods
Cloning of TmYjeQ. The sequence encoding TmYjeQ was amplified by the PCR from T. maritima genomic DNA (American Type Culture Collection) using Deep Vent DNA Polymerase (New England Biolabs, Beverly, MA). The resulting PCR product was purified and prepared for ligation-independent cloning (6) by treatment with T4 DNA polymerase in the presence of 1 mM dTTP for 30 min at 37°C. The prepared DNA was then mixed with a pB4 vector for 5 min at room temperature and transformed into DH5α. This pB4 vector was designed in our laboratory to express the target protein together with an N-terminal His6 tag-maltose-binding protein fusion containing a tobacco etch virus protease cleavage site. Clones were screened by plasmid DNA analysis and were transformed into BL21(DE3)/pSJS1244 for protein expression (7).
Protein Expression, Purification, and Crystallization. A selenomethionine derivative of the protein was expressed in a methionine auxotroph, E. coli strain B834(DE3)/pSJS1244 (7), grown in PASM medium [W. Studier, Brookhaven National Laboratory (Upton, NY), personal communication] supplied with selenomethionine (8). Cells were disrupted by microfluidization (Microfluidics, Newton, MA) in 50 mM Hepes, pH 7.0/300 mM NaCl/1.0 mM PMSF/10 μg/ml DNase/0.1 μg/ml antipain/1 μg/ml chymostatin/0.5 μg/ml leupeptin/0.7 μg/ml pepstatin A, and cell debris were pelleted by centrifugation at 10,000 rpm for 20 min in a Sorvall centrifuge. The supernatant was then spun in a Beckman ultracentrifuge Ti45 rotor at 35,000 rpm for 30 min at 4°C. The fusion protein was affinity-purified by using two 5-ml HiTrap chelating HP columns (GE Healthcare, Piscataway, NJ) in order on an ÅKTA Explorer chromatography system (GE Healthcare, Piscataway, NJ). The fusion proteins was bound to the column in 50 mM Hepes, pH 7.0/300 mM NaCl and was eluted in a gradient of 10–400 mM imidazole with 13 column volumes. Fractions were pooled and dialyzed overnight at room temperature against 50 mM Hepes, pH 7.0/0.1 M NaCl/5 mM 2-mercaptoethanol/10 mM imidazole in the presence of a tobacco etch virus protease. After centrifugation, the supernatant was applied onto a 5-ml HiTrap metal chelating (Ni2+) column. The cleaved recombinant protein was found in the flow-through. Dynamic light scattering (DynaPro 99, Proterion, Piscataway, NJ) showed a single monodisperse peak, indicating homogeneity of the protein. Further purification was performed by using anion-exchange chromatography. The protein was eluted in 20 mM Tris·HCl, pH 7.5/300 mM NaCl. SDS/PAGE showed one band of ≈35 kDa, corresponding to the molecular weight of TmYjeQ. The final protein contains six glycine residues at the N terminus, designed to help tobacco etch virus proteolysis. The protein was concentrated to 8.1 mg/ml for crystallization.
Screening for crystallization conditions was performed by using the sparse-matrix method (9) with several screens from Hampton Research (Laguna Niquel, CA) and Wizard Screen (deCODE genetics, Bainbridge Island, WA). The crystallization robot “Hydra Plus-One” (Matrix Technologies, Hudson, NH) was used to set the screens by using the sitting-drop vapor diffusion method at room temperature.
In the optimized crystallization conditions, 1 μl of the protein solution was mixed with 1 μl of the well solution containing 0.2 M KCl, 0.1 M Tris·HCl (pH 8.0), 15% polyethylene glycol 3350, and 10 mM guanidine HCl by using the hanging-drop vapor diffusion method. A thick plate-shaped crystal grew in 1 week to approximate dimensions of 0.2 × 0.1 × 0.03 mm3.
Data Collection and Reduction. Polyethylene glycol 3350 concentration of the well solution was increased to 30% and the hanging drop was allowed to equilibrate overnight. One microliter of the reservoir solution was added to the hanging drop and incubated overnight for equilibration before being flash-frozen in liquid nitrogen and exposed to x-rays. X-ray diffraction data sets were collected at single wavelengths at the Macromolecular Crystallography Facility, beamline 5.0.2, at the Advanced Light Source at the Lawrence Berkeley National Laboratory by using an Area Detector System (Poway, CA) Quantum 4 charge-coupled device detector placed 280 mm from the sample. The oscillation range per image was 1.0° with no overlap between two contiguous images. X-ray diffraction data were processed and scaled by using denzo and scalepack from the hkl program suite (10). The synchrotron data were collected to 2.8 Å. Data statistics are summarized in Table 1. The crystal belongs to the primitive monoclinic space group C2, with unit cell parameters of a = 51.90 Å, b = 137.23 Å, c = 80.93 Å, and β = 106.0°.
Table 1. Statistics of the peak-wavelength single-wavelength anomalous dispersion data set.
| Data set | Peak |
|---|---|
| Wavelength, Å | 0.97930 |
| Resolution, Å | 50.0 to 2.80 |
| Redundancy | 8.4 (8.7)* |
| Unique reflections | 29,551 (1,478) |
| Completeness, % | 100 (100) |
| I/σ | 13.7 (2.6) |
| Rsym,† % | 11.2 (81.2) |
Numbers in parentheses refer to the highest-resolution shell, which is 2.85 to 2.80 Å for all the peak-wavelength data set.
Rsym = Σhkl Σi| Ihkl, i - 〈I〉hkl|/Σ|Ihkl|.
Structure Determination and Refinement. Twenty-one of 24 possible Se atom positions were located by using the program solve (11) with the figure of merit (FOM) of 0.30 at 2.9-Å resolution. The initial single-wavelength anomalous dispersion phases were further improved by solvent flattening with three-fold noncrystallographic symmetry matrices by using the program resolve (11). The best interpretable map was found from 20.0- to 2.9-Å resolution data with the FOM of 0.57. The three molecular models in the asymmetric unit were built by using the program o (12).
The preliminary model was then refined by using the program cns (13). The reflections in this data set between 20.0 and 2.8 Å were included throughout the refinement calculations. Ten percent of the data were randomly chosen for free R factor cross validation. The refinement statistics are shown in Table 2. Isotropic B factors for individual atoms were initially fixed to 20 Å2 and were refined in the last stages. The 2 Fo – Fc and Fo – Fc maps were used for the manual rebuilding between refinement cycles and for the location of solvent molecules. Atomic coordinates have been deposited in the Protein Data Bank (PDB; ID code 1U0L).
Table 2. Crystal parameters and refinement statistics.
| Parameters | Statistics |
|---|---|
| Space group | C2 |
| Cell dimensions | a = 142.82 Å, b = 114.80 Å, c = 77.09, β = 105.72 |
| Volume fraction of protein, % | 57.6 |
| Vm Å3/Da | 3.01 |
| Total no. of residues | 834 |
| Total non-H atoms | 6,806 |
| No. of water molecules | 56 |
| Average temperature factors | |
| Protein, Å2 | 59.1 |
| Solvent, Å2 | 40.1 |
| GDP and Zn ion, Å2 | 44.6 |
| Resolution range of reflections used, Å2 | 20.0 to 2.8 |
| Amplitude cutoff, σ | 0.0 |
| R factor, % | 21.8 |
| Free R factor, % | 28.4 |
| Stereochemical ideality | |
| Bond, Å | 0.007 |
| Angle, o | 1.46 |
| Improper, o | 0.86 |
| Dihedral, o | 23.1 |
Results
Quality of the Model and Overall Structure. The final models include three molecules in the asymmetric unit. All three models include 278 of 295 residues. Each monomer contains one GDP molecule and one zinc atom, although it was not added during purification and crystallization. The final models have been refined at 2.8-Å resolution to a crystallographic R factor of 21.8% and free R factor of 28.4% (Fig. 2). The averaged B factors for main-chain and side-chain atoms are 55.6 and 61.6 Å2, respectively. In the TmYjeQ models, two N-terminal, two C-terminal residues, and a loop between the H4 and β12 (residues 191–203) are undefined in the electron density map. Table 2 summarizes the refinement statistics as well as model quality parameters. All residues lie in the allowed region of the Ramachandran plot produced with the program procheck (14).
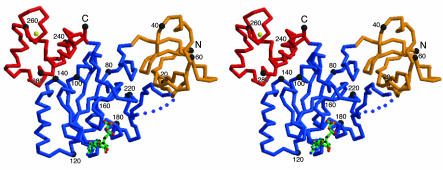
Fig. 2.
A stereo drawing of a Cα trace of TmYjeQ with bound GDP. Each domain is colored according to Fig. 1. Every 20th residue is numbered and is represented by a dot. The bound GDP is represented as a ball-and-stick model. The N (residue Leu-3) and C termini (residue Arg-293) and the secondary elements are labeled. The figure was generated by using the program molscript (31).
The Cα trace of the atomic model of TmYjeQ is shown in Fig. 2. The monomer has approximate dimensions of 75 × 55 × 25 Å3. TmYjeQ consists of three domains: an N-terminal five-stranded β-barrel domain, a central α/β domain, and a C-terminal all-α domain. The three monomers in the asymmetric unit form a triangular quaternary structure. The self-rotation function also revealed the presence of a noncrystallographic threefold symmetry along the C axis with few degrees off. However, results from analytical size exclusion column (data not shown) together with dynamic light scattering showed that the TmYjeQ is a monomer in solution. Because E. coli YjeQ showed a maximum stimulation of GTPase activity at a 1:1 stoichiometry with the ribosome (5), the biologically active state of TmYjeQ may be also a monomeric form.
Circularly Permuted GTPase Domain. The GTPase domain (residues 67–231) is located in the center of the protein. The core of the GTPase domain is a contiguous six-stranded β-sheet of a topology of –1x, –1x, +3x, +2x, –1 (15) or a topology of 3↑2↑1↑4↑6↑5↓ (ref. 16 and Fig. 2). The fold of TmYjeQ resembles other TRAFAC GTPase family members with the characteristic Rossman fold. The interesting feature of TmYjeQ is that the GTPase domain is circularly permuted compared with other TRAFAC GTPases as expected from the sequence analysis (3). The comparison of the GTPase domain of TmYjeQ with c-Ha-ras p21 protein (17–20) clearly shows the occurrence of permutation with the unusual order of characteristic GTPase loops of the G4(N/TKxD)-G5[(T/G)(C/S)A]-G1(Walker A, P-loop)-G2(T)-G3 (Walker B) pattern (Fig. 3) as opposed to the regular G1-G2-G3-G4-G5 pattern seen in most GTPases (3). A lot of circularly permuted proteins have been recognized, mainly by sequence comparison, comparing their three-dimensional structures, and searching the database with the program sheba (21).
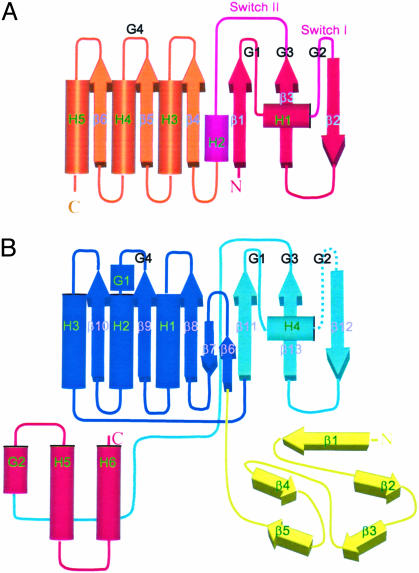
Fig. 3.
A topology diagram of ras protein and TmYjeQ. The 310 (G)- and α-helices are represented by cylinders and β-strands are represented by thick arrows. Secondary structure elements and characteristic GTPase loops are labeled. (A) Ras protein. The color is changed from red to scarlet. The switch I and II regions are pink. (B) TmYjeQ. Each domain is colored according to Fig. 1. The color of the GTPase domain is changed from dark blue to light blue.
N-terminal OB-fold Domain. The N-terminal domain of TmYjeQ revealed two three-stranded antiparallel β-sheets where the first strand is shared by both sheets often found in a Greek key motif, the OB fold (22). As shown in Fig. 2, β-sheets pack orthogonally, forming a five-stranded β-barrel arranged in a 1↑2↓3↑5↑4↓1↑ topology. The OB-fold domain is a compact structural motif frequently used for nucleic acid recognition originally named for its oligonucleotide/oligosaccharide-binding properties. The highly conserved residues, Arg-33 and Gly-46, of the TmYjeQ subfamily are characteristic sequences of the OB fold. One noteworthy property of the TmYjeQ OB-fold domain is its simplicity. Usually, the size of OB folds ranges from 70 to 150 aa in length. Therefore, ≈66 residues of the TmYjeQ OB fold classifies itself as one of the smallest OB folds due to the absence of long insertions among loops usually found in other OB-fold proteins. Such a small OB-fold domain is also found in NusA, a protein known to interact with nonspecific mRNA (23).
When the OB-fold domain of TmYjeQ is aligned with that of tRNA-bound aspartyl tRNA synthetase (rms deviation of 3.3 Å for 64 Cα atoms; PDB ID code 1B16), the positive surface of TmYjeQ is aligned with the bound tRNA surface of aspartyl tRNA synthetase. The positively charged putative RNA-binding surface of TmYjeQ is centered on β2, β3, and Loop L1, corresponding to the substrate-binding surface of other OB-fold domains (22).
C-terminal Unique Zinc-Finger Domain. The C-terminal domain (residues 233–295) of TmYjeQ is composed of one 310-helix, two α-helices, and long loops. Interestingly, three cysteines (Cys-250, Cys-255, and Cys-263) and one histidine (His-257) in this domain are conserved in the sequence alignment (Fig. 1). The psi-blast search with the C-terminal sequence showed that the C(F/h) (F/Y)xxCxHxx(E/D)xxC motif is solely found in the YjeQ subfamily. The crystal structure revealed that these residues are coordinated to zinc ions, forming a zinc-finger motif (Fig. 4). Zinc fingers are protein domains in which the tetrahedrally coordinated zinc contributes to the structural stability of domain. However, the dali (24) search of the C-terminal domain shows a remote structural similarity to a portion of a DNA-repair protein, rad51 fragment (rms deviation of 2.6 Å for 48 Cα atoms; Z = 2.6, PDB ID code 1B22), which does not contain a zinc-finger motif. Therefore, the zinc-finger motif of TmYjeQ is structurally unique to perform its specific function. In general, structurally diverse zinc fingers among proteins perform a broad range of functions in various cellular processes, such as transcription and translation and in interaction modules to various substrates (25).
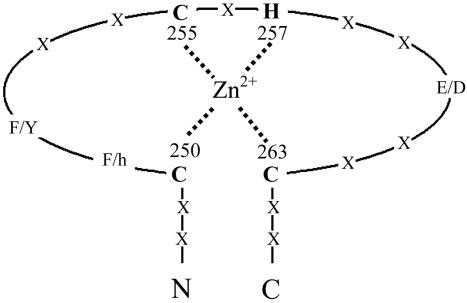
Fig. 4.
A schematic drawing of a zinc-finger motif. The schematic view of the zinc-finger motif is drawn with the labeling of three cysteines and one histidine. The distances between zinc to coordinating atoms are within 2.1 to ≈ 2.4 Å. The two phenylalanines, Phe-251 and Phe-252, involved in a trimer formation, are also indicated. X, any amino acids; h, hydrophilic amino acids.
One more interesting feature of the C-terminal domain is the presence of highly conserved Phe-252 binding with a hydrophobic patch of the OB-fold domain of another molecule in the asymmetric unit. Together with Phe-251, Phe-252 contributes to the formation of a trimeric TmYjeQ in the asymmetric unit. Considering the high affinity of E. coli YjeQ to the ribosome, the crystal structure suggests that highly conserved Phe-252 is one of the key binding residues recognizing DNA, RNA, or is involved in protein–protein interaction. The sequence comparison (Fig. 1) shows highly conserved negatively charged Glu-260 and highly conserved positively charged residues instead of hydrophobic residues in the position of Phe-251 of TmYjeQ (Fig. 4). In summary, this region is a good candidate for the potential species-specific recognition site necessary for proper molecular function.
Structural Comparison Between the TmYjeQ GTPase Domain and ras Protein. The structural studies of apo, GDP, and GTP analog-bound ras protein complexes elucidated that GTP binding and its hydrolysis is associated with conformational changes of two polypeptide chain segments (Fig. 5), switch I (residues 30–38) and switch II (residues 60–76) (17–20). In ras protein, switch I (containing the G-2 loop) coordinates the γ-phosphate moiety of a GTP analog and a bound Mg2+ in the GTP analog complexes (11). Switch II, encompassing the G-3 loop, also interacts with the GTP γ-phosphate moiety in ras protein. The major structural differences between TmYjeQ and ras protein are found in the switch I and II regions. The switch I (loop between α2 and β12) of TmYjeQ is not visible in the crystal structure, implying an extremely flexible nature of the segment unlike that of ras protein (19). The switch II region of TmYjeQ is composed of a long loop connected to the C-terminal zinc-finger domain. Therefore, conformational changes of the switch II region in TmYjeQ can be directly propagated to the downstream zinc-finger domain. However, the spatial counterpart of the helical region (helix α2) of switch II in ras protein are the strands β6 and β7 connecting the GTPase and the OB-fold domains in TmYjeQ, resulting from the circular permutation (Figs. 3 and 5). Because strands β6 and β7 of TmYjeQ are next to the switch II region, some communication between the N-terminal OB fold and the switch II region can be expected.
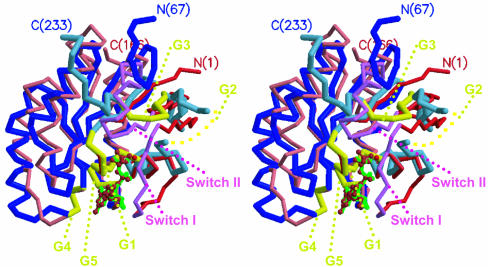
Fig. 5.
A stereo drawing of superposition of the TmYjeQ GTPase domain and ras protein. The same colors are used for the TmYjeQ GTPase domain and ras protein as in Fig. 3. The five GTPase loops are dark yellow. GDP in TmYjeQ is green and magnesium ion and phosphoaminophosphonic acid-guanylate ester in ras protein (PDB ID code 1CTQ) is scarlet.
Interestingly, permutation of the GTPase domain of TmYjeQ occurs in the functionally critical switch II region. This finding implies that circular permutation is one of the ways of keeping the basic molecular function of proteins but diversifying the cellular function of proteins by changing some protein–protein or domain–domain interactions. Similar results were found in the phosphoserine phosphatase family where the circular permutation also helps the diversification of the cellular function of the family by insertion of more than one domain without altering their basic molecular function (26, 27).
Predicted Domain Flexibility. As shown in Fig. 2, there is no contact between the OB-fold and the zinc-finger domains. The averaged B factors of the OB fold, the GTPase, and the zinc-finger domains are 88.6, 46.3, and 60.3 Å2, respectively. The uneven distribution of the domain averaged B factors indicates the presence of a rigid body motion of the domains through connecting hinge loops. As expected from the averaged B factors, the interaction of the GTPase domain with the zinc-finger domain is relatively stronger than with the OB-fold domain. There are one salt bridge (Arg-243–Glu-106) and two salt bridge networks (Arg-278–Asp-99–Tyr-137 and Glu-288–Tyr-96–Lys-100) between the GTPase and the zinc-finger domains. On the contrary, there is only one salt bridge network (Lys-212–Glu-20–Tyr-218) between the OB-fold and the GTPase domains. None of the salt bridges are composed of conserved residues (Fig. 1). In addition, few hydrophobic interactions are detectable between domains. Because the interactions between domains are weak, their interactions can be also easily abolished by a change in surrounding cellular conditions like pH and salt during interaction with the ribosome.
The solvent-accessible surface areas buried by the domain interfaces (per domain) are ≈700 Å2 (between the OB-fold and the GTPase domains) and ≈800 Å2 (between the GTPase and the zinc-finger domains). Therefore, the large contact area and weak interaction of domains suggest that conformational changes induced by binding and hydrolysis of GTP or interaction with other substrates can impact largely and propagate easily to neighboring domains due to the domain properties. In ras protein, the conformational difference induced by γ-phosphate spans >40 Å. Considering the structural feature of TmYjeQ, the conformational changes induced by γ-phosphate may be much larger because of the presence of domain flexibility. Some of the proteins such as RF1 (28), RF2 (29), and NusA (23) also require a large domain flexibility for proper function with the ribosome or RNA polymerase.
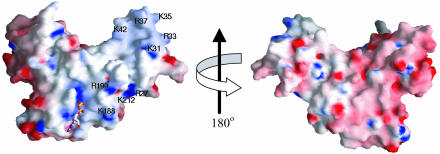
Structural Features Related to Biochemical Functions. N-terminal truncation variants of E. coli YjeQ revealed that the predicted OB-fold region was essential for ribosome binding and GTPase stimulation (5). The crystal structure of TmYjeQ is consistent with this biochemical data. First, the electrostatic surface potential of the GTPase and the N-terminal OB-fold domains in TmYjeQ shows a continuous positive potential surface required for RNA binding (Fig. 6). Second, as discussed above, the structural and surface properties of the two domains infer communication between the OB-fold and the GTPase domains. Therefore, the binding of DNA or RNA to the OB fold of TmYjeQ could modulate the GTPase activity of the GTPase domain.
Fig. 6.
The electrostatic surface potential of TmYjeQ. (A) A molecular surface is created by using the program grasp (red, negative; blue, positive; white, uncharged; ref. 32). The positively charged residues contributing to the putative RNA-binding surface are labeled. (B) The figure was drawn after a 180° rotation of A around the y axis.
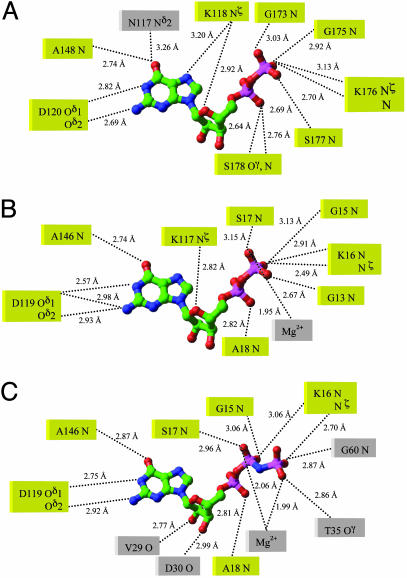
E. coli YjeQ is an essential GTPase and is a factor in ribosome function participating in a guanine nucleotide-dependent interaction (4, 5). The intrinsic GTPase activity was confirmed by using the catalytically impaired (Ser221Ala) variant of E. coli YjeQ, resulting in no stimulation of the GTPase activity by the 30S subunit (5). Comparison of bound GDP of TmYjeQ with that of ras protein reveals that the bound state of both GDPs is quite similar. Many active-site residues and backbone nitrogen atoms involved in bound substrates are highly conserved as shown in Fig. 7. Therefore, regions around the G-1, G-4, and G-5 loops interacting with the GDP moiety are structurally similar in both TmYjeQ and ras protein. However, the regions involved in interaction with the γ-phosphate of GTP are quite different between TmYjeQ and ras protein. Because the γ-phosphate moiety of GTP is expected to interact with the G-2 and G-3 loops, large conformational changes of the switch I and II regions of TmYjeQ should follow during GTP hydrolysis, as shown in ras protein (18). Because the switch I region encompassing the G-2 loop involved in binding magnesium ion is flexible in the GDP-bound form of TmYjeQ, magnesium ion is not detected in the structure, unlike GDP-bound ras protein (18). The GDP binding without the magnesium ion has also been reported in eRF3 from Saccharomyces pombe (30).
Fig. 7.
A schematic drawing of the environmental residues around the guanine nucleotide. (A) GDP in the TmYjeQ GTPase domain. (B) GDP in ras protein (PDB ID code 4Q21). (C) Phosphoaminophosphonic acid-guanylate ester in ras protein (PDB ID code 1CTQ). The residues in the same position during substrates binding are yellow.
All of these biochemical data, together with the crystal structure, imply that the flexible switch I region of TmYjeQ must experience conformational changes to stimulate the GTPase activity during GTP binding and its hydrolysis, perhaps from the unstructured loop in GDP bound form to a structured loop in the GTP bound form.
In summary, the crystal structure of TmYjeQ clearly shows structural features that support and explain the biochemical data of an E. coli homologue (4, 5): GTPase activity, intrinsic RNA-binding ability, and the correlation between the two activities. The circular permutation in GTPase domain provides a structural basis for understanding the coupling of GTPase domain and the OB-fold domain, which is likely to be involved in binding of RNA in ribosome, affecting translation. Thus, the crystal structure of TmYjeQ provides a structural framework for understanding the functions of all members of this family.
Acknowledgments
We thank Barbara Gold for cloning, Marlene Henriquez and Bruno Martinez for expression studies and cell paste preparation, and John-Marc Chandonia for bioinformatics search of the gene. We also thank the staff at the Advanced Light Source, which is supported by the Director, Office of Science, Office of Basic Energy Sciences, Materials Sciences Division, of the U.S. Department of Energy, under Contract DE-AC03-76SF00098 at the Lawrence Berkeley National Laboratory. This work was supported by National Institutes of Health Grant GM 62412.
Abbreviations: TmYjeQ, the YjeQ protein from Thermotoga maritima; PDB, Protein Data Bank; OB fold, oligonucleotide/oligosaccharide-binding fold.
Data deposition: The atomic coordinates have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 1U0L).
References
- 1.Leipe, D. D., Wolf, Y. I., Koonin, E. V. & Aravind, L. (2002) J. Mol. Biol. 317, 41–72. [DOI] [PubMed] [Google Scholar]
- 2.Arigoni, F., Talabot, F., Peitsch, M., Edgerton, M. D., Meldrum, E., Allet, E., Fish, R., Jamotte, T., Curchod, M. L. & Loferer, H. (1998) Nat. Biotechnol. 16, 851–856. [DOI] [PubMed] [Google Scholar]
- 3.Sprang, S. R. (1997) Annu. Rev. Biochem. 66, 639–678. [DOI] [PubMed] [Google Scholar]
- 4.Daigle, D. M., Rossi, L., Berghuis, A. M., Aravind, L., Koonin, E. V. & Brown, E. D. (2002) Biochemistry 41, 11109–11117. [DOI] [PubMed] [Google Scholar]
- 5.Daigle, D. M. & Brown, E. D. (2004) J. Bacteriol. 186, 1381–1387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Aslanidis, C. & de Jong, P.J. (1990) Nucleic Acids Res. 20, 6069–6074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kim, R., Sandler, S. J., Goldman, S., Yokota, H., Clark, A. J. & Kim, S.-H. (1998) Biotech. Lett. 20, 207–210. [Google Scholar]
- 8.Leahy, D. J., Hendrickson, W. A., Aukhil, I. & Erickson, H. P. (1992) Science 258, 987–991. [DOI] [PubMed] [Google Scholar]
- 9.Jancarik, J. & Kim, S.-H. (1991) J. Appl. Crystallogr. 24, 409–411. [Google Scholar]
- 10.Otwinowski, Z. & Minor, W. (1997) Methods Enzymol. 276, 307–326. [DOI] [PubMed] [Google Scholar]
- 11.Terwilliger, T. C. & Berendzen, J. (1999) Acta Crystallogr. D 55, 849–861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jones, A. & Kleywegt, G. (1997) Methods Enzymol. 277, 173–208. [DOI] [PubMed] [Google Scholar]
- 13.Brunger, A. T., Adams, P. D., Clore, G. M., DeLano, W. L., Gros, P., Grosse-Kunstleve, R. W., Jiang, J. S., Kuszewski, J., Nilges, M., Pannu, N. S., et al. (1998) Acta Crystallogr. D 54, 905–921. [DOI] [PubMed] [Google Scholar]
- 14.Laskowski, R. A., MacArthur, M. W., Moss, D. S. & Thornton, J. M. (1993) J. Appl. Crystallogr. 26, 283–291. [Google Scholar]
- 15.Richardson, J. S. (1981) Adv. Protein Chem. 34, 167–339. [DOI] [PubMed] [Google Scholar]
- 16.Zhang, C. & Kim, S.-H. (2000) J. Mol. Biol. 299, 1075–1089. [DOI] [PubMed] [Google Scholar]
- 17.Tong, L. A., de Vos, A. M., Milburn, M. V., Jancarik, J., Noguchi, S., Nishimura, S., Miura, K., Ohtsuka, E. & Kim, S.-H. (1989) Nature 337, 90–93. [DOI] [PubMed] [Google Scholar]
- 18.Milburn, M. V., Tong, L., deVos, A. M., Brunger, A., Yamaizumi, Z., Nishimura, S. & Kim, S.-H. (1990) Science 247, 939–945. [DOI] [PubMed] [Google Scholar]
- 19.Brunger, A.T., Milburn, M. V., Tong, L., deVos, A. M., Jancarik, J., Yamaizumi, Z., Nishimura, S., Ohtsuka, E. & Kim, S.-H. (1990) Proc. Natl. Acad. Sci. USA 87, 4849–4853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kim, S.-H., Privé, G. G. & Milburn, M. V. (1993) in Handbook of Experimental Pharmacology, eds. Dickey, B. F. & Birmbauer, L. (Springer, Berlin) Vol. 108/I, pp. 177–194. [Google Scholar]
- 21.Jung, J. & Lee, B. (2001) Protein Sci. 10, 1881–1886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Theobald, D. L., Mitton-Fry, R. M. & Wuttke, D.S. (2003) Annu. Rev. Biophys. Biomol. Struct. 32, 115–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shin, D. H., Nguyen, H. H., Jancarik, J., Yokota, H., Kim, R. & Kim, S.-H. (2003) Biochemistry 42, 13429–13437. [DOI] [PubMed] [Google Scholar]
- 24.Holm, L. & Sander, C. (1997) Nucleic Acids Res. 25, 231–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Krishna, S. S., Majumdar, I. & Grishin N. V. (2003) Nucleic Acids Res. 31, 532–550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shin, D. H., Roberts, A., Jancarik, J., Yokota, H., Kim, R., Wemmer, D. E. & Kim, S. H. (2003) Protein Sci. 12, 1464–1472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang, W., Cho, H. S., Kim, R., Jancarik, J., Yokota, H., Nguyen, H. H., Grigoriev, I. V., Wemmer, D. E. & Kim S.-H. (2002) J. Mol. Biol. 319, 421–431. [DOI] [PubMed] [Google Scholar]
- 28.Shin, D. H., Brandsen, J., Jancarik, J., Yokota, H., Kim, R. & Kim, S.-H. (2004) J. Mol. Biol. 341, 227–239. [DOI] [PubMed] [Google Scholar]
- 29.Vestergaard, B., Van, L. B., Andersen, G. R., Nyborg, J., Buckingham, R. H. & Kjeldgaard, M. (2001) Mol. Cell 8, 1375–1382. [DOI] [PubMed] [Google Scholar]
- 30.Kong, C., Ito, K., Walsh, M. A., Wada, M., Liu, Y., Kumar, S., Barford, D., Nakamura, Y. & Song, H. (2004) Mol. Cell 14, 233–245. [DOI] [PubMed] [Google Scholar]
- 31.Kraulis, P. J. (1991) J. Appl. Crystallogr. 24, 946–950. [Google Scholar]
- 32.Nicholls, A. (1991) Proteins 11, 281–296. [DOI] [PubMed] [Google Scholar]