Abstract
Broken chromosomes healed by de novo addition of a telomere are a major class of genome rearrangements seen in Saccharomyces cerevisiae and similar to rearrangements seen in human tumors. We have analyzed the sequences of 534 independent de novo telomere additions within a 12-kb region of chromosome V. The distribution of events mirrored that of four-base sequences consisting of the GG, GT, and TG dinucleotides, suggesting that de novo telomere additions occur at short regions of homology to the telomerase guide RNA. These chromosomal sequences restrict potential registrations of the added telomere sequence. The first 11 nucleotides of the addition sequences fell into common families that included 91% of the breakpoints. The observed registrations suggest that the 3′ end of the TLC1 guide RNA is involved in annealing but not as a template for synthesis. Some families of added sequences can be accounted for by one cycle of annealing and extension, whereas others require a minimum of two. The same pattern emerges for sequences added onto the most common addition sequence, indicating that de novo telomeres are added and extended by the same process. Together, these data indicate that annealing is central to telomerase registration, which limits telomere heterogeneity and resolves the problem of synthesizing Rap1 binding sites by a nonprocessive telomerase with a low-complexity guide RNA sequence.
Genomic instability is characteristic of many types of cancer (1). The extensive genome instability seen has suggested that cancer cells might acquire genetic defects that destabilize the genome leading to accumulation of genome rearrangements that activate oncogenes and inactivate tumor suppressor genes. This idea has gained support through the study of cancer susceptibility syndromes associated with increased genome rearrangements (2-5). Terminal deletions are frequently seen in tumor cells (1) and are present in ≈10% of inherited genetic diseases associated with chromosomal aberrations (6). To gain insights into the types of genetic defects that might destabilize the genome, we developed a Saccharomyces cerevisiae assay for genetic analysis of the accumulation gross chromosomal rearrangements (GCRs) that have one breakpoint within a 12-kb region of chromosome V (7). This assay has been used to identify numerous genes and pathways that suppress the accumulation of GCRs (7-15). Although rare in wild-type yeast strains, a frequent rearrangement in mutants with high rates of genome instability is a terminally deleted chromosome V with a new telomere added at the broken end (de novo telomere addition).
How telomerase maintains and synthesizes telomeres has been investigated in many organisms including S. cerevisiae (16). These studies identified proteins other than telomerase that protect telomeres from end joining reactions and activating checkpoints as well as target telomerase and facilitate telomere synthesis. Because of the heterogeneous nature of S. cerevisiae telomere sequences (17-19), most insights into how telomerase synthesizes telomeres have come from studies in which telomerase is used to extend oligonucleotide substrates that can anneal to telomerase RNA, TLC1 (20), and sequencing of bulk telomeres (21, 22). Other proteins affect telomere length; among these is the Pif1 DNA helicase (23). How telomerase synthesizes de novo telomeres at the ends of DNAs that do not contain telomeres is not well understood. Genetic studies have shown that de novo telomere addition require telomerase, Ku, and Cdc13, and is partially inhibited by Pif1 (10, 24-27). However, little is known about the properties that govern de novo telomere addition target sites. A previous study of telomeres added at the site of a HO-endonuclease-induced double strand break suggested that a telomere-like seed sequence was required as an organizer so that telomerase could add telomere sequences at distant sites (28); however, these studies were hampered by the small number of events analyzed. Here we have studied the sequences of 534 independent de novo telomeres and, based on their analysis, define the nature of de novo telomere addition targets and propose a mechanism for the de novo synthesis of telomeres.
Methods
The first and last identifiable breakpoints were identified from the sequence of chromosome V (www.yeastgenome.org) by using custom software. The last identifiable breakpoint was at the first mismatch between the sequenced isolate and database sequence. The first identifiable telomere nucleotide was derived from the last identifiable position as follows. From the T after the breakpoint, the position was moved in a 5′ direction if these sequences were telomeric repeats (TG1-3 or G1-3). The process was repeated if the position ended on a T. The junction sequence, which could be from either the telomere or chromosome V, is the sequence between the first and last identifiable nucleotides. Breakpoint feature statistics were derived by using the reference yeast genome sequence and the position of the breakpoints determined above.
Results
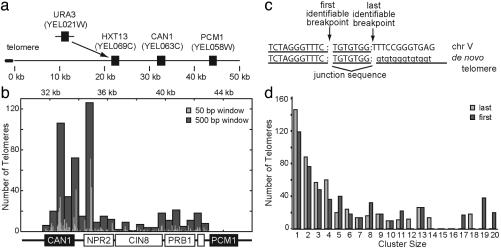
Previously, we identified 534 de novo telomere additions in a GCR assay (Fig. 1a) using a degenerate PCR approach to amplify the breakpoints for sequencing (7-15). Only one isolate was taken from each unique culture to avoid siblings of unique events. These telomere additions (Table 1, which is published as supporting information on the PNAS web site) are located throughout a 12-kb target region from CAN1 to the first essential gene PCM1 (Fig. 1b). Their distribution has hot spots and a bias against the centromeric side of the region, the significance of which is unclear. Defining the breakpoint by either the first identifiable telomere nucleotide or the last identifiable chromosome V nucleotide (Fig. 1c) reveals that each observed site is associated with approximately five events (Fig. 1d). By contrast, random targeting predicts one event per site on average.
Fig. 1.
Nonrandom distribution of de novo telomere additions. (a) The assay was generated by replacing HXT13 with URA3 in haploid strains, and GCRs were isolated by selection against CAN1 and URA3. Breakpoints must occur within or between CAN1 and the most centromeric essential gene, PCM1.(b) Histogram in which the breakpoints for the 534 de novo telomere additions isolates are displayed along chromosome V as the number of breakpoints present in 50-bp (light gray) and 500-bp (dark gray) windows. (c) Breakpoint sequences can be divided into three parts: sequences that are unambiguously chromosomal, sequences that could be chromosomal or telomere-derived (junction sequences), and sequences that are unambiguously telomeric. The first identifiable telomere nucleotide is the position between 1 and 2, and the last identifiable chromosomal nucleotide breakpoint is the position between 2 and 3. Three percent of telomeres are added to non-GT targets, so there is no junction sequence and the first and last identifiable positions are identical. (d) The 534 de novo telomere addition breakpoints are nonrandom. Cluster size is the number of times a specific site was targeted by de novo telomere addition, and the number of breakpoints found in each cluster size is plotted. When analyzed by last identifiable nucleotide (light gray), the average cluster has 4.9, with a minimum of one breakpoint and a maximum of 18; when analyzed by the first identifiable nucleotide (dark gray), the average is 6.5 and the range is from 1 to 20 breakpoints per nucleotide. A Poisson distribution predicts an average cluster size of 1.02 assuming each nucleotide is an equally likely target.
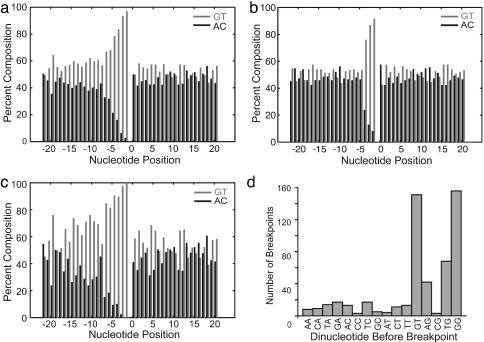
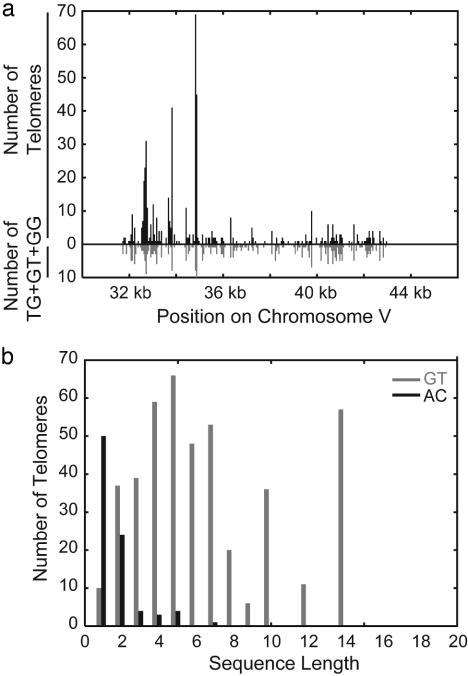
The breakpoint sequences have a 5- to 6-nt TG-rich bias (Fig. 2a). The subset of targets used only once shows a shorter bias (Fig. 2b), whereas targets used multiple times have a longer bias (Fig. 2c). The dinucleotides at the target sites show a preference for GG, GT, and TG found in normal telomeres and the reverse complement to TLC1 (Fig. 2d). The distribution of positions of four contiguous nucleotides composed of only GG, GT, or TG dinucleotides matches the distribution of telomere additions (Fig. 3a), whereas the distribution of positions of shorter or longer such sequences did not match as well (data not shown). Similarly, the most common targets are stretches of two to seven G or T nucleotides (Fig. 3b). Longer stretches of TG-rich DNA occur less often in chromosome V, but are used more frequently (Table 2, which is published as supporting information on the PNAS web site); such sequences may be more efficient or they may act as multiple adjacent targets. The most frequently target, the 14-mer 5′-GGGTGTTGTTGTGG, was involved in 50 events, and telomere additions occurred throughout this sequence.
Fig. 2.
TG bias at the sites of de novo telomere additions. The percentage of G + T (light gray) and A + C (dark gray) nucleotides present at each position relative to the site of de novo telomere addition as defined by the last identifiable nucleotide is graphed for all additions (n = 534) (a), additions at sites used only once (n = 146) (b), and additions at sites used four or more times (n = 243) (c). (d) Histogram of the individual dinucleotide 5′ to last identifiable breakpoint nucleotide method shows a bias toward GG, GT, and TG, but not TT. No such bias exists for histograms generated from the first identifiable breakpoint.
Fig. 3.
Targets sites are short GT-rich sequences. (a) The number of telomere additions in 50-bp windows (black bars above chromosome) between CAN1 and PCM1 compared to the number of four-base sequences made up of two adjacent TG, GT, or GG dinucleotides (e.g., four contiguous bases, gray bars below the chromosome). (b) The number of telomere additions at sequences containing solely G/T (gray) or A/C (black) nucleotides as a function of sequence length. The length includes bases that would be truncated if the breakpoint fell in the middle of the runs of G/T or A/C nucleotides. Despite the preference for G/T-rich stretches, the numbers of different G/T-rich and A/C-rich stretches between CAN1 and PCM1 are roughly identical and are close to distributions that would be predicted by random distribution (data not shown).
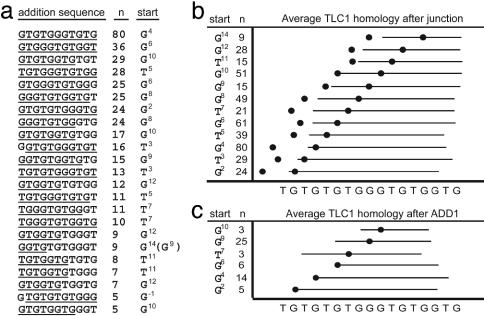
To analyze how breakpoint sequences direct telomere additions, breakpoints were examined for homology to each other and TLC1. Of the 534 telomere additions, 80% could be placed into 23 groups for which the first 11 nucleotides was observed five or more times (Fig. 4a); those used two or more times accounted for 91% of the telomeres seen. The most frequent addition sequence (5′-GTGTGGGTGTG) corresponds to 11 of the 17 nucleotides of the template region of the TLC1 RNA and has been called ADD1 (28). Each 11-nt addition sequence could be registered with the TLC1 template. The position influences the average homology preceding and following the breakpoint and reveals a number of features of telomere additions (Fig. 4b). First, each of the 11-nt sequences contains a substantial contiguous stretch of nucleotides that are complementary to the TLC1 RNA, regardless of the start point. Second, registrations starting in the latter half of the TLC1 homology have junction sequences that closely match (underlined) TLC1 (e.g., GAAGA:GTGG: G10TGTGGTGTGT). Third, registrations in the first half of TLC1 have little or no homology to TLC1 (underlined) in the junction sequence (e.g., GAAGA:GTGG:G4TGTGGGTGTG). However, in all such cases it was possible to identify an alternative registration to the second half of TLC1 such that there was contiguous homology between TLC1 and the region before the breakpoint (e.g., GAAGA:GTGG:G10TGTGGGTGTG); this did not allow contiguous alignment between TLC1 and the entire addition sequence (see below). Fourth, the TLC1 homologies for the addition sequences starting in the first half of the TLC1 homology often only extend until the second and third TG repeats of TLC1 (nucleotides 3-6), and tend not to include the first TG repeat (nucleotides 1-2) (Fig. 4b). Fifth, sequences added at the end of ADD1 are a subset of the sequence families added after the last breakpoint nucleotide (Fig. 4c; e.g., ADD1 followed by ADD1 GTGGA:G:GTGTGGGTGTGGTGTGGGTGTG); this suggests that telomeres are extended by sequential cycles of the same process. Overall, these results suggest that de novo telomere addition involves exact copying of TLC1 RNA and that the TLC1 template can be divided into different functional regions.
Fig. 4.
Addition sequence registration indicates precise TLC1 registration. (a) Twenty-three different 11-nt telomere addition sequences are observed five or more times after the last identifiable chromosome V breakpoint nucleotide and involve 80% (426 of 534) of all breakpoints. The longest homology to the TLC1 RNA template region is underlined. The most common sequence, seen 80 times, is the ADD1 sequence (28). The first nucleotide in TLC1 homology added after the breakpoint is indicated in the start column; for GGTGTGGGT, the sequence can either register with G14 or G9; however, the last junction nucleotide is T, suggesting that G14 is most likely the correct. (b) Telomere additions from a were grouped by start position. The right circle is the last identifiable breakpoint, which is placed by using the start position relative to the TLC1 homology. The left circle is the average position of the first identifiable breakpoint for all telomeres in the group. The horizontal line is the average TLC1 homology for all telomeres both before and after the last identifiable breakpoint. (c) The sequence families added after ADD1 are illustrated as in b and correspond to six of the families illustrated in b. The first nucleotide found after ADD1 is indicated by the circle, and the solid line represents the average length of homology with TLC1. It was possible to assign 56 of the 80 sequences; of the remaining, two aligned starting at nucleotide -1, one aligned starting at nucleotide -3, and the remaining 21 could not be assigned because insufficient sequence was available.
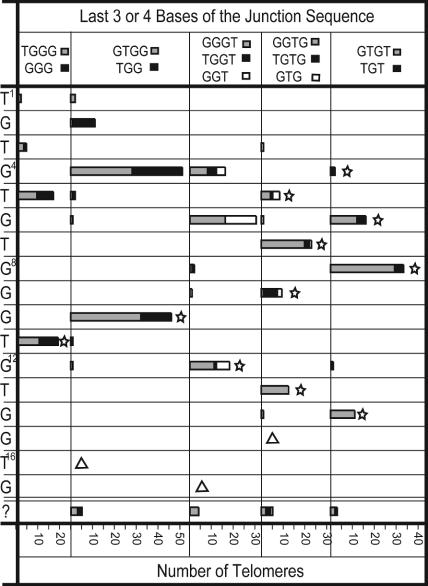
The telomere additions show considerable heterogeneity. No single target sequence directs a single telomere addition sequence, even when added to the same site (Table 3, which is published as supporting information on the PNAS web site) or when added to the junction sequence GTGG (Table 4, which is published as supporting information on the PNAS web site) previously suggested to force telomere registration (28). Despite this, the target sequence obviously dictates allowable TLC1 alignments (Table 1), which can be demonstrated by grouping telomere additions by the last three or four bases of the junction sequence (Fig. 5). For each group, registrations with TLC1 were assigned as for Fig. 4, which suggested a number of features. First, ≈50% of the telomere additions after GGG, TGG, and GGT can be explained by one round of annealing and simple copying, whereas the other 50% cannot. The second group, which includes ADD1 additions, has registrations before T7 with no homology between TLC1 and the junction sequence and all show another registration that includes the junction sequence. Thus, they can be explained by two cycles of annealing-extension-dissociation in which the first round of extension terminates before addition of G15 and/or T16 of the TLC1 homology, preventing them from placement in the first group. Furthermore, these groups are consistent, with only 50% of the TGGGTGT sequences in bulk telomeres being followed by GGT (21, 22). Second, if the 50% termination rate holds for all junction sequences, then registrations before T7 are mostly likely caused by multiple annealing-synthesis-dissociation cycles involving other parts of TLC1, suggesting that (TG)n sequences require multiple cycles and that the first half of the TLC1 homology (the 3′ end of the template) is used in annealing and not commonly for copying, similar to ciliate telomerases (29). Third, common addition sequences (Fig. 4a) can be accounted for by these addition distributions, even if S. cerevisiae telomerase is nonprocessive. Overall, these results support the idea that the mechanism of de novo telomere addition involves multiple cycles of annealing with three to five nucleotides at the 3′ end of the TLC1 template followed by relatively low-processivity synthesis (20, 30).
Fig. 5.
Heterogeneity in de novo telomere additions suggests both one- and two-step mechanisms. Telomere additions with at least three T or G nucleotides before the last identifiable breakpoint were analyzed by the potential position of the first nucleotide after the breakpoint within the TLC1 homology; each histogram is positioned at this first nucleotide. Registration was determined by using only sequence after the last identifiable breakpoint (“?” were not interpretable). Stars indicate addition registrations predicted based on annealing between TLC1 and the end of the junction sequence (sequence between the first and last breakpoint nucleotides; see Fig. 1c). Triangles indicate addition registrations that would be predicted based on annealing TLC1 with the junction sequence ends but are not observed. Surprisingly, many additions with GGG, TGG, and GGT before the last identifiable breakpoint are observed to initiate at positions lacking homology with the junction sequences (histograms without stars); however, these cases can be explained by two annealing-extension-disassociation cycles in which each annealing position is still controlled by three to four bases of homology of the 3′ end with the TLC1 template. A specific example is illustrated in Fig. 6.
Although many mutations cause substantial changes in the rate of telomere additions, we have not observed any effects on target site selection or target length. Targeting is independent of rearrangement rate (Fig. 7, which is published as supporting information on the PNAS web site) and grouping based on similar genetic defects (i.e., checkpoint defects, recombination defects, and pif-m2 mutations) (data not shown). Sufficient numbers of breakpoints exist to examine the effects of the pif1-m2, tel1, mec1, rad9, rad52, rdh54, sgs1, rfc5-1, lig4, and cac1 mutation backgrounds through paired genotypes (Fig. 8, which is published as supporting information on the PNAS web site). Despite the fact that pif1-m2 and tel1 affect telomere length, these mutations have little effect on targeting. New telomeres are added closer to a double-stranded break in pif1-m2 (24), and our data suggest that telomere additions avoid Pif1 inhibition randomly rather than by using longer TG-rich targets. Telomere additions in mec1, rad52, and rdh54 strains tend to occur on the CAN1 side of the breakpoint region. The TG-bias length is slightly shorter in the rad52 and rdh54 strains, possibly because of the elimination of a competing repair pathway, but this not observed in lig4 strains. The chromatin-assembly factor cac1 mutation (12) also causes a slightly shorter length of TG-bias at the breakpoint. However, all of these effects are subtle, as would be expected from the fact that mutations in telomerase specificity genes were not included in this study. The GCR assay is, however, capable of measuring altered specificities, as shown with yku80-135i that requires longer TG-rich targets (25).
Discussion
The 534 events resolve into patterns that provide a number of insights into apparently heterogeneous de novo telomere additions (17-19). Moreover, the chromosomal sequence provides a start position that allows for specifying functional regions of the TLC1 template, which is problematic for experiments that only sequence bulk telomeres (21, 22). Finally, these results indicate the central role of annealing in controlling registration, which resolves the apparent contradiction of common addition sequences (Fig. 4) and long TLC1-like stretches in bulk telomeres (21, 22) with the lack of processivity of telomerase (20).
De novo telomere additions were targeted to short three- to five-base sequences consisting of GG, GT, and TG dinucleotides that resembled telomere sequences, with longer sequences acting as hot spots. These addition targets are consistent with those suggested to be within the vicinity of telomere “organizing” sequences (24, 28). However, no organizers are within 241 kb of the region studied here (24), indicating that organizers are not essential for de novo telomere additions, in contrast to previous proposals (28). The selected targets may be the result of multiple specificities determined by short sequences that can anneal with TLC1 and possibly contain minimal Cdc13-binding sites (10, 31) and Est1 recognition sites. Our data cannot resolve whether broken target DNAs are resected to leave a target sequence at the very end of a 3′ overhang or whether annealing can occur at internal sequences followed by cleavage by the endonucleolytic activity of telomerase itself (20, 32, 33); however, only a small percentage target fragments are without TG-rich ends.
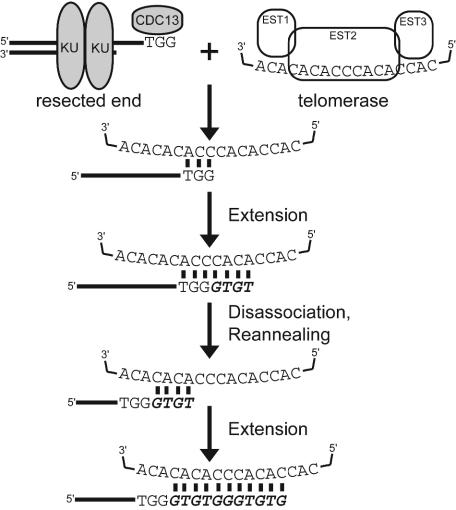
The importance of annealing in determining TLC1 registration is demonstrated by the fact that every telomere-like chromosome V target falls into an alignment with TLC1 that allows for direct homology (Fig. 4). A simple model of telomere synthesis accounts for common families of specific addition sequences. Some require only a single cycle of annealing and extension, whereas others like ADD1 (Fig. 6) require two annealing-extension-disassociation cycles. Moreover, two cycles are the minimum for generating ADD1. Common sequences, like ADD1, tend to use the favored annealing positions revealed by analysis of the registration of addition to each telomere-like chromosome V target (Fig. 5), could potentially be generated by many annealing-extension-disassociation cycles, and could even occur in different cell cycles. At an extreme, adding one nucleotide in each synthesis cycle by using only the most preferred annealing site for each new end could generate ADD1. This model contrasts with the initial single cycle mechanism for ADD1 synthesis (28); like our data, the original ADD1 additions lack homology between the target and TLC1 that allow single cycle synthesis. Our model accounts for other published de novo telomere additions at other genomic regions and integrated pBR322 sequences (24, 28), supporting its generality.
Fig. 6.
Proposed mechanism for de novo telomere addition illustrated by addition of ADD1 onto a TGG breakpoint by two cycles of annealing and synthesis. In vitro studies indicate telomerase requires a 3′ single-stranded substrate (16) that could be revealed by resection of the broken chromosome V (44). Initial annealing of TGG at the preferred location allows up to five bases in the first annealing-synthesis-dissociation cycle; synthesis of more than five bases would not generate an ADD1 addition sequence. Dissociation after either 10G10 or 10GT11 are added will generate new ends that will preferentially reanneal to the initial annealing registration (Fig. 5) and therefore give the appearance of high processivity through this region when analyzing only bulk telomeric sequences. On the other hand, dissociation of longer fragments generated by addition of 10GTG12, 10GTGT13, 10GTGTG14, or 10GTGTGG15 will generate new ends that will preferentially allow reannealing to a second, common registration on the TLC1 template (only the specific case of 10GTGT13 addition in the first cycle is illustrated). Synthesis in the second registration is sufficient to add final nucleotides of the 11-nt ADD1 sequence. The robustness of reannealing of potential intermediate sequences to the first and second registrations on the TLC1 template explains the high frequency of ADD1 additions (Fig. 4a).
These results provide important clues as to how the TLC1 template is used. The 3′ end of the TLC1 template appears to primarily anneal to the target and does not template synthesis. Synthesis frequently does not copy the 5′ end of the template, which may be due to low processivity combined with annealing preferences on the guide RNA. Thus, the portion of the TLC1 involved in templating synthesis appears to be between T7 and G17, indicating that multiple cycles of annealing and synthesis are required to generate long (TG)n sequences. The proposal that regions of TLC1 (corresponding to TGTGTGTGGGTGTGGTG) are not available for annealing (21) is not borne out by the patterns of registrations of added telomeres to the breakpoints (Fig. 4). That proposal assumes that annealing is unimportant, (TG)n sequences are synthesized at the 5′ end of the template, and processivity is higher in vivo than in vitro (20). In contrast, the data leading to these conclusions (21) can be explained by using our model involving poor processivity, multiple annealing-synthesis-dissociation cycles, and a crucial role for annealing in controlling TLC1 registration. If the 3′ end were available for synthesis and the central regions of the template were not available for annealing, then S. cerevisiae telomerase would be substantially different from ciliate telomerases (29). By contrast, our results suggest that yeast telomerase is much more similar to other telomerases. The dramatic heterogeneity of S. cerevisiae telomeres is a result of poor telomerase processivity (20) and multiple potential annealing sites within TLC1; however, our results suggest that annealing moderates telomere randomness and allows synthesis of sites for the Rap1 telomere-binding protein (22).
The three to five nucleotides of homology at these 534 chromosomal healing events closely resembles the short homologies identified in the small number of de novo telomere additions sequenced in other eukaryotes with less degenerate telomeres, including humans (34-36), mice (37), Plasmodium (38, 39), and wheat (40, 41). Importantly, in many cancer cells, telomerase is reactivated after chromosomal rearrangements induced by telomere dysfunction (42) and, in combination with other defects that lead to broken and ultimately rearranged chromosomes, this could drive de novo telomere additions. Indeed, terminally deleted chromosomes with telomeres at their ends have been observed in the karyotypes of cancer cells (43).
Supplementary Material
Acknowledgments
We thank Elizabeth Blackburn for suggesting that we analyze the relationship between de novo telomeres and TLC1 RNA and Meng-Er Huang and Tom Petes for comments on the manuscript. This work was supported by National Institutes of Health Grant GM26017 (to R.D.K.) and a postdoctoral fellowship from the Damon Runyon Cancer Research Foundation and the Robert Black Charitable Trust (to C.D.P.).
Abbreviation: GCR, gross chromosomal rearrangement.
References
- 1.Mitelman, F. (1991) Catalog of Chromosome Aberrations in Cancer (Wiley-Liss, New York).
- 2.Kolodner, R. & Marsischky, G. (1999) Curr. Opin. Genet. Dev. 9, 89-96. [DOI] [PubMed] [Google Scholar]
- 3.Thompson, L. & Schild, D. (2002) Mutat. Res. 509, 49-78. [DOI] [PubMed] [Google Scholar]
- 4.Mohaghegh, P. & Hickson, I. (2002) Int. J. Biochem. Cell Biol. 34, 1496-1501. [DOI] [PubMed] [Google Scholar]
- 5.Friedberg, E. (2001) Nat. Rev. Cancer 1, 22-33. [DOI] [PubMed] [Google Scholar]
- 6.Borgaonkar, D. S. (1989) Chromosomal Variation in Man (Liss, New York).
- 7.Chen, C. & Kolodner, R. (1999) Nat. Genet. 23, 81-85. [DOI] [PubMed] [Google Scholar]
- 8.Myung, K., Datta, A., Chen, C. & Kolodner, R. (2001) Nat. Genet. 27, 113-116. [DOI] [PubMed] [Google Scholar]
- 9.Myung, K., Datta, A. & Kolodner, R. (2001) Cell 104, 397-3408. [DOI] [PubMed] [Google Scholar]
- 10.Myung, K., Chen, C. & Kolodner, R. (2001) Nature 411, 1073-1076. [DOI] [PubMed] [Google Scholar]
- 11.Myung, K. & Kolodner, R. (2002) Proc. Natl. Acad. Sci. USA 99, 4500-4507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Myung, K., Pennaneach, V., Kats, E. & Kolodner, R. (2003) Proc. Natl. Acad. Sci. USA 100, 6640-6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Myung, K. & Kolodner, R. (2003) DNA Rep. 2, 243-258. [DOI] [PubMed] [Google Scholar]
- 14.Pennaneach, V. & Kolodner, R. (2004) Nat. Genet. 36, 612-617. [DOI] [PubMed] [Google Scholar]
- 15.Huang, M., Rio, A., Nicolas, A. & Kolodner, R. (2003) Proc. Natl. Acad. Sci. USA 100, 11529-11534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Blackburn, E. H. (1992) Annu. Rev. Biochem. 61, 113-129. [DOI] [PubMed] [Google Scholar]
- 17.Cohn, M., McEachern, M. & Blackburn, E. (1998) Curr. Genet. 33, 83-91. [DOI] [PubMed] [Google Scholar]
- 18.Shampay, J., Szostak, J. & Blackburn, E. (1984) Nature 310, 154-157. [DOI] [PubMed] [Google Scholar]
- 19.McEachern, M. & Blackburn, E. (1994) Proc. Natl. Acad. Sci. USA 91, 3453-3457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cohn, M. & Blackburn, E. (1995) Science 269, 396-400. [DOI] [PubMed] [Google Scholar]
- 21.Foerestemann, K. & Lingner, J. (2001) Mol. Cell. Biol. 21, 7277-7286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ray, A. & Runge, K. (2001) Nucleic Acid Res. 29, 2382-2394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schulz, V. & Zakian, V. (1994) Cell 76, 145-155. [DOI] [PubMed] [Google Scholar]
- 24.Manghas, J., Alexander, M., Sandell, L. & Zakian, V. (2001) Mol. Cell. Biol. 12, 4078-4089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stellwagen, A., Haimberger, Z., Veatch, J. & Gottschling, D. (2003) Genes Dev. 17, 2384-2395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Diede, S. & Gottschling, D. (1999) Cell 99, 723-733. [DOI] [PubMed] [Google Scholar]
- 27.Zhou, J., Monson, E., Teng, S., Schulz, V. P & Zakian, V. A. (2000) Science 289, 771-774. [DOI] [PubMed] [Google Scholar]
- 28.Kramer, K. & Haber, J. (1993) Genes Dev. 7, 2345-2356. [DOI] [PubMed] [Google Scholar]
- 29.Autexier, C. & Grieder, C. (1994) Genes Dev. 8, 563-575. [DOI] [PubMed] [Google Scholar]
- 30.Singer, M. & Gottschling, D. (1994) Science 266, 404-409. [DOI] [PubMed] [Google Scholar]
- 31.Anderson, E., Halsey, W. & Wuttke, D. (2003) Biochemistry 42, 3751-3758. [DOI] [PubMed] [Google Scholar]
- 32.Collins, K. & Greider, C. (1993) Genes Dev. 7, 1364-1376. [DOI] [PubMed] [Google Scholar]
- 33.Melek, M., Green, E. & Shippen, D. (1996) Mol. Cell. Biol. 16, 3437-3445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lamb, J., Harris, P., Wilkie, A., Wood, W., Dauwerse, J. & Higgs, D. (1993) Am. J. Hum. Genet. 52, 668-676. [PMC free article] [PubMed] [Google Scholar]
- 35.Wilkie, A., Lamb, J., Harris, P. C., Finney, R. & Higgs, D. (1990) Nature 346, 868-872. [DOI] [PubMed] [Google Scholar]
- 36.Flint, J., Craddock, C., Villegas, A., Bentley, D., Williams, H., Galanello, R., Cao, A., Wood, W., Ayyub, H. & Higgs, D. (1994) Am. J. Hum. Genet. 55, 505-512. [PMC free article] [PubMed] [Google Scholar]
- 37.Sprung, C., Reynolds, G., Jasin, M. & Murnane, J. (1999) Proc. Natl. Acad. Sci. USA 96, 6781-6786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mattei, D. & Scherf, A. (1994) Mutat. Res. 324, 115-120. [DOI] [PubMed] [Google Scholar]
- 39.Schraf, A., Carter, R., Petersen, C., Alano, P., Nelson, R., Aikawa, M., Mattei, D., Silva, L. d. & Leech, J. (1992) EMBO J. 11, 2293-2301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tsujimoto, H., Yamada, T. & Sasakuma, T. (1997) Proc. Natl. Acad. Sci. USA 94, 3140-3144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tsujimoto, H., Usami, N., Hasegawa, K., Yamada, T., Nagaki, K. & Sasakuma, T. (1999) Mol. Gen. Genet. 262, 851-856. [DOI] [PubMed] [Google Scholar]
- 42.Maser, R. & DePinho, R. (2002) Science 297, 565-569. [DOI] [PubMed] [Google Scholar]
- 43.Kawai, K., Viars, C., Arden, K., Tarin, D., Urquidi, V. & Goodison, S. (2002) Genes Chromosomes Cancer 34, 1-8. [DOI] [PubMed] [Google Scholar]
- 44.White, C. & Haber, J. (1990) EMBO J. 9, 663-673. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.