Abstract
Siglecs are a recently discovered family of animal lectins that belong to the Ig superfamily and recognize sialic acids (Sias). CD33-related Siglecs (CD33rSiglecs) are a subgroup with as-yet-unknown functions, characterized by sequence homology, expression on innate immune cells, conserved cytosolic tyrosine-based signaling motifs, and a clustered localization of their genes. To better understand the biology and evolution of CD33rSiglecs, we sequenced and compared the CD33rSiglec gene cluster from multiple mammalian species. Within the sequenced region, the segments containing CD33rSiglec genes showed a lower degree of sequence conservation. In contrast to the adjacent conserved kallikrein-like genes, the CD33rSiglec genes showed extensive species differences, including expansions of gene subsets; gene deletions, including one human-specific loss of a novel functional primate Siglec (Siglec-13); exon shuffling, generating hybrid genes; accelerated accumulation of nonsynonymous substitutions in the Sia-recognition domain; and multiple instances of mutations of an arginine residue essential for Sia recognition in otherwise intact Siglecs. Nonsynonymous differences between human and chimpanzee orthologs showed uneven distribution between the two β sheets of the Sia-recognition domain, suggesting biased mutation accumulation. These data indicate that CD33rSiglec genes are undergoing rapid evolution via multiple genetic mechanisms, possibly due to an evolutionary “arms race” between hosts and pathogens involving Sia recognition. These studies, which reflect one of the most complete comparative sequence analyses of a rapidly evolving gene cluster, provide a clearer picture of the ortholog status of CD33rSiglecs among primates and rodents and also facilitate rational recommendations regarding their nomenclature.
Interactions between glycan chains and their receptors are important in many biological processes. Such receptors can be intrinsic or extrinsic to the organism bearing the glycans (1). Host–pathogen interactions and host defense are two interrelated examples of biological processes wherein the importance of glycan–receptor interactions is well recognized. Cell-surface glycans (in particular, peripherally located sugars) and their receptors (often called lectins) are of interest both for their biomedical importance and their intriguing evolutionary history, reflecting the front line of the evolutionary “arms race” with pathogens, where rapid evolution is thought to take place by means of the “Red Queen Effect” (2, 3).
Sialic acids (Sias) are a class of acidic sugars found mostly at the outermost position of glycan chains on cell surfaces of deuterostomes (vertebrates and some higher invertebrates). The most numerous intrinsic Sia receptors in vertebrates are the Siglecs, a family of lectins of the Ig superfamily (Fig. 1 and ref. 4). Siglec molecules have multiple extracellular Ig-like domains (ranging from 2 to 17, each encoded by a single exon), followed by a single-pass transmembrane domain and a cytoplasmic tail. Although genes for 11 functional Siglecs and 1 Siglec-like protein are present in the human genome, only 8 are known in the mouse genome (5). This species difference largely reflects the CD33-related Siglecs (CD33rSiglecs), a subgroup defined by their mutual sequence similarity and clustered gene localization. CD33 (also known as Siglec-3) was the first to be discovered among this subgroup of Siglecs (originally as a cell-surface marker of human myeloid cells), hence the group was named after it. Seven functional Siglec genes (CD33/Siglec-3 and Siglec-5–10) and a Siglec-like gene (Siglec-L1, here renamed Siglec-XII) reside within an ≈0.5-Mb region of human chromosome 19q13.3-q13.4, whereas only four Siglec genes occur in the syntenic region of mouse chromosome 7B2 (5).
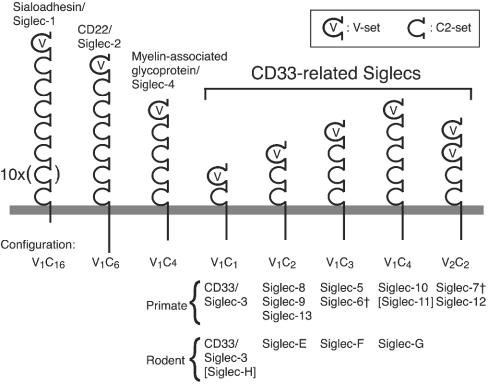
Fig. 1.
Schematic representation of Siglecs in primates and rodents. Siglecs have one V-set domain (a domain similar to Ig's variable region) and 1–16 C2-set domains (domains similar to Ig's constant region), followed by transmembrane and cytoplasmic domains. Genes for Sialoadhesin/Siglec-1, CD22/Siglec-2, and myelin-associated glycoprotein/Siglec-4 are located outside of the Siglec gene cluster in both primates and rodents. Clear orthologs have been established for each of these genes between human and mouse. Most of the genes for CD33rSiglec subfamily are in the Siglec cluster described here, with the exception of primate Siglec-11 and rodent Siglec-H, whose genes are outside of the gene cluster (indicated with square brackets). CD33rSiglecs are further classified into five subgroups (V1C1, V1C2, V1C3, V1C4, and V2C2), based on the number of V- and C2-set Ig-like domains. The basic configuration of the V2C2 subgroup (Siglec-7 and 12/XII) is V1 + V1C2, and the V1C2 part is highly similar to other Siglecs with V1C2 configuration and thus can be considered a part of the V1C2 subgroup. Although the primate SIGLEC6 gene has V1C3 configuration similar to that of SIGLEC5, the exon coding for a potential third C2-set domain is inactivated. Similarly, the primate SIGLEC7 gene has the exon coding for a potential second V-set domain inactivated. These are indicated with †.
Most CD33rSiglecs are expressed on cells involved in innate immunity, such as monocytes, granulocytes, macrophages, and natural killer cells (4, 6). A functional role for CD33rSiglecs in regulating innate immunity is suggested by this expression pattern and by tyrosine-based signaling motifs in their cytoplasmic tails, including a canonical ITIM motif (7). Indeed, antibody crosslinking of some CD33rSiglecs can dampen cell activation, arrest proliferation, or induce apoptosis (8, 9).
Sia recognition specificity has been modified in human CD33rSiglecs, due to loss of a Sia called N-glycolylneuraminic acid (Neu5Gc). This evolutionary change occurred in the human lineage after our last common ancestor with the chimpanzee and bonobo (10, 11), allowing recognition of N-acetylneuraminic acid, which, being a precursor to Neu5Gc accumulates at higher levels in humans (12). These data favor the hypothesis that CD33rSiglecs function primarily as self-recognition receptors for endogenous Sia ligands. However, they could also be involved in recognizing the many bacterial pathogens expressing Sias on their cell surfaces (13, 14). Regardless of which of these possibilities is ultimately proven correct, we hypothesize that CD33rSiglecs are evolving rapidly, to meet the challenge either of rapidly changing pathogens or of changing host Sia-expression patterns.
To date, there have been few reports of large-scale genomic comparisons of orthologous sequences from multiple vertebrate species (15, 16). Here we compare the CD33rSiglec gene cluster in chimpanzee, baboon, rat, mouse, and human. This analysis indicates that CD33rSiglecs, in fact, are evolving rapidly via multiple genetic mechanisms, and that selective pressure is apparently applied most heavily on the first Ig-like domain (Ig1), which is essential for Sia recognition.
Methods
Bacterial Artificial Chromosome (BAC) Isolation, Mapping, and Sequencing. BAC clones containing genomic regions encompassing the chimpanzee, baboon, and rat Siglec gene clusters were isolated and mapped from BACPAC Resource libraries (www.chori.org/bacpac) by using an established universal probe hybridization strategy (15, 17) and were subjected to standard shotgun sequencing (18, 19) by the National Institutes of Health Intramural Sequencing Center (www.nisc.nih.gov) to an advanced draft stage (see individual GenBank entries and Supporting Text, which is published as supporting information on the PNAS web site, for details and methods for assembly of sequence sets, pairwise sequence alignment, and gene prediction/annotation).
Analysis of Multispecies Conserved Sequences (MCSs). A multiple sequence alignment was generated with refine, the multiple alignment program associated with multipipmaker (20), and a 730-kb subregion of this alignment (representing near-complete coverage in all species) was analyzed for MCSs by previously established methods (21). The subregion was further divided into Siglec-containing and non-Siglec-containing portions (based on the coordinates of the Siglec genes) to make comparisons on the distribution of MCS. See Supporting Text to view the defined regions and MCSs.
Molecular Phylogenetic Analyses. Multiple nucleotide or amino acid sequence alignments were generated with clustal x (ftp://ftpigbmc.u-strasbg.fr/pub/ClustalX) (22) and were manually adjusted. NEXUS output files were then analyzed with paup*4.0 (Sinauer, Sunderland, MA). Both distance-based (neighbor-joining) and maximum-parsimony methods were used, yielding trees with very similar topology. Ancestral nucleotide sequences of human and chimpanzee Siglec orthologs (Ig1-coding exon only) were reconstructed by a maximum parsimony method.
Statistical Analysis of Ortholog Sequence Differences. Synonymous (Sd) and nonsynonymous (Nd) differences, synonymous (S) and nonsynonymous (N) sites, and proportions of synonymous (pS) and nonsynonymous (pN) sequence differences between orthologs were calculated with dnasp 3.53 (www.ub.es/dnasp; see ref. 23), which uses the Nei–Gojobori method (24). We first calculated these numbers exon by exon between human and chimpanzee Siglec orthologs. We then calculated the respective sum of these numbers (Σ Sd, Σ Nd, Σ S, and Σ N) in all Ig1-coding exons (i.e., exon 2 of Siglec-3, -5, -6, and -10 and exon 1 of Siglec-7, -8, -9, and -12). [Exons 1 and 2 of Siglec-3, -5, -6, and -10 encode for a signal peptide and Ig1, respectively, whereas exon 1 of Siglec-7, -8, -9, and -12 encodes for both.] This process is similar to concatenating all human CD33rSiglec Ig1-coding exons and comparing that with similarly concatenated chimpanzee sequence, then calculating Sd, Nd, S, and N. Hence we call it concatenation for convenience. A similar process was repeated for other exons and other pair of species (human–baboon and chimp–baboon). Fisher's exact test or the χ2 test was used to analyze the statistical significance of differences between observed and expected distributions.
Mapping Amino Acid Differences in Human–Chimpanzee CD33rSiglec Ortholog Pairs onto the Human Siglec-7 First Ig-Like Domain. The amino acid sequences of all human and chimpanzee CD33rSiglecs (except for chimpanzee Siglec-13) were aligned with clustal x and manually adjusted. Based on this alignment, amino acid differences were projected onto the corresponding position of the human Siglec-7 amino acid sequence, of which the crystal structure is available (see ref. 25 and Protein Data Bank ID code 1O7V).
See Supporting Text for cloning of chimpanzee Siglec-13, generating wild-type and mutant chimpanzee Siglec-5 H119R and baboon Siglec-6 L111R and Sia-binding assay.
Results
Comparative Sequencing. Overlapping BACs encompassing the chimpanzee, baboon, and rat genomic regions orthologous to the human CD33rSiglec cluster on chromosome 19q13.3-q13.4 were isolated and sequenced. Assembly yielded near-complete sequences from the chimpanzee (858 kb), baboon (703 kb), and rat (556 kb). The assembled advanced draft sequence quality was high, providing 98–99% coverage for each species. Systematic comparisons in conjunction with the already available human (ref. 26; chr19:55978691–56924690 from the April 2003 freeze) and mouse (ref. 27; chr7:31900001–32500000 from the February 2003 freeze) sequences revealed essentially all known genes in the region.
Overview of the Sequenced Region. In all five species, the kallikrein (KLK) gene cluster falls at the 5′ side of the Siglec gene cluster (Fig. 2 and Fig. 4, which is published as supporting information on the PNAS web site). Notably, although the KLK cluster has the same number and order of genes in all five species, the Siglec cluster in rodents and primates shows marked differences, with the former taxa having only four Siglec genes each and the latter ranging from seven (in the baboon) to nine (in the chimpanzee).
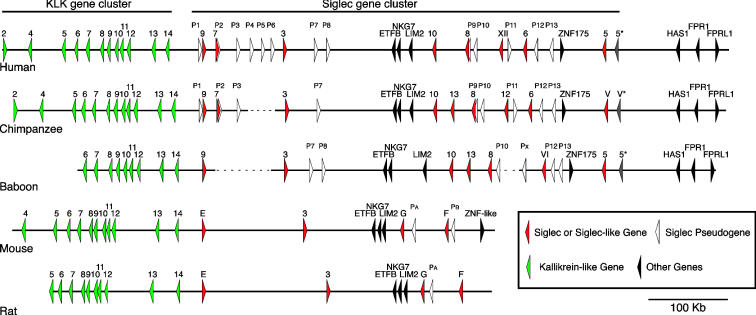
Fig. 2.
Comparisons of the Siglec gene cluster in human, chimpanzee, baboon, rat, and mouse. Order and arrangement of KLK-like genes (green triangles), the Siglec genes (red triangles), and pseudogenes (white triangles), as well as other genes (black triangles) in five genomes. The dotted lines represent regions (>30 kb in length) present in human genomes but absent in the chimpanzee and baboon genomes.
Siglec Nomenclature. The Siglec nomenclature used here (Table 1) is based on the current study, prior literature, our unpublished observations, and available genomic sequences, as well as continuing discussions led by A.V. among an international panel of interested scientists. Because human Siglecs were the first to be fully characterized due to the availability of complete genomic sequence, the numbering system originated therein. Additional non-human Siglecs are numbered systematically as one moves away from the human lineage. Because the last functional Siglec identified in the human genome was Siglec-11, the functional chimpanzee ortholog of the original human Siglec-L1 is herewith named chimpanzee Siglec-12. By agreement within the international group, Siglecs lacking the essential arginine required for Sia recognition are numbered by using corresponding roman numerals. Thus, human Siglec-L1 (the ortholog of chimpanzee Siglec-12 lacking the essential arginine) is renamed Siglec-XII. Likewise, other Siglecs lacking the essential arginine are assigned a corresponding roman numeral to the functional ortholog in related taxa (chimpanzee Siglec-V and baboon Siglec-VI). The additional Siglec gene in the chimpanzee and baboon CD33rSiglec clusters, which appears to be deleted in humans (see below), is named Siglec-13. There is an additional potential Siglec gene at the 3′ end of the Siglec cluster in primate genomes labeled 5* (or V*) in Fig. 2, which shows extensive sequence identity (>99%) with a segment of SIGLEC5. Although there is a cDNA sequence corresponding to this potential gene (GenBank accession no. AY358369), protein expression and sialic acid binding have yet to be demonstrated.
Table 1. Probable orthologous correspondences of CD33-related Siglecs in the five mammalian species compared in this study.
| Human | Chimpanzee | Baboon | Mouse | Rat |
|---|---|---|---|---|
| CD33rSiglecs found within the Siglec gene cluster | ||||
| CD33/Siglec-3 | CD33/Siglec-3 | CD33/Siglec-3 | CD33/Siglec-3 | CD33/Siglec-3 |
| Siglec-5 | *Siglec-V | Siglec-5 | Siglec-F | Siglec-F |
| Siglec-6 | Siglec-6 | *Siglec-VI | ||
| Siglec-7 | Siglec-7 | NF | NF | NF |
| Siglec-8 | Siglec-8 | Siglec-8 | NF | NF |
| Siglec-9 | Siglec-9 | Siglec-9 | Siglec-E | Siglec-E |
| Siglec-10 | Siglec-10 | Siglec-10 | Siglec-G | Siglec-G |
| *Siglec-XII | Siglec-12 | NF | NF | NF |
| NF | Siglec-13 | Siglec-13 | NF | NF |
| CD33rSiglecs found outside of the Siglec gene cluster | ||||
| Siglec-11 | Siglec-11 | ? | NF | NF |
| NF | NF | ? | Siglec-H | *Siglec-H |
These data are based on the current paper, our unpublished observations, past literature, and other genomic information available on-line to date (including chimpanzee and rat high-throughout genomic sequences in the GenBank database). Criteria used to assign orthologs include sequence similarity, map location, gene structure, and phylogenetic relationships. NF, the corresponding Siglec was not found in available data in this species; *, Siglec-like molecules missing the arginine residue required for optimal Sia recognition; ?, published genomic information to date is not sufficient to determine status.
Because there were fewer rodent CD33rSiglecs, and orthologous correspondence is not easily established (see below), they have been given an alphabetical nomenclature.
The Siglec Cluster Has Fewer MCSs. Using recently established methods (21), 750 MCSs representing the top 5% of most conserved sequence were identified (Table 2; see Supporting Text for detailed information on the overall distribution of MCSs in the genomic region under study). We divided this region into either Siglec- or non-Siglec-containing subregions (representing 415 and 315 kb, respectively; see Supporting Text). Despite the fact that the Siglec-containing subregion is larger, it contains less than half the number of MCS bases compared to the non-Siglec-containing subregion (12,390 vs. 24,917 bases, respectively). Low conservation in the Siglec-containing subregion, i.e., rapid evolution of the Siglec genes, is also observed when looking specifically at this subregion's coding and untranslated sequence as well (see Supporting Text).
Table 2. Distribution of MCSs in the sequenced region.
| Total | Siglec coding | Non-Siglec coding | |
|---|---|---|---|
| Sequence length | 730,000 nt (100%) | 415,000 nt (56.8%) | 315,000 nt (43.2%) |
| Number of bases contained in the MCSs | 37,307 nt (100%) | 12,390 nt (33.2%) | 24,917 nt (66.8%) |
The difference between Siglec-coding and non-Siglec-coding subregions in the ratio of MCS bases per total sequence length was statistically significant (P < 0.001 by the χ2 test).
Deletion of Siglec Genes or Pseudogenes. The baboon Siglec cluster (see Fig. 2) lacks orthologs of SIGLEC7 and SIGLEC12. However, the baboon genome has a remnant of SIGLEC7, and the duplication separating SIGLEC7 from SIGLEC12/XII may have predated the split of the monkey–ape lineages, judging from the topology of the phylogenetic tree reconstructed by using sequences of the C-terminal portion of Siglecs (Fig. 3C). Thus, the genes for Siglec-7 and Siglec-12 appear to have been lost in the ancestry of baboons after the split of the Old World monkey and ape lineages.
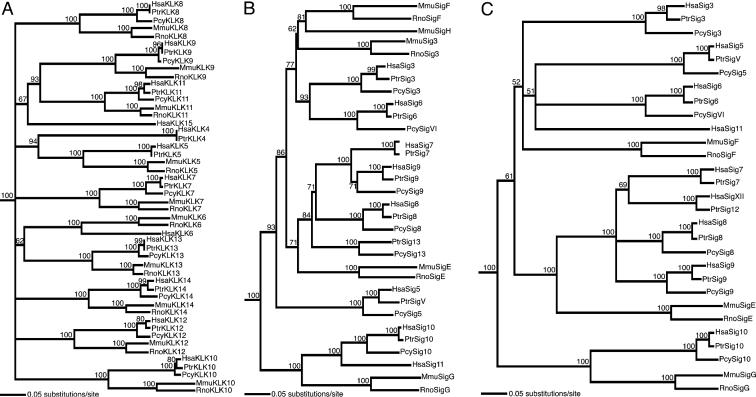
Fig. 3.
Molecular phylogenetic analyses of KLK-like molecules and Siglecs. Although human Siglec-11 and mouse Siglec-H genes are outside the cluster sequenced here, they were included in this analysis for comparison. (A) KLK-like molecules (full-length; 357 aa). Human KLKs1–3 were used together as an outgroup (not shown). Chimpanzee and baboon KLK6 were not included in the analysis because of insufficient sequence data for this gene. (B) Siglec N-terminal regions (signal peptide, Ig1 and Ig2, and linker peptide between Ig2 and Ig3; 291 aa). Primate Siglec-12/XII was excluded because it has two V-set domains and cannot be aligned with other Siglecs. (C) Siglec C-terminal regions (transmembrane domain and cytosolic tail; 184 aa). Rodent Siglec-3, Siglec-H, and primate Siglec-13 were excluded because these have much shorter cytoplasmic domains than other Siglecs. Amino acid sequences were used for calculating the distance matrix and reconstruction of the phylogenetic trees by using the neighbor-joining method. The bootstrap support value for each internode (as percent for 1,000 replications) is indicated above it. Siglec-4 was used as an outgroup for the trees in B and C. Note that Siglec-4 is not a CD33rSiglec but shows overall structural similarity to them, especially at the C-terminal region.
A previously unknown Siglec gene is present in both the chimpanzee and baboon genomes but is apparently deleted from the human genome (Fig. 2). We found that the chimpanzee version recognizes Sia when expressed as a recombinant protein (data not shown), confirming that it is a bona fide Siglec, hence designated as Siglec-13.
Another difference is that the segment containing Siglec pseudogenes P4–P6 or P3–P6 in the human genome is missing in chimpanzee and baboon genomes, respectively (Fig. 2). Because the overall segment does not show extensive sequence similarity to any other part of the human genome, it is unlikely to have been a very recent duplication in the ancestry of human after the split from the chimpanzee/bonobo clade. Rather, it is more likely to have been deleted in the ancestry of the chimpanzee and of the baboon independently. Further studies are needed to address this issue.
Expansion and Diversification of the CD33rSiglec Subgroup in Primates. Genes for the KLK-like proteins (KLK4–KLK14) are well conserved, and orthologs (as judged from genomic location in the five species) show the expected branch topology in the molecular phylogenetic tree, i.e., mouse joins to rat, human to chimpanzee, and the human–chimpanzee clade to baboon (Fig. 3A). Thus, all of the duplications of KLK genes in the sequenced region predated divergence of rodent and primate lineages, and these genes have not undergone processes that homogenize paralogous sequences (such as gene conversion) or lineage-specific loss or gain afterward.
Putative Siglec orthologs among the three primates or between the two rodents, as judged from genomic locations, also show the expected branch topology (Figs. 2 and 3 B and C). However, with the exception of primate Siglec-10 and rodent Siglec-G clades, clear-cut orthologous correspondence between primate and rodent Siglecs cannot be established and in some cases is incongruent with the putative orthology based on the map position and gene structure.
Primate Siglec-10 and rodent Siglec-G clades show the phylogenetic relationship consistent with their presumed orthology, as judged from genomic location. Possible gene duplication or gene conversion involving SIGLEC10, giving rise to SIGLEC11 in primates (located outside of the genomic region being studied in this paper), was discussed in a previous study (28).
Although Siglec-E is the only rodent Siglec with three Ig-like domains, the three primates have three to five such Siglecs (Siglec-7, -8, -9, -12, and -13), suggesting that gene duplications involving this subgroup of Siglecs happened in primate lineage after the divergence of primate and rodent lineages. Species- or lineage-specific deletion of some of the genes may have followed, as explained above.
Primate and rodent CD33/Siglec-3 should be orthologous, judging from the gene location and structure (CD33/Siglec-3 is the only Siglec with two Ig-like domains in the cluster), and the same applies to the primate and rodent Siglecs with four Ig-like domains (primate Siglec-5 and -6 and rodent Siglec-F). However, these fail to show the expected phylogenetic relationship (Fig. 3B). The observed tree topology may be explained by the high degree of homoplasy (convergent or parallel nucleotide substitution) among these genes, partial gene duplication followed by reciprocal recombination with another Siglec gene, or partial gene conversion, among other possibilities.
Examination of incongruities between phylogenetic trees reconstructed from different parts of the Siglec molecules (Fig. 3 B and C and Fig. 5, which is published as supporting information on the PNAS web site), along with analyses of gene structures, genomic locations, and comparison of intron sequences, suggest the occurrence of exon/domain shuffling and generation of hybrid genes among CD33rSiglecs (see Supporting Text and Fig. 6, which are published as supporting information on the PNAS web site, for an example). However, providing proof for every such possible case is beyond the scope of this paper.
Rapid Accumulation of Nonsynonymous Substitutions in the First Ig-Like Domain of Siglecs. Our prior comparisons of human, chimpanzee, bonobo, and gorilla Siglec-9 sequences indicated domain-specific rapid evolution in the first Ig-like domain, with a functional consequence in humans, i.e., accommodation of the binding site for the loss of Neu5Gc, the preferred ligand of chimpanzee Siglec-9 (12). Using the current dataset, we performed similar comparisons of human, chimpanzee, and baboon CD33rSiglec orthologs. As summarized in Table 4, which is published as supporting information on the PNAS web site, there are indeed examples where the pN/pS ratio is >1 (such as the human–chimpanzee Siglec-5 pair). However, in no case did Fisher's exact test reject the null hypothesis that the higher nonsynonymous substitution frequency compared to the synonymous ones is due to chance. To achieve a more robust statistical analysis, we concatenated all CD33rSiglec Ig1-coding exon in a species and compared it with a counterpart in another species (see Methods for details). χ2 tests did not support the hypothesis that pN is significantly higher than pS in the Ig1 of CD33rSiglecs. We also calculated lineage-specific substitution rates via reconstruction of ancestral Ig1-coding exon sequence for each human–chimpanzee ortholog pair and counting the differences between the ancestral and extant sequences. In both human and chimpanzee lineages, pN/pS in the concatenated Ig1-coding exons was >1 (human, 2.15; chimpanzee, 1.78), i.e., Ig1 of Siglecs accumulated nonsynonymous substitution at a higher rate than synonymous ones in both lineages. However, the null hypothesis pN = pS was not rejected by the χ2 test in either case.
Nevertheless, we observed pN ≥ pS in the Ig1 of all three primate pairs compared in this study. In striking contrast, we found that pN was consistently much smaller than pS in the similarly concatenated Ig2- and Ig3-coding exons of CD33rSiglecs (Table 3), indicating that these domains are under purifying selection. Furthermore, comparison of pN in the Ig1-versus either Ig2- or Ig3-coding exons by a χ2 test revealed statistically significant differences (P < 0.05) in almost all cases. This is in contrast to near-constant pS in all these domains. Thus, the frequency of fixation of nonsynonymous substitution is significantly and consistently higher in the Ig1 than in adjacent domains of Siglecs, reflecting domain-specific rapid evolution of the Ig1, which happens to contain the Sia recognition site.
Table 3. Differential evolution rates of exons encoding Ig domains of siglecs.
| Exon | Sd | Nd | S | N | pN/pS |
|---|---|---|---|---|---|
| Human—chimpanzee comparison | |||||
| Ig1 | 7 | 42 | 784.25 | 2431.75 | 1.935 |
| Ig2 | 17 | 19 | 582.92 | 1631.08 | 0.399 |
| Ig3 | 14 | 11 | 494.74 | 1332.26 | 0.292 |
| Human—baboon comparison | |||||
| Ig1 | 47.34 | 150.66 | 576.26 | 1781.74 | 1.029 |
| Ig2 | 37 | 46 | 434.25 | 1227.75 | 0.440 |
| Ig3 | 33.25 | 44.75 | 353.08 | 942.92 | 0.504 |
| Chimpanzee—baboon comparison | |||||
| Ig1 | 57.84 | 181.16 | 670.74 | 2065.26 | 1.017 |
| Ig2 | 46.5 | 49.5 | 511.5 | 1426.5 | 0.382 |
| Ig3 | 36.75 | 50.25 | 416.41 | 1131.09 | 0.503 |
For analysis of the difference between pN and pS in each concatenated Ig-like domain, a 2 × 2 contingency table [consisting of (Sd, S-Sd) and (Nd, N-Nd)] was made for each domain and analyzed by the χ2 test. For analysis of the difference in pN between two Ig-like domains, a 2 × 2 contingency table [consisting of (Nd, N-Nd) in two domains] was made for each comparison and analyzed by the χ2. Similar analyses were performed for pS, as well.
Uneven Distribution of Amino Acid Changes in the Ig1. To gain insight into the nature and dynamics of the rapid accumulation of non-synonymous changes in the Ig1 of CD33rSiglecs, we mapped the amino acid differences between the human–chimpanzee Siglec ortholog pairs onto the known crystal structure of Siglec-7 (25). This analysis revealed several hotspots (Fig. 7, which is published as supporting information on the PNAS web site; secondary structural assignment is according to ref. 25): (i) the N-terminal region leading to the β-strand A; (ii) loop C-C′; (iii) loop C′-D; and (iv) the β-strand F, which harbors the essential arginine. Of these, regions i, ii, and iv may be relevant to glycan recognition, as suggested by previous in vitro mutagenesis studies (25, 29, 30). This result suggests that rapid evolution of the Ig1 of CD33rSiglecs may be related to the modification of these molecules to accommodate ongoing changes in host sialylation patterns.
There is also a trend that one of the two faces (including C-C′ and C′-D loops) of Ig1 is burdened by more amino acid changes than the other (Fig. 7 and Data Set 1, which are published as supporting information on the PNAS web site). It is also noteworthy that the other face carries an N-linked glycan site (Fig. 7), potentially making it inaccessible to other macromolecules (this N-glycosylation site is conserved in all CD33rSiglecs, except in Siglec-8 and 12/XII). If certain pathogens use Siglecs as cellular receptors (31), amino acid changes on the accessible face of the molecule may also be selected for, further driving the evolution of Ig1.
Species-Specific Changes in the Essential Arginine Residue Involved in Sia Recognition. All functional Siglecs have a conserved essential arginine residue in the Ig1 that is required for optimal Sia recognition (4). We previously noted that human Siglec-XII has a cysteine residue at this position, thus abrogating Sia recognition (32). Because the chimpanzee ortholog cSiglec-12 has arginine at this position and is bound preferentially to Neu5Gc in vitro, this replacement of arginine may be a rare event that was selected for following the human loss of Neu5Gc expression (32). Surprisingly, we found that the baboon Siglec-6 and chimpanzee Siglec-5 orthologs also have the essential arginine residue changed to leucine (R111L) and histidine (R119H), respectively. Furthermore, both these changes attenuate interaction with Sia-containing glycans by ≈80% in binding assays by using recombinant proteins (data not shown; they are therefore named baboon Siglec-VI and chimpanzee Siglec-V according to the currently proposed nomenclature).
Discussion
Here we report high-quality sequences of the chimpanzee, baboon, and rat CD33rSiglec gene clusters and, together with available human and mouse sequences, perform detailed multispecies sequence comparisons. These analyses yielded important insights about the evolution of these genes, including strong support for our hypothesis that CD33rSiglecs are undergoing rapid evolution via multiple genetic mechanisms, particularly in the Ig1 Sia-binding domains. This domain may be evolving rapidly to: (i) keep up with the constant evolution of bacterial pathogens, some of which carry Sias and are recognized by Siglecs; and/or (ii) keep up with changes in the host Sia-expression profile, which itself is evolving rapidly to escape other pathogens that recognize Sias; and/or (iii) evade as-yet-unknown pathogens that use Siglecs themselves as receptors. Definitive evaluation of these possibilities requires detailed analyses of the sialylation patterns of the species studied here, as well as detailed glycan-recognition specificity studies of all of the Siglecs under consideration.
Recent lower-resolution comparative studies of the initial draft sequence of the chimpanzee genome indicate that the CD33rSiglec gene cluster may be one of the most rapidly evolving loci in the entire genome (Tarjei Mikkelsen, personal communication). Although sequence identity between human and chimpanzee KLK-coding sequences (KLK4,5,7∼14) is 99.2% [7,866 nucleotides (nt)], it is 98.5% (12,267 nt) for Siglec-coding sequences (CD33/Siglec-3, Siglec-5∼10, and Siglec-12) and 98.2% (3,253 nt) for Ig1-coding sequences. There are some other examples of immunity-related clustered gene families [e.g., MHC, killer-cell inhibitory receptors (KIRs), and defensins] that also appear to be evolving rapidly (33–35). With the KIR genes, there is also evidence of domain shuffling (36). However, unlike some of these molecules, the CD33rSiglecs have so far not shown extensive sequence polymorphism within the human population (12). Thus, the process of speciation itself may be solely responsible for the observed between-species differences of CD33rSiglecs. The observed between-species differences reported here are also concordant with our recent finding that humans show distinct patterns of terminal sialylation differing in multiple tissues from those of chimpanzee, bonobo, gorilla, or orangutan (37).
This work also raises several issues for future exploration. First, additional sequence data from other mammalian orders (e.g., artiodactyls and carnivores) could allow reconstruction of ancestral rodent–primate Siglec sequences. Second, detailed comparison of glycan-binding specificities of Siglec orthologs in closely related species (such as the three primates examined here) could help elucidate the basis for rapid evolution of these genes. Third, it remains to be seen whether intrinsic or extrinsic Sia ligand changes are the evolutionary driving force behind the rapid evolution of the Ig1 of Siglecs.
Supplementary Material
Acknowledgments
We thank members of the National Institutes of Health Intramural Sequencing Center Comparative Sequencing Program for generating sequences reported here, particularly J. Idol, V. Maduro, S.-Q. Lee-Lin, M. Portnoy, A. Prasad, R. Blakesley, A. Young, G. Bouffard, B. Maskeri, J. McDowell, and P. Thomas. We also thank Beth Nickerson and Richard Gibbs (Baylor College School of Medicine, Houston) for initial collaborative efforts, and Morris Goodman, Tasha Altheide, and Toshi Hayakawa for helpful comments. This work was supported by U.S. Public Health Service Grants R01GM-32373 and P01 HL57345 (to A.V.) and by the National Human Genome Research Institute, National Institutes of Health for comparative sequencing (to E.D.G.).
Abbreviations: Ign, nth Ig-like domain of a Siglec; BAC, bacterial artificial chromosome; KLK, kallikrein; MCSs, multispecies conserved sequences; N, no. of nonsynonymous sites; Nd, no. of nonsynonymous differences; pN, nonsynonymous substitution rate (Nd/N); pS, synonymous substitution rate (Sd/S); S, no. of synonymous sites; Sd, no. of synonymous differences; Sia, sialic acid; Neu5Gc, N-glycolylneuraminic acid; CD33rSiglecs, CD33-related Siglecs.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database [accession nos. AC130782, AC130783, AC132069, AC133392, AC134953, and AC140096 (chimpanzee); AC130188, AC130272, AC130273, AC130786, and AC137058 (baboon); and AC134313, AC135541, and AC137057 (rat)].
References
- 1.Varki, A. (1993) Glycobiology 3, 97–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gagneux, P. & Varki, A. (1999) Glycobiology 9, 747–755. [DOI] [PubMed] [Google Scholar]
- 3.Baum, J., Ward, R. H. & Conway, D. J. (2002) Mol. Biol. Evol. 19, 223–229. [DOI] [PubMed] [Google Scholar]
- 4.Crocker, P. R. & Varki, A. (2001) Trends Immunol. 22, 337–342. [DOI] [PubMed] [Google Scholar]
- 5.Angata, T., Hingorani, R., Varki, N. M. & Varki, A. (2001) J. Biol. Chem. 276, 45128–45136. [DOI] [PubMed] [Google Scholar]
- 6.Angata, T. & Brinkman-Van der Linden, E. (2002) Biochim. Biophys. Acta 1572, 294–316. [DOI] [PubMed] [Google Scholar]
- 7.Ravetch, J. V. & Lanier, L. L. (2000) Science 290, 84–89. [DOI] [PubMed] [Google Scholar]
- 8.Vitale, C., Romagnani, C., Puccetti, A., Olive, D., Costello, R., Chiossone, L., Pitto, A., Bacigalupo, A., Moretta, L. & Mingari, M. C. (2001) Proc. Natl. Acad. Sci. USA 98, 5764–5769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nutku, E., Aizawa, H., Hudson, S. A. & Bochner, B. S. (2003) Blood 101, 5014–5020. [DOI] [PubMed] [Google Scholar]
- 10.Chou, H. H., Takematsu, H., Diaz, S., Iber, J., Nickerson, E., Wright, K. L., Muchmore, E. A., Nelson, D. L., Warren, S. T. & Varki, A. (1998) Proc. Natl. Acad. Sci. USA 95, 11751–11756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chou, H. H., Hayakawa, T., Diaz, S., Krings, M., Indriati, E., Leakey, M., Paabo, S., Satta, Y., Takahata, N. & Varki, A. (2002) Proc. Natl. Acad. Sci. USA 99, 11736–11741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sonnenburg, J. L., Altheide, T. K. & Varki, A. (2004) Glycobiology 14, 339–346. [DOI] [PubMed] [Google Scholar]
- 13.Angata, T. & Varki, A. (2002) Chem. Rev. 102, 439–470. [DOI] [PubMed] [Google Scholar]
- 14.Vimr, E. & Lichtensteiger, C. (2002) Trends Microbiol. 10, 254–257. [DOI] [PubMed] [Google Scholar]
- 15.Thomas, J. W., Touchman, J. W., Blakesley, R. W., Bouffard, G. G., Beckstrom-Sternberg, S. M., Margulies, E. H., Blanchette, M., Siepel, A. C., Thomas, P. J., McDowell, J. C., et al. (2003) Nature 424, 788–793. [DOI] [PubMed] [Google Scholar]
- 16.Boffelli, D., McAuliffe, J., Ovcharenko, D., Lewis, K. D., Ovcharenko, I., Pachter, L. & Rubin, E. M. (2003) Science 299, 1391–1394. [DOI] [PubMed] [Google Scholar]
- 17.Thomas, J. W., Prasad, A. B., Summers, T. J., Lee-Lin, S. Q., Maduro, V. V., Idol, J. R., Ryan, J. F., Thomas, P. J., McDowell, J. C. & Green, E. D. (2002) Genome Res. 12, 1277–1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wilson, R. K. & Mardis, E. R. (1997) Genome Analysis: A Laboratory Manual, eds. Birren, B., Green, E. D., Klapholz, S., Myers, R. M. & Roskams, J. (Cold Spring Harbor Lab. Press, Cold Spring Harbor, NY), Vol. 1, pp. 397–454. [Google Scholar]
- 19.Green, E. D. (2001) Nat. Rev. Genet. 2, 573–583. [DOI] [PubMed] [Google Scholar]
- 20.Schwartz, S., Elnitski, L., Li, M., Weirauch, M., Riemer, C., Smit, A., Green, E. D., Hardison, R. C. & Miller, W. (2003) Nucleic Acids Res. 31, 3518–3524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Margulies, E. H., Blanchette, M., Haussler, D. & Green, E. D. (2003) Genome Res. 13, 2507–2518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jeanmougin, F., Thompson, J. D., Gouy, M., Higgins, D. G. & Gibson, T. J. (1998) Trends Biochem. Sci. 23, 403–405. [DOI] [PubMed] [Google Scholar]
- 23.Rozas, J. & Rozas, R. (1999) Bioinformatics 15, 174–175. [DOI] [PubMed] [Google Scholar]
- 24.Nei, M. & Gojobori, T. (1986) Mol. Biol. Evol. 3, 418–426. [DOI] [PubMed] [Google Scholar]
- 25.Alphey, M. S., Attrill, H., Crocker, P. R. & Van, A. D. M. (2003) J. Biol. Chem. 278, 3372–3377. [DOI] [PubMed] [Google Scholar]
- 26.International Human Genome Sequencing Consortium (2001) Nature 409, 860–921. [DOI] [PubMed] [Google Scholar]
- 27.International Mouse Genome Sequencing Consortium (2002) Nature 420, 520–562. [DOI] [PubMed] [Google Scholar]
- 28.Angata, T., Kerr, S. C., Greaves, D. R., Varki, N. M., Crocker, P. R. & Varki, A. (2002) J. Biol. Chem. 277, 24466–24474. [DOI] [PubMed] [Google Scholar]
- 29.May, A. P., Robinson, R. C., Vinson, M., Crocker, P. R. & Jones, E. Y. (1998) Mol. Cell 1, 719–728. [DOI] [PubMed] [Google Scholar]
- 30.Yamaji, T., Teranishi, T., Alphey, M. S., Crocker, P. R. & Hashimoto, Y. (2002) J. Biol. Chem. 277, 6324–6332. [DOI] [PubMed] [Google Scholar]
- 31.Vanderheijden, N., Delputte, P. L., Favoreel, H. W., Vandekerckhove, J., Van Damme, J., van Woensel, P. A. & Nauwynck, H. J. (2003) J. Virol. 77, 8207–8215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Angata, T., Varki, N. M. & Varki, A. (2001) J. Biol. Chem. 276, 40282–40287. [DOI] [PubMed] [Google Scholar]
- 33.Vilches, C. & Parham, P. (2002) Annu. Rev. Immunol. 20, 217–251. [DOI] [PubMed] [Google Scholar]
- 34.Hughes, A. L. (2002) Immunol. Rev. 190, 161–168. [DOI] [PubMed] [Google Scholar]
- 35.Flajnik, M. F. & Kasahara, M. (2001) Immunity 15, 351–362. [DOI] [PubMed] [Google Scholar]
- 36.Rajalingam, R., Parham, P. & Abi-Rached, L. (2004) J. Immunol. 172, 356–369. [DOI] [PubMed] [Google Scholar]
- 37.Gagneux, P., Cheriyan, M., Hurtado-Ziola, N., Van der Linden, E. C., Anderson, D., McClure, H., Varki, A. & Varki, N. M. (2003) J. Biol. Chem. 278, 48245–48250. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.