Abstract
Glutaredoxin (Grx) belongs to the thioredoxin fold superfamily and catalyzes glutathione-dependent oxidoreductions. The recently discovered mitochondrial and nuclear Grx (Grx2) differs from the more abundant cytosolic Grx (Grx1) by its higher affinity toward S-glutathionylated proteins and by being a substrate for thioredoxin reductase. Here, we have successfully established a method to silence the expression of Grx2 in HeLa cells by using short interfering RNA to study its role in the cell. Cells with levels of Grx2 <3% of the control were dramatically sensitized to cell death induced by doxorubicin/adriamycin and phenylarsine oxide but did not show signs of a general increase in oxidative damage with respect to carbonylation and glutathionylation. The ED50 for doxorubicin dropped from 40 to 0.7 μM and for phenylarsine oxide from 200 to 5 nM. However, no differences were detected after treatment with cadmium, a known inhibitor of Grx1. These results indicate a crucial role of Grx2 in the regulation of the mitochondrial redox status and regulation of cell death at the mitochondrial checkpoint.
The cellular thiol homeostasis is maintained by the thioredoxin (Trx) and the glutaredoxin (Grx) systems. Trx and Grx belong to a family of thiol disulfide oxidoreductases that share a common fold, known as the Trx-fold, and a common active-site motif, Cys-X-X-Cys (1). Trx is kept in the reduced state by the seleno-enzyme Trx reductase (TrxR) with electrons from NADPH. Oxidized Grx is reduced by two molecules of glutathione (GSH), yielding oxidized glutathione (GSSG). GSSG is subsequently reduced by glutathione reductase, also with NADPH as electron donor. Trxs and Grxs catalyze protein disulfide reductions by a dithiol mechanism involving both active-site thiols. In addition, Grxs catalyze reactions by a monothiol mechanism, requiring only the more N-terminal active-site cysteine. This mechanism is a prerequisite for the unique ability of Grxs to reduce disulfides formed between protein thiols and GSH (deglutathionylation) or to participate in their formation (glutathionylation) (2).
Trxs and Grxs are ubiquitous proteins present in a large number of isoforms in different species ranging from viruses to higher eukaryotes (3). Two Trx and two Grx systems have been identified so far in mammalian cells. Trx1 and TrxR1 are located in the cytosol, whereas Trx2 and TrxR2 are targeted to mitochondria (4, 5). These systems cover a wide range of functions (6), and knockout of either Trx1 or Trx2 causes embryonic lethality in mice (7, 8). Grx1 is localized in the cytosol and is involved in a series of biological processes, i.e., ribonucleotide reduction (9), dehydroascorbate reduction (10), cellular differentiation (11), regulation of transcription factor activity (12–14), Cd-induced apoptosis (15), and recovery from oxidative stress (16, 17). The second human dithiol Grx (Grx2) is targeted to either mitochondria or nuclei by an alternative splicing mechanism (18, 19). Apart from different import sequences, encoded by alternative first exons, the isoforms share a common 125-residue Grx domain encoded by exons II–IV. Human Grx2 differs significantly from other Grxs. It is slightly larger, 16 kDa (processed) instead of 9–12 kDa and contains, in contrast to the Grx consensus sequence Cys-Pro-Tyr-Cys, a serine instead of the proline in its active site. This difference, however, is responsible for a remarkably high affinity of Grx2 toward glutathionylated substrates, in particular protein GSH disulfides (20). Furthermore, Grx2 can receive electrons from TrxR, supporting both the dithiol and the Grx-specific monothiol mechanisms (20). This unique electron pathway could be of major importance during stress conditions. Under these conditions the [GSH]/[GSSG] ratio is decreased and/or the intracellular pH falls, and thus, the active site in Grxs cannot be reduced by GSH (9, 21). These findings in vitro suggested an important role for Grx2 in protection and recovery of cells from oxidative stress. Therefore, the aim of this study was to establish a method to silence the expression of Grx2 in a human cell line by using short interfering RNA (siRNA) to study its role in the cell.
Experimental Procedures
General Materials. Disposable cell culture materials were purchased from Techno Plastic Products (Trasadingen, Switzerland). Chemicals were purchased from Sigma, unless otherwise stated, and were of analytical grade or better.
Cell Cultures. HeLa cells were cultivated in DMEM (Invitrogen) supplemented with 10% heat inactivated FBS (PAA, Pasching, Austria), 2 mM glutamine (PAA), and 100 units·ml–1 penicillin/streptomycin (PAA), unless otherwise stated. The cells were incubated at 37°C in a 90% humidified atmosphere containing 5% CO2.
Probe Preparation. Cells for ELISA, Western blotting, and enzymatic assays were harvested by trypsinization and centrifugation in a microcentrifuge (1,600 × g for 10 min). The supernatant was removed, and the cells were lysed in 50 μl of lysis buffer [10 mM Tris, pH 7.5/10 mM NaCl/3 mM MgCl2/0.1% Nonidet P-40/1× fold complete protease inhibitor mixture (Roche Applied Science)]. After a 15-min incubation at room temperature, the samples were quickly frozen. After thawing, the samples were centrifuged (30,000 × g for 3 min), and the protein content in the supernatant was measured against a known BSA standard (0.01–0.8 mg·ml–1) at 595 nm by using Bradford reagent (BioRad) in 96-well plates.
Western Blotting. SDS/PAGE was run by using Ready-Gels and the Protean 3 Cell (Bio-Rad) according to the manufacturer's instructions. Proteins from SDS gels (8–16% gradients) were transferred to TransBlot transfer membranes (Bio-Rad) by wet transfer in 10 mM Mops (pH 9.0), 10% methanol, and 0.1% SDS at 100 V for 2 h. The membrane was blocked in PBST (137 mM NaCl/3 mM KCl/6.5 mM Na2HPO4/1.5 mM KH2PO4/0.5% Tween) containing 1% BSA and 5% nonfat dry milk powder (Bio-Rad). Rabbit anti-Grx2 Abs (C. Johansson and A.H., unpublished results) were used at a dilution of 1:2,000, and the mouse mAb specific for GSH moieties on proteins (101-A, Virogen, Watertown, MA) at 1:500 dilution. Peroxidase-coupled anti-rabbit or anti-mouse Abs (Dako) were used to identify antigen Ab conjugates. Ab binding was measured by using Western Lightning Chemiluminescence Reagent Plus (PerkinElmer) according to the manufacturer's instructions.
ELISA. Quantification of Grx2 and Grx1 was done by using specific sandwich ELISAs (22). In brief, 96-well plates were coated with affinity-purified Abs (3 μg·ml–1 for Grx1 and 2 μg ml–1 for Grx2) overnight at 4°C. After blocking with 10 mg·ml–1 BSA, standards of 0.05–50 ng·ml–1 and diluted cell extracts were allowed to react with the Abs overnight at 4°C. The conjugates were identified by using affinity-purified biotinylated Abs and alkaline phosphatase-conjugated streptavidine (Dako). Quantification was done after staining with P-nitrophenyl phosphate (Sigma) at 405 nm in a multiplate reader (VersaMax, Molecular Devices).
Immunocytochemistry. Cells were seeded on glass plates to a density of 20–40% 7 h before fixation with 4% formaldehyde at room temperature for 30 min. Mitochondria were stained with 50 nM MitoTracker red (Molecular Probes) at 37°C for 30 min before fixation. Anti-Grx2 Abs were preincubated for 2 h at room temperature at 1:10 dilution with 0.5 mg of BSA in 10 μl of PBS containing 0.3% Triton X-100. The Abs were diluted 1:50 and incubated with the fixed cells in a moisture chamber at 4°C overnight. After washing with PBS 3 × 5 min, cells were incubated with the secondary Ab (1:250, Alexa Fluor 488 goat anti-rabbit IgG, Molecular Probes) for 60 min at room temperature. After 3 × 5min washing with PBS, the slides were mounted and analyzed by using the fluorescence microscope Olympus BX60. Images were collected with a C4742–95-10SC digital camera (Hamamatsu Photonics, Hamamatsu City, Japan).
siRNA. siRNAs were produced by in vitro transcription by using the Silencer siRNA Construction kit (Ambion). The following oligonucleotides were designed encoding the desired siRNA strands: target sequence 1 (TS1), AAGCATCCGCCTATACAATCTCC TGTCTC (sense) and AAAGATTGTATAGGCGGATGCCCTGTCTC (antisense); TS15, AACAGTTCCACCACTTTATAGCCTGTCTC (sense) and AACTATAAAGTGGTGG AACTGCCTGTCTC (antisense); and TS28, AATGAAGCCTATGAGTGTCAGCCTGTCTC (sense) and AACTGACACTCATAGGCTTCACCTGTCTC (antisense). As control an unspecific “scrambled” (scr) siRNA was designed by using the following oligonucleotides: scr, AACATTCACTCAGGTCATCAGCCTGTCTC (sense) and AACTGATGACCTG AGTGAATGCCTGTCTC (antisense). Before transfection (1 day), cells were seeded in six-well plates and transfection was done at a cell confluency of 50–70% by using Oligofectamin (Invitrogen) according to the manufacturer's protocol. Briefly, 20 nM of the 21-nt double-stranded RNA and 12 μl of Oligofectamin was added to supplemented DMEM to yield a total volume of 2.5 ml. After 6 h of incubation, an equal volume of medium was added to the cells. The diluted transfection mix was removed 24 h after transfection. The cells were washed twice with PBS and cultured in supplemented DMEM, which was changed twice per day. Cells were harvested at different time points after transfection, and the amount of target protein was analyzed by ELISA to confirm knockdown.
Viability Assays. Cell viability was determined by using the Cell Proliferation kit II (Roche Applied Science, Mannheim, Germany). In brief, cells were seeded in 96-well plates to a density of 50%, 24 h before treatment. The cells were washed with PBS and incubated with various substances in 150 μl of medium for 24 h. The amount of viable cells was analyzed in a microplate reader at 490 nm. As additional control, single measurements were done by using the accumulation of neutral red in living cells but not in dead cells (23). After 24 h of treatment, cell layers were incubated with supplemented DMEM containing 50 μg·ml–1 neutral red for 3 h to allow accumulation. The stained cells were washed three times with PBS and lysed by using 50% ethanol and 1% acetic acid. The resulting lysates were analyzed spectrophotometrically at 524 nm in a spectrophotometer. Untreated cells and empty vials served as 100% and 0% standards for both methods.
Carbonylation Assay. Protein carbonyl content was determined as described in ref. 24. The protein concentrations of the cell extracts were adjusted to 1 μg·μl–1 before incubation with 3 vol of derivatization solution (10 mM 2,4-dinitrophenylhydrazine/6 M guanidine-HCl/0.5 M potassium phosphate, pH 2.5). After 30 min at room temperature, the probes were neutralized with 2 M Tris (base) and applied to a Bio-Dot microfiltration apparatus (Bio-Rad) equipped with a TransBlot transfer membrane (BioRad) soaked in PBS. After blotting, the membrane was washed five times with PBS and blocked with PBS containing 1% BSA and 5% nonfat dry milk powder (Bio-Rad) and thereafter incubated overnight with 1:1,000 diluted antidinitrophenyl Abs (Dako) in PBST at 4°C. Alkaline phosphatase-conjugated anti-rabbit Abs (Dako) were used to identify antigen Ab conjugates, and nitroblue tetrazulium chloride/5-bromo-4-chloro-3-indolylphosphate 4-toludine (Roche) was used for staining.
Results
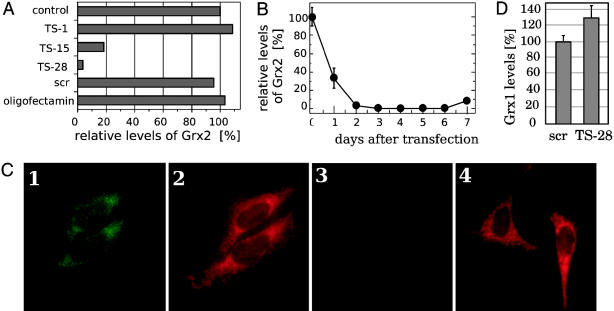
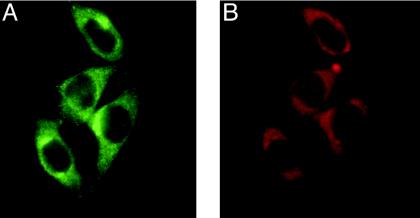
siRNA-Mediated Knockdown of Human Grx2 in Cultured Cells. siRNA duplexes provide a tool for the study of gene functions in vivo. We have used in vitro-transcribed siRNAs specific for the mRNA of human Grx2 for the transient transfection of HeLa cells. After transfection (4 days), the cells were assayed for Grx2 levels by ELISA. Of three tested siRNA target sequences (siRNA-TS), two were effective in decreasing protein levels, whereas one had no effect (Fig. 1 A and B). Neither the transfection procedure itself, nor the transfection of HeLa cells with a control of unspecific siRNA (siRNA-scr), had an effect on Grx2 levels (Fig. 1 A). siRNA-TS28 was found to be the most effective, and was used for all subsequent experiments. The kinetics of the decrease in Grx2 levels by siRNA-TS28 are shown in Fig. 1B. At 4 days after transfection, the protein levels were below the ELISA detection limit (0.05 ng·ml–1), corresponding to levels of <3% of the initial values (≈3ng·mg–1). The level stayed low until day 5 and increased again on day 6. On day 7 the protein level was back to ≈30% of the initial value. In addition, immunocytochemistry was used to confirm specific loss of the protein upon transfection with siRNA-TS28. As shown in Fig. 2, Grx2-specific staining in untreated cells costains well with MitoTracker red staining, indicating that Grx2 is located mainly in mitochondria in HeLa cells. Fig. 1C, images 1 and 3, demonstrates the specific loss of Grx2 in siRNA-TS28, but not in siRNA-scr-transfected cells. As a consequence, all subsequent experiments were performed between days 3.5 and 5.5 by using siRNA-TS28.
Fig. 1.
Specific knockdown of Grx2. (A) Levels of Grx2 in cells treated with different siRNAs specific for Grx2 (TS1, TS15, and TS28). Untreated HeLa cells (control), cells transfected with an unspecific siRNA (scr), and cells treated with only transfectant (oligofectamin) were used as controls. The levels were determined by ELISA; 100% corresponds to 2.9 mg·ml–1.(B) Kinetic of the decline in Grx2 levels transfected with siRNA-TS28 as determined by ELISA. (C) Immunocytochemical detection of Grx2 in HeLa cells (images 1 and 3) and mitochondrial staining with MitoTracker red (images 2 and 4). Images 1 and 2 show HeLa transfected with the unspecific siRNA-scr. Images 3 and 4 show cells transfected with the Grx2-specific siRNA-TS28. (D) Relative levels of Grx1 in cells treated with siRNA-scr and siRNA-TS28; 100% Grx1 corresponds to 80 ng·mg–1.
Fig. 2.
Mitochondrial localization of Grx2 in HeLa cells. (A) Immunocytochemical detection of Grx2 in HeLa cells. (B) Mitochondrial staining with MitoTracker red.
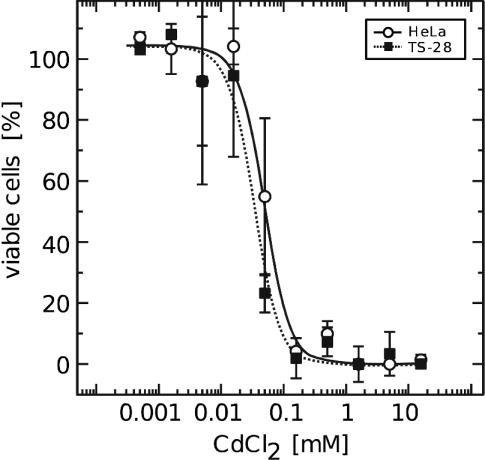
The lack of Grx2 did not negatively influence the viability of the cells when propagated under optimal conditions, including frequent exchange of the growth medium. These medium changes prevent accumulation of oxidized proteins in the cell, for instance accumulation of oxidized Trx1 in cells propagated in old medium (A.H., unpublished results). The knockdown of Grx2 did not significantly alter the levels of cytosolic Grx1 (Fig. 1D), and even when the cells were treated with Cd, a known inhibitor of Grx1, no significant difference in viability was detected (Fig. 3).
Fig. 3.
Viability of untreated and Grx2 knockdown HeLa cells after treatment with Cd. Cell viability was determined by using Roche's cell proliferation and viability kit II. The cells were seeded to a density of 50% in 96-well plates (≈4 × 103 cells per well) 24 h before treatment with Cd for 24 h. The curves represent the averages of four different experiments. ○ and straight lines, untreated HeLa cells; ▪ and dotted lines, HeLa cells at 4 days after transfection with the human Grx2-specific siRNA-TS28.
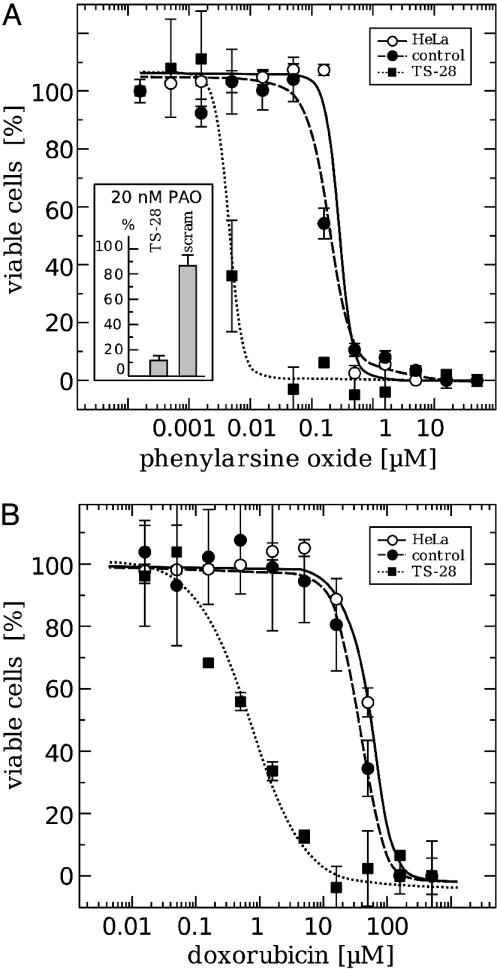
Sensitivity Toward Phenylarsine Oxide (PAO) and Doxorubicin (Dox). PAO targets vicinal dithiols and thereby both the Trx and Grx systems (25). Dox is an anticancer drug known to intercalate with DNA, but also induces reactive oxygen species (ROS) (26, 27) by using electrons from mitochondrial complex I (28). Here we have analyzed how the lack of Grx2 affects the cell viability when challenged with these compounds (Fig. 4). Cells were treated at 4 days after transfection with different concentrations of the compounds for 24 h and were subsequently analyzed for viability. Cells lacking Grx2 showed a highly increased sensitivity toward both PAO and Dox (Fig. 4). The effective dose, defined as the concentration yielding 50% viability (ED50), decreased for PAO from ≈200 nM to 5 nM and for Dox from 40 μMto0.7 μM when cells were depleted of Grx2.
Fig. 4.
Viability of untreated, control, and Grx2 knockdown HeLa cells after treatment with Dox (A) and PAO (B). Cell viability was determined by using Roche's cell proliferation and viability kit II, and the results were confirmed by the analysis of neutral red accumulation in the cells (Inset). The cells were seeded to a density of 50% in 96-well plates (≈4 × 103 cells per well) 24 h before treatment with Dox or PAO for another 24 h. The curves represent the averages of four different experiments. ○ and straight lines, untreated HeLa cells. HeLa cells 4 days after transfection with an unspecific siRNA (• and dashed lines). HeLa cells 4 days after transfection with the human Grx2-specific siRNA-TS28 (▪ and dotted lines).
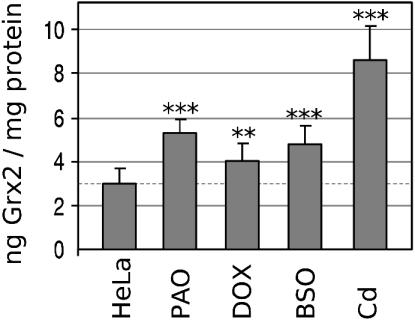
Grx2 Levels in Response to Treatment with Cd, PAO, and Dox. The decreased viability of Grx2–cells upon Dox and PAO treatment prompted us to investigate whether the endogenous levels of Grx2 in untransfected cells are changed, and therefore of importance, in the natural defense against these compounds. HeLa cells were treated with 0.1 nM PAO, 10 μM Dox, 20 μM Cd, or 100 μM buthionine sulfoximine (BSO). We have included BSO, an inhibitor of GSH synthesis, to investigate the changes in Grx2 levels upon depletion of its electron donor. The cells were treated for 24 h before measurement of Grx2 levels. As shown in Fig. 5, treatment with all of these compounds led to a small but significant increase in cellular Grx2 concentration. Untreated cells exhibited a Grx2 level of 2.9 ± 0.7 ng/mg total protein. Cd treatment increased this value 2.9-fold to 8.6 ± 1.5 ng·mg–1, treatment with PAO 1.8-fold to 5.4 ± 0.6 ng·mg–1, Dox treatment 1.4-fold to 4.1 ± 0.7 ng·mg–1, and the depletion of GSH by BSO increased the Grx2 level 1.6-fold to 4.8 ± 0.8 ng mg–1.
Fig. 5.
Levels of Grx2 in HeLa cells after treatment with PAO, Dox, BSO, and Cd. HeLa cells at a density of 50–70% were treated for 24 h with 0.1 nM PAO, 10 μM Dox, 100 μM BSO, or 20 μM Cd. The levels of Grx2 were determined by ELISA. The SDs are indicated. Significance of the difference between the means of the control group and the individual test group was calculated by using Student's t test. The following number of experiments were performed: n = 12, control group; n = 3, PAO, Dox, BSO, and Cd. No, P > 0.05; *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.005.
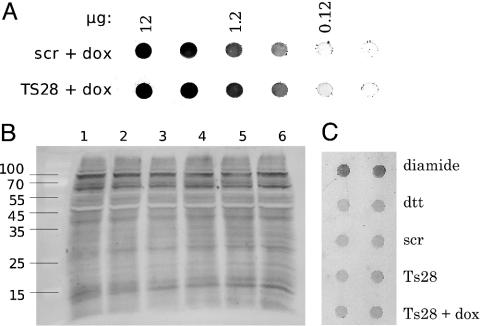
Oxidative Stress. Oxidative stress, defined as the amount of damage induced by ROS and other free radical species, leads to oxidative modifications of biomolecules. One such modification is the introduction of carbonyl groups to proteins. Analysis of the carbonyl content of proteins in cells lacking Grx2, compared with untreated and control cells transfected with the unspecific siRNA-scr, showed no significant difference (data not shown). In addition, cells lacking Grx2 were treated with Dox, a drug known to induce free radicals in mitochondria, for 1 or 24 h. These cells exhibited a slightly increased protein carbonyl content compared with controls (Fig. 6A), but this effect did not reach the extent of carbonylation induced by peroxide treatment of HeLa cells (data not shown). Protein glutathionylation has been discussed to be a result of oxidative stress and a mechanism to protect protein thiols from more serious irreversible oxidations. The fact that Grxs are highly specific for the reduction of protein-GSH disulfides prompted us to analyze protein glutathionylation in vivo in our cells by using an Ab specific for GSH moieties on proteins. As shown in Fig. 6 B and C, the lack of Grx2 did not cause a detectable increase in protein glutathionylation, neither did treatment of the Grx2–cells with Dox.
Fig. 6.
Oxidative modifications of proteins in untreated, control, and Grx2 knockdown HeLa cells. Protein carbonylation (A) and glutathionylation (B and C) in HeLa cells, HeLa cells transfected with an unspecific siRNA (scr), and HeLa cells transfected with the human Grx2-specific siRNA-TS28 (TS28). (A) Dot blot of total cell extracts after treatment with 0.2 μM Dox for 24 h. (B) Western blot of cell extracts (40 μg per sample) by using antiglutathione moiety Abs specific for protein-mixed disulfides with glutathione. Lane 1, Untreated HeLa cells; lane 2, transfected with an unspecific siRNA; lane 3, lane 2 treated with 25 μM Dox for 1 h; lane 4, transfected with human Grx2-specific siRNA-TS28; lane 5, lane 4 treated with 25 μM Dox for 1 h, and lane 6, untreated HeLa cells. (C) Dot blot of total cell extracts (10 μg per sample). Treatment with 50 μM diamide for 1 h was used as positive control. The same extract treated with 10 mM DTT for 30 min was used as negative control. siRNA-TS28-transfected cells were additionally treated with 25 μM Dox for 1 h before harvesting.
Discussion
Oxidative stress caused by excess production of ROS or lack of antioxidant capacity affects virtually all cellular processes. Several studies have implied an importance of Grxs during oxidative conditions. To investigate the influence of Grx2 on cellular oxidative homeostasis and on the cells' ability to handle oxidative stress, Grx2 was knocked down by RNA interference. This form of posttranscriptional gene silencing is mediated by short double-stranded RNAs that induce highly sequence-specific degradation of the mRNA (29). Because of the specificity for mRNA containing the corresponding sequence, the utilization of this mechanism has become a powerful tool for the analysis of gene function in vivo (30). Here, we have established a method to lower the expression of Grx2 in HeLa cells by using transient transfection with short double-stranded RNA. The protein levels, determined by a highly sensitive ELISA, were decreased to levels <3% compared with untransfected and siRNA-scr-transfected control cells at day 3 after transfection. In addition, immunostaining of Grx2 was undetectable in Grx2–cells (Fig. 1). This result is in agreement with earlier observations for HeLa cells, where the maximum amount of silencing is usually seen ≈72 h after transfection (31). The remarkably high efficiency and the specificity of the knockdown allowed us to investigate how the lack of Grx2 may affect the cells. The decreased levels of Grx2 per se did not influence the proliferation rate of the cells nor their viability when propagated under optimal conditions. Also, the level of cytosolic Grx1 remained unaffected in the knockdown cells. However, our results suggest a prominent role of Grx2 during oxidative conditions, when the levels of free radicals, ROS, and GSSG in the cell are elevated.
There is increasing evidence that the formation of mixed disulfides of protein thiols with GSH is a key event in the redox regulation of cellular processes. Grxs are highly specific for the reduction of mixed disulfides between protein thiols and GSH (32, 33), suggesting a common mechanism of redox regulation with proteins being S-glutathionylated upon oxidation of the GSH pool and deglutathionylated by Grxs. Glutathionylation alters the biological function of several important processes and proteins such as actin polymerization (34), GAPDH (35), protein tyrosine phosphatase 1B (36), creatine kinase (37), c-Jun (38), NF-κB subunit p50 (39), caspase-3 (40), HIV protease (41), and cAMP-dependent protein kinase (42). In many cases Grxs have been shown to restore the biological activity (34–36, 41). Although Grx2 has a particular high affinity for protein GSH disulfides (20), we were unable to detect a general increase in total protein glutathionylation in the knockdown cells. This finding might have been expected because Grx2 is located mainly in mitochondria. However, our results do not exclude that Grx2 could be essential for the regulation of specific proteins by affecting the reversible formation of mixed disulfides with GSH. The anti:GSH-moiety Ab used in this study is not suitable for the detection of glutathionylation of low abundant proteins in cell extracts. The major source of reactive oxygen in the mitochondrial matrix is the NADH ubiquinone oxidoreductase/complex I. Two proteins on the matrix side of complex I have been reported to be subject to S-glutathionylation. Formation of these mixed disulfides upon oxidation of the mitochondrial GSH pool correlated with an increased ROS production that was reversed by reduction of the mixed disulfides (43). Other proteins of the energy metabolism in mitochondria show alterations in response to the GSH status as well. The Krebs cycle enzyme α-ketoglutarate dehydrogenase was shown to be inactivated upon treatment of mitochondria with H2O2, leading to a decline in the rate of state 3 NADH-linked respiration (44). Restoration of the activity by the cytosolic Grx1, but not Trx, indicated that glutathionylation modulated the enzymatic activity (45). These findings suggest a potential role for Grx2 in the regulation of respiration and ROS levels in response to the redox status by reversible glutathionylation. Grx2 has also been shown to reduce small molecular weight compounds like dehydroascorbate and CoA disulfides (20), and this activity might add to its detoxifying potential.
Grxs have been implied in the regulation of cell death in previous studies. Acute Cd exposure of cells leads to inhibition of Grx1 and initiates apoptosis (15). Grx1 also protects cells from apotosis induced by hydrogen peroxide by regulating the redox state of Akt (46). Escherichia coli Grx2 and human Grx1 have been shown to protect cerebellular granulae neurons from dopamine-induced apoptosis, involving the Ras-phosphoinositide 3-kinase and Jun N-terminal kinase pathways (47, 48). On the other hand, apoptosis signal-regulating kinase 1 is inactive when bound to reduced Grx1 (49), and oxidation of Grx1 causes release of apoptosis signal-regulating kinase 1 and hence initiation of apoptosis. Therefore, we were interested in whether knockdown of Grx2 could influence the cell viability after treatment with various substances. The Grx2 knockdown cells were highly sensitized to treatment with PAO, with a 40-fold drop in ED50. In addition, Grx2 was 1.8-fold up-regulated in response to a low-dose PAO treatment in untransfected cells. PAO complexes vicinal dithiols and targets thereby both the Trx and the Grx systems. PAO also gives rise to the formation of high levels of superoxide and hydrogen peroxide and induces apoptosis (25, 50). As mentioned, Cd is an inhibitor for Grx1 and an inducer of programmed cell death (15), but in contrast to PAO, cells with silenced expression of Grx2 did not show increased cell death when treated with Cd. Because PAO binds to the selenolthiol group of TrxR more strongly than it does to dithiol groups (57), it will most likely lead to the loss of the Trx systems. Cells lacking Grx2 will, in that case, not be able to complement the loss of one system with the other. The up-regulation of Grx2 after Cd treatment indicates that Grx2, as Grx1, might also be inhibited by the heavy metal, but it does not seem to be a target for its cell toxic effects.
Dox is an important anticancer drug used in the therapy of human tumors. Its therapeutic effect is based on direct DNA damage and disturbed DNA repair. Its use in therapy is, however, limited by a potentially lethal cardiomyopathy (51). Dox has a quinone-like structure that can be reduced to a semiquinone free radical (26). This radical can be transferred to thiols or molecular oxygen, subsequently causing oxidative modifications of nucleic acids, lipids, and proteins. These modifications, in turn, may lead to DNA cleavage and inhibition of mitochondrial bioenergetics, thus causing apoptosis (52). Treatment with antioxidants like N-acetylcysteine protects cells from cardiomyopathy (53) and attenuation of toxicity has been detected in cells overexpressing manganese superoxide dismutase, Trx1, or Grx1 (54–56). Here, we have shown that silencing of Grx2 expression strongly amplifies the sensitivity of HeLa cells toward Dox, with the ED50 dropping ≈60-fold from 40 to 0.7 μM. The question about the mechanism of the increase in sensitivity toward PAO and Dox remains to be investigated, but the finding clearly demonstrates that Grx2 is a crucial part of the cellular defense system against free radicals and redox-regulation of apoptosis. Moreover, overexpression of Grx2 prevents from induction of mitochondria-apoptosis by various substances including Dox (M.E., unpublished data).
In conclusion, we have developed a sensitive and efficient siRNA method for knockdown of Grx2. In untreated HeLa cells, this knockdown does not significantly alter the overall levels of carbonylation or glutathionylation. However, knock-down caused a remarkable increase in sensitivity toward cell death induced by Dox and PAO. Modification of sensitive thiol groups in the cell is sufficient to initiate cell death, and our results suggest that Grx2 is required to restore the redox status of one or more specific target to prevent or modulate cell death responses. Grx2 clearly represents a potential drug target. How Grx2 modulates the mitochondrial response to oxidative stress has to be answered next.
Acknowledgments
We thank Lena Ringdén for excellent secretarial work. This investigation was supported by grants from the Knut and Alice Wallenberg Foundation, the Swedish Cancer Society (961 and 4648-BO2-01VA), and the Wenner-Gren Foundation.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: BSO, buthionine sulfoximine; Dox, doxorubicin; Grx, glutaredoxin; PAO, phenylarsine oxide; ROS, reactive oxygen species; scr, scrambled, siRNA, short interfering RNA; Trx, thioredoxin; TrxR, Trx reductase; TS, target sequence; GSH, glutathione; GSSG, oxidized GSH.
References
- 1.Holmgren, A. (1989) J. Biol. Chem. 264, 13963–13966. [PubMed] [Google Scholar]
- 2.Fernandes, A. P. & Holmgren, A. (2004) Antioxid. Redox Signal. 6, 63–74. [DOI] [PubMed] [Google Scholar]
- 3.Vlamis-Gardikas, A. & Holmgren, A. (2002) Methods Enzymol. 347, 286–296. [DOI] [PubMed] [Google Scholar]
- 4.Spyrou, G., Enmark, E., Miranda-Vizuete, A. & Gustafsson, J. (1997) J. Biol. Chem. 272, 2936–2941. [DOI] [PubMed] [Google Scholar]
- 5.Miranda-Vizuete, A., Damdimopoulos, A. E., Pedrajas, J. R., Gustafsson, J. A. & Spyrou, G. (1999) Eur. J. Biochem. 261, 405–412. [DOI] [PubMed] [Google Scholar]
- 6.Arnér, E. S. J. & Holmgren, A. (2000) Eur. J. Biochem. 267, 6102–6109. [DOI] [PubMed] [Google Scholar]
- 7.Matsui, M., Oshima, M., Oshima, H., Takaku, K., Maruyama, T., Yodoi, J. & Taketo, M. M. (1996) Dev. Biol. 178, 179–185. [DOI] [PubMed] [Google Scholar]
- 8.Nonn, L., Williams, R. R., Erickson, R. P. & Powis, G. (2003) Mol. Cell. Biol. 23, 916–922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Holmgren, A. (1979) J. Biol. Chem. 254, 3672–3678. [PubMed] [Google Scholar]
- 10.Wells, W. W., Xu, D. P., Yang, Y. F. & Rocque, P. A. (1990) J. Biol. Chem. 265, 15361–15364. [PubMed] [Google Scholar]
- 11.Takashima, Y., Hirota, K., Nakamura, H., Nakamura, T., Akiyama, K., Cheng, F. S., Maeda, M. & Yodoi, J. (1999) Immunol. Lett. 68, 397–401. [DOI] [PubMed] [Google Scholar]
- 12.Nakamura, T., Ohno, T., Hirota, K., Nishiyama, A., Nakamura, H., Wada, H. & Yodoi, J. (1999) Free Radical Res. 31, 357–365. [DOI] [PubMed] [Google Scholar]
- 13.Bandyopadhyay, S., Starke, D. W., Mieyal, J. J. & Gronostajski, R. M. (1998) J. Biol. Chem. 273, 392–397. [DOI] [PubMed] [Google Scholar]
- 14.Hirota, K., Matsui, M., Murata, M., Takashima, Y., Cheng, F. S., Itoh, T., Fukuda, K. & Yodoi, J. (2000) Biochem. Biophys. Res. Commun. 274, 177–182. [DOI] [PubMed] [Google Scholar]
- 15.Chrestensen, C. A., Starke, D. W. & Mieyal, J. J. (2000) J. Biol. Chem. 275, 26556–26565. [DOI] [PubMed] [Google Scholar]
- 16.Wang, G. M., Raghavachari, N. & Lou, M. F. (1997) Exp. Eye Res. 64, 693–700. [DOI] [PubMed] [Google Scholar]
- 17.Xing, K. Y. & Lou, M. F. (2002) Exp. Eye Res. 74, 113–122. [DOI] [PubMed] [Google Scholar]
- 18.Lundberg, M., Johansson, C., Chandra, J., Enoksson, M., Jacobsson, G., Ljung, J., Johansson, M. & Holmgren, A. (2001) J. Biol. Chem. 276, 26269–26275. [DOI] [PubMed] [Google Scholar]
- 19.Gladyshev, V. N., Liu, A., Novoselov, S. V., Krysan, K., Sun, Q. A., Kryukov, V. M., Kryukov, G. V. & Lou, M. F. (2001) J. Biol. Chem. 276, 30374–30380. [DOI] [PubMed] [Google Scholar]
- 20.Johansson, C., Lillig, C. H. & Holmgren A. (2004) J. Biol. Chem. 279, 7537–7543. [DOI] [PubMed] [Google Scholar]
- 21.Åslund, F., Berndt, K. D. & Holmgren, A. (1997) J. Biol. Chem. 272, 30780–30786. [DOI] [PubMed] [Google Scholar]
- 22.Lundberg, M., Fernandes, A. P., Kumar, S. & Holmgren, A. (2004) Biochem. Biophys. Res. Commun. 319, 801–809. [DOI] [PubMed] [Google Scholar]
- 23.Lindl, T. (2002) in Cell and Tissue Culture (Spectrum Akademischer, Heidelberg), pp. 238–239.
- 24.Levine, R. L., Williams, J. A., Stadtman, E. R. & Shacter, E. (1994) Methods Enzymol. 233, 346–357. [DOI] [PubMed] [Google Scholar]
- 25.Pallis, M., Bradshaw, T. D., Westwell, A. D., Grundy, M., Stevens, M. F. & Russell, N. (2003) Biochem. Pharmacol. 66, 1695–1705. [DOI] [PubMed] [Google Scholar]
- 26.Bachur, N. R., Gordon, S. L. & Gee, M. V. (1977) Mol. Pharmacol. 13, 901–910. [PubMed] [Google Scholar]
- 27.Gutierrez, P. L., Gee, M. V. & Bachur, N. R. (1983) Arch. Biochem. Biophys. 223, 68–75. [DOI] [PubMed] [Google Scholar]
- 28.Davies, K. J., Doroshow, J. H. & Hochstein, P. (1983) FEBS Lett. 153, 227–230. [DOI] [PubMed] [Google Scholar]
- 29.Tuschl, T. (2001) Chem. Biochem. 2, 239–245. [Google Scholar]
- 30.Dykxhorn, D. M., Novina, C. D. & Sharp, P. A. (2003) Nat. Rev. Mol. Cell Biol. 4, 457–467. [DOI] [PubMed] [Google Scholar]
- 31.Elbashir, S. M., Harborth, J., Weber, K. & Tuschl, T. (2002) Methods 26, 199–213. [DOI] [PubMed] [Google Scholar]
- 32.Gravina, S. A. & Mieyal, J. J. (1993) Biochemistry 32, 3368–3376. [DOI] [PubMed] [Google Scholar]
- 33.Yoshitake, S., Nanri, H., Fernando, M. R. & Minakami, S. (1994) J. Biochem. 116, 42–46. [DOI] [PubMed] [Google Scholar]
- 34.Wang, J., Boja, E. S., Tan, W., Tekle, E., Fales, H. M., English, S., Mieyal, J. J. & Chock, P. B. (2001) J. Biol. Chem. 276, 47763–47766. [DOI] [PubMed] [Google Scholar]
- 35.Lind, C., Gerdes, R., Schuppe-Koistinen, I. & Cotgreave, I. A. (1998) Biochem. Biophys. Res. Commun. 247, 481–486. [DOI] [PubMed] [Google Scholar]
- 36.Barett, W. C., DeGnore, J. P., König, S., Fales, H. M., Keng, Y. F., Zhang, Z. Y., Yim, M. B. & Chock, P. B. (1999) Biochemistry 38, 6699–6705. [DOI] [PubMed] [Google Scholar]
- 37.Reddy, S., Jones, A. D., Cross, C. E., Wong, P. S. Y. & van der Vliet, A. (2000) Biochem. J. 347, 824–827. [PMC free article] [PubMed] [Google Scholar]
- 38.Klatt, P., Pineda-Molina, E., de Lacoba, M. G., Padilla, A. C., Martinez-Galisteo, E., Barzena, J. A. & Lamas, S. (1999) FASEB J. 13, 1481–1490. [DOI] [PubMed] [Google Scholar]
- 39.Pineda-Molina, E., Klatt, P., Vazquez, J., Martina, A., de Lacoba, M. G., Perez-Sala, D. & Lamas, S. (2001) Biochemistry 40, 14134–14142. [DOI] [PubMed] [Google Scholar]
- 40.Zech, B., Wilm, M., van Eldik, R. & Brühne, B. (1999) J. Biol. Chem. 274, 20931–20936. [DOI] [PubMed] [Google Scholar]
- 41.Davis, D. A., Newcomb, F. M., Starke, D. W., Ott, D. E., Mieyal, J. J. & Yarchoan, R. (1997) J. Biol. Chem. 272, 25935–42590. [DOI] [PubMed] [Google Scholar]
- 42.Humphries, K. M., Juliano, C. & Taylor, S. S. (2002) J. Biol. Chem. 277, 43505–43511. [DOI] [PubMed] [Google Scholar]
- 43.Taylor, E. R., Hurrell, F., Shannon, R. J., Lin, T. K., Hirst, J. & Murphy, M. P. (2003) J. Biol. Chem. 278, 19603–19611. [DOI] [PubMed] [Google Scholar]
- 44.Nulton-Persson, A. C. & Szweda, L. I. (2001) J. Biol. Chem. 276, 23357–23361. [DOI] [PubMed] [Google Scholar]
- 45.Nulton-Persson, A. C., Starke, D. W., Mieyal, J. J. & Szweda, L. I. (2003) Biochemistry 42, 4235–4242. [DOI] [PubMed] [Google Scholar]
- 46.Murata, H., Ihara, Y., Nakamura, H., Yodoi, J., Sumikawa, K. & Kondo, T. (2003) J. Biol. Chem. 278, 50226–50233. [DOI] [PubMed] [Google Scholar]
- 47.Daily, D., Vlamis-Gardikas, A., Offen, D., Mittelmann, L., Melamed, E., Holmgren, A. & Barzilai, A. (2001) J. Biol. Chem. 276, 1335–1344. [DOI] [PubMed] [Google Scholar]
- 48.Daily, D., Vlamis-Gardikas, A., Offen, D., Mittelmann, L., Melamed, E., Holmgren, A. & Barzilai, A. (2001) J. Biol. Chem. 276, 21618–21626. [DOI] [PubMed] [Google Scholar]
- 49.Song, J. J., Rhee, J. G., Suntharalingam, M., Walsh, S. A., Spitz, D. R. & Lee, Y. J. (2002) J. Biol. Chem. 277, 46566–46575. [DOI] [PubMed] [Google Scholar]
- 50.McStay, G. P., Clarke, S. J. & Halestrap, A. P. (2002) Biochem. J. 367, 541–548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Singal, P. K. & Iliskovic, N. (1998) N. Engl. J. Med. 339, 900–905. [DOI] [PubMed] [Google Scholar]
- 52.Wallace, K. B. (2003) Pharmacol. Toxicol. 93, 105–115. [DOI] [PubMed] [Google Scholar]
- 53.Doroshow, J. H., Locker, G. Y., Ifrim, I. & Myers, C. E. (1981) J. Clin. Invest. 68, 1053–1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yen, H. C., Oberley, T. D., Vichitbandha, S., Ho, Y. S. & St. Clair, D. K. (1996) J. Clin. Invest. 98, 1253–1260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Shioji, K., Kishimoto, C., Nakamura, H., Masutani, H., Yuan, Z., Oka, S. & Yodoi, J. (2002) Circulation 106, 1403–1409. [DOI] [PubMed] [Google Scholar]
- 56.Meyer, E. B. & Wells, W. W. (1999) Free Radical Biol. Med. 26, 770–776. [DOI] [PubMed] [Google Scholar]
- 57.Johansson, L., Chen, C., Thorell, J. O., Fredriksson, A., Stone-Elander, S., Gafvelin, G. & Arnér, E. S. J. (2004) Nat. Methods, in press. [DOI] [PubMed]