Abstract
In the developing pancreas, the basic helix–loop–helix (bHLH) protein Neurogenin3 (Ngn3) specifies which precursor cells ultimately will become endocrine cells and initiates the islet differentiation program. NeuroD1, a closely related bHLH protein and a downstream target of Ngn3, maintains the differentiation program initiated by Ngn3. We have developed an in vitro model of Ngn3-dependent differentiation by infecting pancreatic duct cell lines with an Ngn3-expressing adenovirus. We found that both Ngn3 and its downstream target NeuroD1 activated the islet differentiation program in these cells by inducing the expression of genes with early roles in the differentiation cascade, as well as genes characteristic of fully differentiated islet cells. Induction of these genes, as exemplified by the insulin1 gene, involved alteration of the local chromatin structure. Interestingly, the subsets of genes activated by Ngn3 and NeuroD1 were not completely overlapping, indicating that these two bHLH proteins serve specific functions in the development of the endocrine pancreas. In addition, microarray gene expression analysis identified a previously uncharacterized group of Ngn3-induced genes with potentially important roles in islet development and function. These studies demonstrate how Ngn3 initiates islet differentiation and provide us with a model for testing methods for producing islet cells for people with diabetes.
During pancreatic development, differentiation of endocrine and exocrine cells from a common endodermal progenitor cell requires the coordinated regulation of specific sets of genes. This process can be envisioned as a hierarchy or cascade of transcription factors that initiate and maintain the distinct gene expression programs that define the various pancreatic cell types (1). Among these factors, the basic helix–loop–helix (bHLH) protein Neurogenin3 (Ngn3) plays a dominant role in the specification of the endocrine/islet cell lineage.
During embryonic development, Ngn3 appears transiently in scattered pancreatic epithelial cells (2, 3). Several lines of evidence indicate that the expression of Ngn3 in these undifferentiated cells directs them to an endocrine cell fate and initiates the program of islet differentiation. First, lineage tracing shows that these transient Ngn3-expressing cells differentiate exclusively into islet cells (4). Second, mice homozygous for a targeted deletion of the ngn3 gene fail to generate any islet cells (5). Third, ectopic expression of Ngn3 drives embryonic endoderm to an endocrine fate (2, 3, 6).
Ngn3 may play a similar role in the generation of new islet cells postnatally. It has been suggested that cells along the pancreatic ducts may act as progenitors for new islet cells in the postnatal period, although recent lineage tracing experiments suggest that the bulk of newly generated beta cells in adult mice result from the replication of preexisting beta cells (7).
Ngn3 initiates islet cell differentiation, but other factors downstream of ngn3 must complete the task. Genetic studies in mice along with gene expression studies have placed several transcription factors in a differentiation cascade downstream of Ngn3 (1), but this approach is time-consuming and has not revealed new members of the cascade. As a result, our understanding of the epistatic relationships among islet differentiation genes remains rudimentary, and we have yet to identify the genes downstream of ngn3 that specify the distinct islet cell subtypes.
To overcome these limitations, we developed an in vitro model of Ngn3-dependent differentiation by infecting mouse pancreatic duct cell lines with a Ngn3-expressing adenovirus. We found that Ngn3 and other proendocrine bHLH factors switch on the islet differentiation program in these duct cells by activating a number of genes, including both early members of the differentiation cascade and genes characteristic of fully differentiated islet cells. In addition, cDNA microarray gene expression analysis detected previously uncharacterized Ngn3-induced genes with potentially important roles in pancreas biology. These studies demonstrate how Ngn3 initiates islet differentiation and provide us with a valuable model system for studying islet cell differentiation.
Materials and Methods
Cell Lines and Cell Culture. Mouse mPAC L20 and mPAC L4S2 cell lines (8), human PANC-1 cells (American Type Culture Collection), and 3T3 cells were cultured in DMEM supplemented with 10% FCS and antibiotics. The human pancreatic duct cells HPDE-E6E7 c7 and c11 (9) were kindly provided by M. Tsao (University of Toronto, Toronto) and were cultured in keratinocyte serum-free medium supplemented with 50 mg/ml bovine pituitary extract, 5 ng/ml epidermal growth factor, and antibiotics. The pancreatic islet cell lines αTC1.6 and βTC3 were cultured in DMEM supplemented with 15% horse serum, 2.5% FCS, and antibiotics.
Construction, Purification, and Infection of Recombinant Adenoviruses. cDNAs encoding human Ngn3, rat Mash1, and mouse NeuroD1 were cloned into the pAC.CMV shuttle vector, and recombinant adenoviruses were constructed by homologous recombination in HEK293 cells as described in ref. 10. The adenovirus expressing mouse MyoD was prepared with the Adeno-X system (Clontech). A control adenovirus expressing bacterial β-galactosidase was a kind gift from C. B. Newgard (Duke University, Durham, NC). For viral infection, cells were incubated with adenoviruses at a multiplicity of infection of 50 for 2 h at 37°C. Then, virus-containing medium was replaced, and cells were cultured for the times indicated.
RNA Isolation and RT-PCR. Total RNA was isolated by using the RNeasy kit (Qiagen, Valencia, CA). First-strand cDNA was prepared from 1 μg of total RNA with the SuperScript II RT kit and random hexamer primers (GIBCO/BRL). One microliter of the cDNA was used as a template for PCR reactions, and all RNA samples were tested in the absence of reverse transcriptase. PCR products were analyzed on agarose gels. All results were confirmed with at least three independent RNA samples.
Twenty-five to 150 ng of cDNA was used as a template for TaqMan quantification. All primer/probe sets were validated with a standard curve by using serial cDNA dilutions. Quantification was performed on an Applied Biosystems Prism 7900HT sequence detection system. Measurement of the expression levels of mouse β-glucuronidase (mGUS) was used as an internal control for cDNA quantity and quality in all assays. Sequences of all primers and probes are available upon request.
Quantitative Chromatin Immunoprecipitation (ChIP) Assay. Approximately 107 L20 cells (transfected with adenovirus encoding either β-galactosidase or NeuroD1) were fixed in 1% formaldehyde for 10 min and processed for ChIP as described in ref. 11. Coimmunoprecipitated insulin promoter fragments were quantified by SYBR Green I-based real-time PCR with primers amplifying the proximal and distal promoter regions as detailed in ref. 12. Rabbit polyclonal antibodies to acetylated-H3 and H3-dimethyl-K4 were from Up-state Biotechnology (Lake Placid, NY).
cDNA Microarrays. Slides carrying ≈18,000 mouse cDNAs were generated by the Mouse Microarray Consortium at the University of California, San Francisco. Of those, 15,000 clones corresponded to mouse ESTs from the National Institute of Aging set of mouse ESTs (13). The remaining clones were amplified from personal stocks of various University of California, San Francisco laboratories. The complete list of clones can be obtained from the Mouse Microarray Consortium Web site, http://mmc.ucsf.edu.
First-strand synthesis was performed with 10 μg of total RNA, dT(16) oligomer, and 5-(3-aminoallyl)-dUDP. The cDNA product was then purified, coupled with cy3 or cy5 dye, hybridized, and washed as described in ref. 14. Microarrays were scanned with an Axon 400B scanner (Axon Instruments, Union City, CA). Data were acquired and analyzed by using genepix pro-4 software (Axon Instruments) and stored and normalized by using the NOMAD microarray database system (http://ucsf-nomad.sourceforge.net). For each pair of samples (Ngn3 vs. βgal), multiple hybridizations were performed, including a reverse dye hybridization to control for potential dye bias. The four best-quality hybridizations, corresponding to three independent infections, were selected to calculate the mean fold change indicated in Table 1.
Table 1. Ngn3-induced genes in mPAC L20 cells.
| GenBank accession no. | Protein name | Fold increase* | SEM | Confirm† |
|---|---|---|---|---|
| Transcription | ||||
| NM_010919 | Nkx2.2 | 16.3 | 4.2 | C |
| NM_008814 | PDX-1 | 14.5 | 7.4 | C |
| MMU76208 | Ngn3 | 12.5 | 3.5 | C |
| AB038696 | Olig-1 | 12.0 | 5.0 | C |
| Y09166 | Neurogenin1 | 11.3 | 2.3 | C |
| NM_007897 | Ebf1 | 10.3 | 5.3 | NT |
| NM_010895 | NeuroD2 | 7.3 | 1.5 | C |
| X74134 | Coup TF1 | 7.0 | 2.4 | C |
| L14610 | RZR beta | 6.7 | 2.1 | NT |
| MMU28068 | NeuroD1 | 6.2 | 0.8 | C |
| S79041 | Gsh2‡ | 6.1 | 2.9 | C |
| AF153046 | Hb9 | 5.5 | 1.6 | NT |
| S80555 | RAR beta | 4.9 | 1.1 | NT |
| BG074244 | Nsbp1 | 4.7 | 1.0 | NT |
| AF260236 | Hes-6 | 4.5 | 0.7 | C |
| XM_123529 | Lhx4 | 4.4 | 1.0 | NT |
| NM_009385 | Nkx2.1 | 4.3 | 0.8 | NT |
| AF086757 | NSCL1 | 4.0 | 1.2 | NT |
| NM_010133 | Engrailed 1 | 3.8 | 1.0 | NT |
| AB031040 | Lhx6.1a | 3.4 | 0.9 | NT |
| U79550 | Slug | 3.2 | 0.7 | NT |
| U67841 | Dlx6 | 2.7 | 0.2 | C |
| AF202039 | Nkx2.4 | 2.6 | 0.6 | NT |
| NM_021459 | Isl-1 | 2.6 | 0.1 | C |
| Signaling | ||||
| NM_010950 | Numb-Like | 10.7 | 2.4 | C |
| NM_011183 | Presenilin2 | 9.5 | 2.7 | NC |
| NM_017218 | ErbB3 | 8.4 | 1.4 | NT |
| X80903 | Delta 1 | 7.9 | 4.1 | C |
| NM_008943 | Presenilin 1 | 6.1 | 1.5 | NT |
| MMU04710 | IGF-II receptor | 5.0 | 0.8 | NT |
| BC010770 | Somatostatin | 4.4 | 1.1 | C |
| D32210 | Notch2 | 4.3 | 1.7 | C |
| M33385 | TrkB | 4.2 | 1.2 | NC |
| L20336 | Dopamine receptor 1a | 4.1 | 0.9 | NT |
| Z48746 | FGF8 | 3.0 | 0.4 | NT |
| NM_010491 | IAPP | 2.6 | 0.4 | C |
| X93581 | TrkA | 2.6 | 0.4 | C |
| Cell organization | ||||
| NM_007667 | Cadherin 8 | 7.2 | 2.1 | NT |
| M31131 | Cadherin N‡ | 5.9 | 1.5 | C |
| NM_009152 | Sema3a | 4.4 | 0.8 | NC |
| AF041083 | Robo 1 | 4.3 | 0.9 | NT |
| NM_008737 | Neuropilin-1‡ | 4.0 | 0.7 | C |
| NM_013657 | Sema3c | 3.6 | 0.7 | NT |
| BG069465 | Antigen CD63 | 3.6 | 0.5 | NT |
| NM_010111 | EphrinB2‡ | 3.0 | 0.2 | NT |
| BG085134 | Antigen CD24a‡ | 2.6 | 0.3 | NT |
| BG076893 | RhoB | 2.6 | 0.5 | NT |
| Protein metabolism | ||||
| BG078497 | Cathepsin L‡ | 4.7 | 1.7 | C |
| BG076444 | p162 subunit of elF3 | 2.5 | 0.5 | NT |
| DNA synthesis | ||||
| BG069854 | DNA repair XRCC1 | 3.0 | 0.6 | NT |
SEM, standard error of the mean.
mPAC L20 cells were treated with AdCMV-βgal or AdCMV-ngn3, and RNA was extracted 48 h later. Data are means of fold induction by Ngn3 relative to β-galactosidase from four hybridizations using RNA from three independent experiments. All genes induced ≥2.5-fold are shown.
Confirmation of the microarray data was performed for a subset of genes by using RT-PCR. C, upregulation was confirmed in at least two of three independent experiments; NC, not confirmed; NT, not tested.
These genes were spotted more than once on the array, and the fold induction shown is the mean of all spots for that gene.
Results
Induction of Endocrine Differentiation Genes by Proendocrine bHLH Factors. Activation of the bHLH protein NeuroD1 by Ngn3 is one of the earliest events in pancreatic islet cell differentiation (15). Therefore, we used neuroD1 gene activation as an indicator of competence to respond to Ngn3. As assessed by RT-PCR, infection with a recombinant adenovirus encoding Ngn3 (AdCMV-ngn3) led to the appearance of neuroD1 transcript in all duct cell lines tested, including mouse mPAC cells (lines L20 and L4S2) and human HPDE-E6E7 (lines c7 and c11) and PANC-1 cells, but not in the fibroblast cell line NIH 3T3 (Fig. 1A).
Fig. 1.
Ngn3 induces early and late endocrine differentiation genes in duct cells. (A) Levels of neuroD1 mRNA were assayed by RT-PCR in mouse duct cell lines mPAC L20 and mPAC L4S2, beta cell line βTC3, and fibroblast cell line NIH 3T3, and human duct cell lines HPDE-E6E7c7, HPDE-E6E7c11, and PANC-1. mRNA was harvested from duct cell lines 48 h after infection with an adenovirus expressing Ngn3 (AdCMV-ngn3) or a control adenovirus expressing β-galactosidase (AdCMV-βgal). (B) RT-PCR for the indicated mRNAs was performed 48 h after infection of mPAC L20 and mPAC L4S2 cells. RNA from βTC3 cells or freshly isolated mouse islets was used in the lane labeled “Positive control.” (C) RNA from mPAC L20 cells was collected at the indicated time points after infection with AdCMV-ngn3. RT-PCR detection of adenovirally expressed human ngn3 (A and C) and β-actin mRNAs is shown as internal controls. The numbers of PCR cycles were 25 for β-actin, 30 for ngn3, and 35–38 for other transcripts.
We next tested whether Ngn3 could activate genes thought to lie further downstream in the endocrine differentiation cascade (1). Ngn3 induced readily measurable levels of pax4 and nkx2.2 mRNAs in both mouse mPAC cell lines (Fig. 1B). The nkx2.2 gene contains at least three promoters that are active in the pancreas, and the nkx2.2 mRNA induced by Ngn3 included exon 1A, but not exon 1B (data not shown), consistent with our previous conclusion that Ngn3 activates the 1A promoter but not the 1B promoter (16). Ngn3 induced modest levels of Pax6 expression in some, but not all, samples tested and did not induce nkx6.1 mRNA in either cell line.
Time-course experiments established the sequence of gene activation events. Appearance of pax4 and nkx2.2 mRNAs occurred earlier (16 and 24 h after virus infection, respectively) than neuroD1 (36 h), indicating that Ngn3 can activate these genes in the absence of NeuroD1 (Fig. 1C). These findings were confirmed by using TaqMan real-time PCR (data not shown). The delayed induction of neuroD1 mRNA by Ngn3 is intriguing because Ngn3 is thought to directly regulate NeuroD1 expression by binding to its promoter (15). Synthesis of additional cofactors may be necessary before activation of NeuroD1 in mPAC cells.
With the exception of glucagon, infection with AdCMV-ngn3 activated the expression of all of the islet hormone genes including islet amyloid polypeptide (IAPP) and grhelin (Fig. 1B and data not shown). In addition, Ngn3 stimulated the expression of the beta cell glucose sensor glucokinase, but not the glucose transporter GLUT-2. Ngn3 also increased levels of the mRNA encoding MafA, a transcription factor recently implicated in regulation of the insulin gene (17). Taken together, these results demonstrate that Ngn3 activates the islet differentiation program in mPAC cells, including both early and late genes.
Several bHLH proteins are expressed during pancreatic development (3). To determine whether these factors share with Ngn3 the ability to drive endocrine-specific gene expression in mPAC cells, we used adenovirus vectors to express NeuroD1, Mash1, and the myogenic bHLH factor MyoD (18) and compared their patterns of downstream target activation with that of Ngn3.
Both NeuroD1 and Mash1 drove endocrine-specific gene expression in mPAC cells, but they activated a more restricted panel of target genes as compared with Ngn3 (Fig. 2C). NeuroD1 shared with its upstream activator Ngn3 the capacity to induce pax4, nkx2.2, insulin, glucokinase, and IAPP mRNAs but failed to activate other Ngn3-downstream genes, namely the islet hormone genes somatostatin and PP. On the other hand, at equivalent viral doses, NeuroD1 induced higher insulin and IAPP mRNA levels than Ngn3. Mash1 activated an even more restricted set of target genes; only strongly inducing pax4, PP, glucokinase, and IAPP gene expression. Relative differences in the expression levels of induced genes were confirmed by real-time TaqMan RT-PCR (Fig. 2D). The observed differences between Ngn3 and NeuroD1 most likely reflect functional differences between these two factors because the percentages of infected cells were similar for both adenoviruses (data not shown).
Fig. 2.
Other bHLH factors also can induce the endocrine differentiation program. mPAC cells were infected with Adeno-βgal, Adeno-ngn3 (as in previous experiments), or with adenoviruses expressing the bHLH proteins NeuroD1, Mash 1, and MyoD, and gene expression was analyzed by RT-PCR 48 h later. (A) Adenovirus-encoded transcripts for bHLH factors were detected. (B) RT-PCR primers for neuroD1 and ngn3 detect the endogenous mouse mRNAs, and the primers for mash1 detect both the endogenous mouse mRNA and the rat adenovirus-encoded mRNA. (C) Transcripts for early and late islet differentiation genes were detected. RNA from βTC3 cells or freshly isolated mouse islets (for glucagon, somatostatin, and PP transcripts) was used for positive controls. PCR cycle numbers were 30 for virally encoded mRNAs, 25 for β-actin, and 35–38 for other mRNAs. (D) Relative expression of the indicated genes in mPAC L20 cells was measured by real-time TaqMan RT-PCR. Results represent mean ± SEM from three independent experiments and are expressed relative to levels attained in ngn3-expressing cells (arbitrarily given the value of 1). For all mRNAs studied, except for PP, values in control mPACs could not be determined (und). mGUS was used as the endogenous control in all TaqMan assays.
The ability to activate the islet differentiation program was specific to the neuroendocrine bHLH proteins, because MyoD activated none of the pancreatic genes except IAPP and only in mPAC L20 cells at low levels (Fig. 2C). It should be noted that we observed some differences between the two mPAC lines in other experiments as well. In most cases, the responses in the two mPAC lines were the same, but mRNA induction levels in L4S2 cells were generally lower than in L20 cells. Higher adenoviral infection efficiency in L20 (80–90%) as compared with L4S2 (40–50%) cells may explain these differences (data not shown), although intrinsic differences between the two cell lines cannot be ruled out.
It should be noted that levels of islet hormone mRNAs, in particular insulin, in treated mPAC cells were significantly lower than in βTC3 or islets. Insulin mRNA levels in NeuroD1-infected mPAC cells were only 0.00013% of levels found in βTC3 and 0.00001% of those found in islets as assayed by TaqMan RT-PCR. Conversely, levels of somatostatin mRNA in ngn3-expressing mPAC cells were definitely closer than insulin to those found in islets (0.14%). Levels of the induced mRNAs, however, were assayed shortly after the initiation of transcription (somatostatin and insulin mRNAs were not detectable until 36–40 h in culture), whereas levels in βTC3 cells or islets were assayed at steady state. This point is particularly important when considering long half-life mRNAs such as insulin (≈100 h). In addition, these results represent an average over a cell population, within which only a small percentage of cells express the gene. For example, only 2–5% of the mPAC cells expressing ngn3 express somatostatin after 48 h in culture (data not shown).
bHLH genes often function in transcriptional activation cascades during differentiation in a variety of tissues. For example, Mash1 lies upstream of Neurogenin1 and NeuroD1 in the olfactory neuron lineage (19). To determine whether the proendocrine function of Mash1 could result from activation of Ngn3 expression, we assessed endogenous ngn3 gene expression in mPAC cells treated with the Mash1-encoding adenovirus. Mash1 strongly induced ngn3 mRNA (Fig. 2B), but neither Ngn3 nor NeuroD1 induced mash1 mRNA. These results suggest that the Mash1-dependent induction of endocrine cell markers could be secondary to the activation of Ngn3 expression. It should be noted, however, that a similar relationship may not exist in vivo, where Mash1 is not required for the expression of Ngn3 in the pancreas (3). Unexpectedly, Ngn3 expression was activated weakly by NeuroD1 and MyoD in mPAC L20 cells, but not in L4S2 cells.
Interestingly, we observed that Ngn3 and NeuroD1 activated their respective endogenous genes in both mPAC lines (Fig. 2B). This autoactivation is consistent with a previous report that neural bHLH proteins can activate their own expression in pluripotent mouse P19 embryonal carcinoma cells (20).
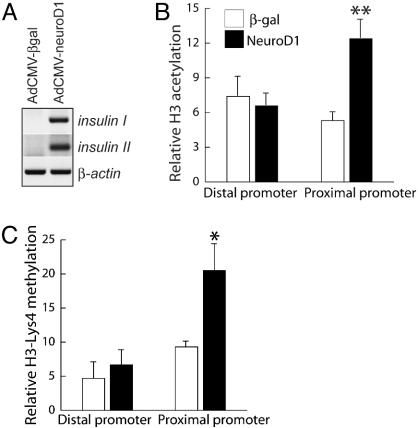
Proendocrine bHLH Factors Induce Chromatin Changes at the insulin1 Locus. Activation of insulin gene transcription in beta cells is associated with characteristic modifications of histone H3, namely H3 acetylation and H3-Lys-4 methylation, at the proximal insulin1 promoter (12). Both modifications are normally associated with open, euchromatic regions of transcriptionally active genes (21). To determine whether similar changes accompany activation of insulin1 gene transcription in NeuroD1transfected mPAC L20 cells, we performed ChIP assays with antibodies toward specifically modified histones. NeuroD1 transfection results in significant (P < 0.05) increases in H3 acetylation and H3 (Lys-4) methylation at the proximal insulin1 promoter region (≈300 bp upstream of the transcriptional start site), but not at the distal promoter region (≈4,800 bp upstream) (Fig. 3). These modifications are similar in magnitude to those observed at the proximal insulin1 promoter in beta cells (12).
Fig. 3.
Histone modifications at the insulin locus in Adeno-NeuroD1-infected cells. mPAC cells were infected with Adeno-βgal (open bars) or Adeno-NeuroD1 (filled bars) and harvested 60 h later. (A) Insulin I and II transcripts were detected by RT-PCR. (B and C) Graphs display data of real-time PCR quantification of distal (–4,653 to –4,812 bp) and proximal (–126 to –296 bp) insulin1 promoter fragments after ChIP with antiacetylated H3 (B) or anti-H3-dimethyl-Lys-4 (C). Results are expressed relative to control (normal rabbit serum in place of specific antibody). Results represent mean ± SEM from at least three independent ChIP assays from three independent cell samples. Adeno-NeuroD1-infected cells showed significantly higher levels of H3 acetylation (*, P < 0.05 by two-tailed Student's t test) and H3-Lys-4 methylation (**, P < 0.001) than Adeno-βgal-infected cells at the proximal, but not the distal, insulin promoter.
Microarray Analysis of Ngn3-Induced Genes. The adenovirus-infected mPAC cells provide a useful model for studying islet differentiation in vitro. We exploited this model to identify target genes of Ngn3. For this purpose, we used microarrays containing ≈18,000 mouse cDNAs and compared gene expression patterns in mPAC L20 cells infected with either AdCMV-βgal (control) or AdCMV-ngn3. All comparisons were performed 48 h after viral infection, meaning that genes differentially expressed in Ngn3-expressing mPAC cells could include genes whose expression was directly or indirectly controlled by Ngn3.
Expression profile analysis identified 51 mRNAs induced by at least 2.5-fold (Table 1) and 86 mRNAs down-regulated at least 3-fold (data not shown) in response to Ngn3. The list includes five genes that we had already identified (ngn3, neuroD1, nkx2.2, somatostatin, and IAPP) (Fig. 1). Nearly 50% of the identified genes code for transcription factors from diverse families, which is not unexpected given the proposed role of Ngn3 as an early transcriptional switch in the endocrine differentiation cascade. Interestingly, 20% of the identified genes code for proteins involved in cell migration and cell-to-cell interactions, many of which have roles in neural growth and axon guidance (i.e., neuropilins, semaphorins, and trk). In addition, some of the genes are involved in signaling cascades with known relevant roles in pancreatic development, such as Notch (2). Although many of the up-regulated genes encode factors already known to play a role in pancreatic biology (i.e., Pdx1, Hb9, and Isl1), participation in pancreatic development has not been previously addressed for many of them.
Of the downstream genes that we identified, we selected 19 for confirmation by RT-PCR. Induction in response to Ngn3 was reproduced in at least two of three independent experiments for all of the genes tested except for three (presenilin2, TrkB, and Sema3a), as indicated in Table 1. Representative RT-PCRs for some of these genes are shown in Fig. 4A.
Fig. 4.
Confirmation by RT-PCR of Ngn3-induced genes identified by microarray analysis. Induction of the indicated genes was analyzed by RT-PCR from RNA isolated from mPAC L20 cells 48 h after infection with AdCMV-βgal or AdCMV-ngn3 (A) or from pancreatic cells (B). In all cases, results were confirmed by using RNA from at least three independent samples.
Next, for some of the confirmed genes, we used RT-PCR to evaluate expression in vivo in the developing mouse pancreas (e12.5–e17.5), adult mouse islets, and islet-derived cell lines (αTC1.6 and βTC3). Transcripts for the bHLH factors Olig1 and Neurogenin1 were barely detectable in embryonic pancreas even after 38 PCR cycles (Fig. 4B). Olig1 belongs to a family of oligodendrocyte lineage-specific bHLH factors (22, 23). Of four related genes, olig2, olig3, and bhlhb5 also were not detected in the pancreas, whereas bhlhb4 was expressed in the embryonic pancreas, but not in adult islets, and was not induced by Ngn3 (data not shown). In contrast, mRNAs encoding the bHLH factors NeuroD2 and Hes6, an inhibitor of Hes1, were present in the embryonic pancreas at all stages studied (Fig. 4B). Interestingly, neuroD2 mRNA was not found in adult islets. All of the other genes studied were detected at significant levels in the pancreas with the exception of the homeobox gene gsh2 (data not shown).
Discussion
The present study demonstrates that expression of Ngn3 is sufficient to trigger the endocrine differentiation program in pancreatic duct cell lines. This program includes both the cascade of genes encoding transcription factors that regulate islet-cell development as well as the genes encoding markers characteristic of mature islet cells, such as insulin. In agreement with prior work in vivo (3), other proendocrine bHLH factors NeuroD1 and, to a lesser extent, Mash1, share with Ngn3 this ability to drive endocrine differentiation in vitro. Activation of target genes, as demonstrated by the ChIP analysis of the insulin1 gene, involves alteration of the local chromatin structure. Finally, microarray analysis identified additional genes downstream of Ngn3.
The transcriptional cascade activated by Ngn3 in mPAC cells closely resembles the one operating in the mouse embryo during pancreatic development. In mPAC cells, Ngn3 induced expression of the transcription factors NeuroD1, Pax4, Pax6, Nkx2.2, and Isl-1, all of which previously have been placed downstream of Ngn3 (1, 5). An important exception was the homeodomain transcription factor Nkx6.1, which lies downstream of Nkx2.2 during normal islet-cell differentiation (24). In mPAC cells, nkx2.2 mRNA appeared as early as 24 h after Ngn3 infection, but nkx6.1 mRNA could not be detected up to 48 h later (data not shown). Similarly, Ngn3 failed to induce the expression of Nkx6.1 in human duct-cell cultures (25). Prior studies have revealed that regulation of Nkx6.1 expression is complex and involves both transcriptional and translational mechanisms (26). Thus, it is possible that Nkx6.1 expression is controlled by Ngn3 and Nkx2.2 posttranscriptionally. Consistent with this idea, we can detect nkx6.1 mRNA at normal levels in late fetal pancreases from Nkx2.2-null mice (data not shown), even though Nkx6.1 protein is absent (24), indicating that translational control of Nkx6.1 expression is crucial during normal pancreatic development.
Early Ngn3 expression in the pancreatic anlage of embryonic mice causes both expanded and precocious differentiation of glucagon-expressing cells (2, 3). These results led to the proposal that alpha cells are the default fate of Ngn3-expressing cells. Accordingly, ectopic Ngn3 also was found to induce, although not exclusively, glucagon expression in chick embryos (6) and retinoic-acid-derived endoderm cells (27). Therefore, it is surprising that Ngn3 induces expression of all of the islet hormones except glucagon in mPAC cells. Similarly, Ngn3 failed to elicit alpha cell differentiation in human duct-cell cultures (25). The mPAC cells may express inhibitory factors or lack positive regulators that allow alpha cell differentiation in the developing pancreas in vivo. In support of this explanation, the POU homeodomain factor Brn4, an important regulator in the determination of alpha cell lineage (28), is not induced by Ngn3 in mPAC cells (data not shown).
The observation that NeuroD1, like Ngn3, switches on the islet cell differentiation program in mPAC cells is not surprising. Indeed, prior work demonstrated that both factors drive endocrine cell differentiation in transgenic mouse models (3). The current observation is that the sets of genes activated by the two factors, including the islet hormone genes, are not identical. Although Ngn3 induces mRNAs for somatostatin and PP and, to a lesser extent, insulin, NeuroD1 predominantly induces insulin mRNA. Consistent with these studies in mPAC cells, NeuroD1-deficient mice have a drastic reduction in the number of beta cells, whereas the delta and PP cell lineages are largely unaffected (29), suggesting that factors other than NeuroD1 down-stream of Ngn3 may be important for the differentiation of the delta and PP cell lineages. Our microarray data identify a number of bHLH and non-bHLH transcription factors induced by Ngn3 that could serve in such roles. Among these, NeuroD2, a bHLH factor closely related to NeuroD1, was strongly induced by Ngn3 and was present in the developing pancreas but was largely absent from adult islets. Although the role of NeuroD2 in neural development has been recently established (30), its function in endocrine differentiation remains unexplored.
Members of the Ngn and NeuroD families of bHLH transcription factors also participate in neural development. Members of these families promote neuronal differentiation from competent precursor neural cells (31, 32), but they also can impose a neuronal fate on nonneural cells (31). Thus, it is possible that expression of Ngn3 (or NeuroD1) may initiate neural as well as endocrine differentiation programs in mPAC cells. In fact, many of the genes that we identified as Ngn3 targets in the mPAC microarray experiments are known neural genes. Although this observation is not surprising, given the recognized parallels between neurons and islet cells (1), the expression of some of these neural genes could result from more relaxed regulatory constraints in the in vitro system or nonspecific effects from excessive transgene expression. The bHLH genes ngn1 and olig-1 and the homeobox gene gsh2, none of which are expressed at significant levels in the pancreas as assessed by RT-PCR (Fig. 4), illustrate the activation by Ngn3 of nonpancreatic neuronal genes in mPAC cells.
The current study does not address whether pancreatic duct cells ever function normally in vivo as progenitors for islet cells or whether normal duct cells can respond to the proendocrine bHLH genes and differentiate into endocrine cells. Mouse mPAC cells have ductal characteristics (expression of cytokeratins) but also express A2B5, a neuroendocrine ganglioside expressed in developing islet cells (8). Thus, mPAC cells could represent a stem-like progenitor or dedifferentiated duct cell. Nevertheless, Ngn3 also triggered endocrine gene expression from human PANC-1 cells (data not shown), which are derived from pancreatic ductal epithelium, suggesting that the capacity to activate the endocrine differentiation program may be a true characteristic of normal pancreatic duct cells. Heremans et al. (25) reported that Ngn3 could drive endocrine differentiation in primary-cultured human pancreatic duct cells. However, because the researchers started with a mixed cell population, it was not possible with certainty to determine which cells responded to Ngn3. In fact, the rather low percentage of differentiated islet cells in the primary duct cultures suggests that very few of the Ngn3-infected cells were responsive to Ngn3. Furthermore, cells with the capacity to differentiate into islet cells in response to ectopically expressed proendocrine genes may not actually function as islet-cell precursors during normal islet cell genesis in vivo. Identification of bona fide islet cell precursors requires careful lineage-tracing analysis in vivo.
One ultimate goal of these studies is the production of insulin-producing cells for people with diabetes. The mPAC cell model system provides us with a unique means to identify genes involved in islet-cell formation and to test the ability of specific genes or gene combinations to drive endocrine differentiation. Knowledge gained from these studies also could be applied to the development of beta cells from human embryonic stem cells. Although recent studies suggest that the generation of insulin-producing cells from embryonic stem cells may not be difficult (33), a better understanding of islet cell differentiation may be important for the development of methods for the production of truly normal beta cells (34, 35).
Acknowledgments
We thank Christina Jamieson and the University of California, San Francisco, Mouse Microarray Consortium for supplying mouse cDNA arrays and advice. This work was supported by National Institutes of Health Grants DK02683 (to R.M.), DK60581 (to R.M.), and DK61245 (to M.G.), cores from National Institutes of Health Grant P30 DK063720 (to M.G.), and Juvenile Diabetes Research Foundation Grant 1-2000-655 (to M.G.) and Fellowship Awards 3-2000-667 (to R.G.) and 3-2000-847 (to C.M.).
Abbreviations: bHLH, basic helix–loop–helix; ChIP, quantitative chromatin immunoprecipitation; IAPP, islet amyloid polypeptide; Ngn3, Neurogenin3.
References
- 1.Wilson, M. E., Scheel, D. & German, M. S. (2003) Mech. Dev. 120, 65–80. [DOI] [PubMed] [Google Scholar]
- 2.Apelqvist, A., Li, H., Sommer, L., Beatus, P., Anderson, D., Honjo, T., Hrabe de Angelis, M., Lendahl, U. & Edlund, H. (1999) Nature 400, 877–881. [DOI] [PubMed] [Google Scholar]
- 3.Schwitzgebel, V. M., Scheel, D. W., Conners, J. R., Kalamaras, J., Lee, J. E., Anderson, D. J., Sussel, L., Johnson, J. D. & German, M. S. (2000) Development (Cambridge, U.K.) 127, 3533–3542. [DOI] [PubMed] [Google Scholar]
- 4.Gu, G., Dubauskaite, J. & Melton, D. (2002) Development (Cambridge, U.K.) 129, 2447–2457. [DOI] [PubMed] [Google Scholar]
- 5.Gradwohl, G., Dierich, A., LeMeur, M. & Guillemot, F. (2000) Proc. Natl. Acad. Sci. USA 97, 1607–1611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Grapin-Botton, A., Majithia, A. & Melton, D. (2001) Genes Dev. 15, 444–454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dor, Y., Brown, J., Martinez, O. I. & Melton, D. A. (2004) Nature 429, 41–46. [DOI] [PubMed] [Google Scholar]
- 8.Yoshida, T. & Hanahan, D. (1994) Am. J. Pathol. 145, 671–684. [PMC free article] [PubMed] [Google Scholar]
- 9.Ouyang, H., Mou, L., Luk, C., Liu, N., Karaskova, J., Squire, J. & Tsao, M. S. (2000) Am. J. Pathol. 157, 1623–1631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Becker, T., Noel, R., Coats, W., Gomez-Foix, A., Alam, T., Gerard, R. & Newgard, C. (1994) Methods Cell Biol. 43, 161–189. [DOI] [PubMed] [Google Scholar]
- 11.Chakrabarti, S. K., James, J. C. & Mirmira, R. G. (2002) J. Biol. Chem. 277, 13286–13293. [DOI] [PubMed] [Google Scholar]
- 12.Chakrabarti, S. K., Francis, J., Ziesmann, S. M., Garmey, J. C. & Mirmira, R. G. (2003) J. Biol. Chem. 278, 23617–23623. [DOI] [PubMed] [Google Scholar]
- 13.Tanaka, T. S., Jaradat, S. A., Lim, M. K., Kargul, G. J., Wang, X., Grahovac, M. J., Pantano, S., Sano, Y., Piao, Y., Nagaraja, R., et al. (2000) Proc. Natl. Acad. Sci. USA 97, 9127–9132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.DeRisi, J. L., Iyer, V. R. & Brown, P. O. (1997) Science 278, 680–686. [DOI] [PubMed] [Google Scholar]
- 15.Huang, H., Liu, M., El-Hodiri, H., Chu, K., Jamrich, M. & Tsai, M. (2000) Mol. Cell. Biol. 20, 3292–3307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Watada, H., Scheel, D. W., Leung, J. & German, M. S. (2003) J. Biol. Chem. 278, 17130–17140. [DOI] [PubMed] [Google Scholar]
- 17.Olbrot, M., Rud, J., Moss, L. G. & Sharma, A. (2002) Proc. Natl. Acad. Sci. USA 99, 6737–6742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sassoon, D. A., Garner, I. & Buckingham, M. (1988) Development (Cambridge, U.K.) 104, 155–164. [DOI] [PubMed] [Google Scholar]
- 19.Cau, E., Gradwohl, G., Fode, C. & Guillemot, F. (1997) Development (Cambridge, U.K.) 124, 1611–1621. [DOI] [PubMed] [Google Scholar]
- 20.Farah, M. H., Olson, J. M., Sucic, H. B., Hume, R. I., Tapscott, S. J. & Turner, D. L. (2000) Development (Cambridge, U.K.) 127, 693–702. [DOI] [PubMed] [Google Scholar]
- 21.Jenuwein, T. & Allis, C. D. (2001) Science 293, 1074–1080. [DOI] [PubMed] [Google Scholar]
- 22.Zhou, Q., Wang, S. & Anderson, D. J. (2000) Neuron 25, 331–343. [DOI] [PubMed] [Google Scholar]
- 23.Lu, Q. R., Yuk, D., Alberta, J. A., Zhu, Z., Pawlitzky, I., Chan, J., McMahon, A. P., Stiles, C. D. & Rowitch, D. H. (2000) Neuron 25, 317–329. [DOI] [PubMed] [Google Scholar]
- 24.Sussel, L., Kalamaras, J., Hartigan-O'Connor, D. J., Meneses, J. J., Pedersen, R. A., Rubenstein, J. L. & German, M. S. (1998) Development (Cambridge, U.K.) 125, 2213–2221. [DOI] [PubMed] [Google Scholar]
- 25.Heremans, Y., Van De Casteele, M., in't Veld, P., Gradwohl, G., Serup, P., Madsen, O., Pipeleers, D. & Heimberg, H. (2002) J. Cell Biol. 159, 303–312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Watada, H., Mirmira, R. G., Leung, J. & German, M. S. (2000) J. Biol. Chem. 275, 34224–34230. [DOI] [PubMed] [Google Scholar]
- 27.Vetere, A., Marsich, E., Di Piazza, M., Koncan, R., Micali, F. & Paoletti, S. (2003) Biochem. J. 371, 831–841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hussain, M. A., Miller, C. P. & Habener, J. F. (2002) J. Biol. Chem. 277, 16028–16032. [DOI] [PubMed] [Google Scholar]
- 29.Naya, F. J., Huang, H. P., Qiu, Y., Mutoh, H., DeMayo, F. J., Leiter, A. B. & Tsai, M. J. (1997) Genes Dev. 11, 2323–2334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Olson, J. M., Asakura, A., Snider, L., Hawkes, R., Strand, A., Stoeck, J., Hallahan, A., Pritchard, J. & Tapscott, S. J. (2001) Dev. Biol. 234, 174–187. [DOI] [PubMed] [Google Scholar]
- 31.Lee, J. E., Hollenberg, S. M., Snider, L., Turner, D. L., Lipnick, N. & Weintraub, H. (1995) Science 268, 836–844. [DOI] [PubMed] [Google Scholar]
- 32.Ma, Q., Kintner, C. & Anderson, D. J. (1996) Cell 87, 43–52. [DOI] [PubMed] [Google Scholar]
- 33.Lumelsky, N., Blondel, O., Laeng, P., Velasco, I., Ravin, R. & McKay, R. (2001) Science 292, 1389–1394. [DOI] [PubMed] [Google Scholar]
- 34.Halban, P. A., Kahn, S. E., Lernmark, A. & Rhodes, C. J. (2001) Diabetes 50, 2181–2191. [DOI] [PubMed] [Google Scholar]
- 35.Rajagopal, J., Anderson, W. J., Kume, S., Martinez, O. I. & Melton, D. A. (2003) Science 299, 363. [DOI] [PubMed] [Google Scholar]