Abstract
Major histocompatibility complex class I molecules present peptides of 8-10 residues to CD8+ T cells. We used 19 predicted proteomes to determine the influence of CD8+ T cell immune surveillance on protein evolution in humans and microbial pathogens by predicting immunopeptidomes, i.e., sets of class I binding peptides present in proteomes. We find that class I peptide binding specificities (i) have had little, if any, influence on the evolution of immunopeptidomes and (ii) do not take advantage of biases in amino acid distribution in proteins other than the concentration of hydrophobic residues in NH2-terminal leader sequences.
The evolution of class I molecules of the MHC enabled the adaptive immune system of jawed vertebrates to detect and destroy intracellular pathogens and malignant cells. Class I molecules bind peptides of 8-11 residues generated intracellularly and carry them to the cell surface for perusal by CD8+ T cells, the major effector arm of antigen-specific cell-mediated immunity. Nearly all cell types constitutively express class I molecules, which in the absence of pathogens are loaded exclusively with cellular peptides. Most of these peptides seem to originate from DRiPs, defective forms of nascent proteins that are rapidly degraded after their synthesis (1, 2). The fact that all cellular proteins can potentially be processed into class I binding peptides raises the question of whether class I specificity has influenced, or has been influenced by, the vertebrate proteome. It is possible, for example, that class I molecules take advantage of structural motifs in proteins. In this case, class I peptide-binding motifs would be expected to be overrepresented in the proteome relative to the random occurrence of such motifs in proteins predicted by amino acid frequencies. Conversely, class I binding motifs might be negatively selected in proteins to reduce binding of self-peptides and simplify the task of tolerance induction. Similarly, it is of interest to determine whether class I specificity has influenced the proteome of pathogens, particularly those under intense selection pressure from TCD8+. Here, we introduce the concept of the immunopeptidome, the set of class I binding peptide ligands contained in a proteome. We examine the predicted immunopeptidomes of a number of vertebrate and nonvertebrate organisms and vertebrate pathogens to determine the influence of class I binding specificity on protein evolution.
Methods
The protein sequences for human and mouse were those predicted from the Celera versions of the two genomes (3, 4). Protein data for several classes of human viruses were collected from GenBank by using the Genomes section of Entrez (5). These included eight types of human herpesviruses (simplex viruses HSV-1 and HSV-2, varicellovirus VZV/HHV-3, lymphocryptovirus EBV/HHV-4, cytomegalovirus HCMV/HHV-5, roseoloviruses HHV-6 and HHV-7, and rhadinovirus HHV-8), with genome sequences varying between 125 kb and 230 kb and containing between 72 and 204 annotated proteins. In addition, protein sequences for the human adenoviruses A, B, C, D, E, and F, and for 87 strains of the human papillomaviruses, were obtained from the same source. These totaled 199 adenovirus proteins and 577 human papillomavirus proteins. Last, we downloaded the protein sequences from HIV-1 and HIV-2 strains from the ftp site of the HIV Database at the Los Alamos National Laboratory and selected only those in entries annotated as complete genomes (82 HIV-1 genomes totaling 686 proteins and 11 HIV-2 genomes with a total of 94 proteins). In addition to viruses, a number of bacterial and eukaryotic pathogens and nonpathogens of humans were also extracted. Protein sequences resulting from near-final annotation of Staphylococcus aureus strain COL were extracted from The Institute of Genomic Research Comprehensive Microbial Resources database (6). Furthermore, the sequences for Mycobacterium tuberculosis H37Rv (7), Methanococcus jannaschii (8), and Caenorhabditis elegans (9) were collected from the Genomes section of Entrez, as described above. Protein sequences inferred based on the most recent Drosophila melanogaster annotation (10) were obtained from Celera's internal database.
To test the possible correlations between the locations of single nucleotide polymorphisms (SNPs) and those of class I epitopes in the proteome, we used the test of homogeneity (P = 0.05, df = 1) for the hypothesis that SNPs are distributed homogeneously in the epitope and nonepitope regions. The tests were performed for each allele individually and for their combination, and separately for synonymous, nonsynonymous, and combined SNPs. We used the set of Celera cSNPs, a curated nonredundant data set consisting of cSNPs collected from the hgmd (11) and dbsnp (12) public databases, or extracted from the Celera proprietary fragment data. This provided us with 47,424 nonsynonymous SNP locations, 27,320 synonymous ones, and 672 positions harboring both SNP types.
To predict T cell ligands, we used the ligand prediction pipeline presented in Florea et al. (13). The pipeline uses a set of four algorithms to predict which peptides are ligands based on examples of known ligands; the results of the four algorithms are combined by using a voting heuristic. High-binding peptides were predicted from these voting-based scores by comparison with known ligands from the mhcpep database that had been classified into high (H), moderate (M), or low (binders) based on IC50 values, where high corresponds to an IC50 value of <50 nM, medium corresponds to an IC50 value between 50 and 500 nM, and low to corresponds to an IC50 value of >500 nM. We classified as high binders those peptides whose voting scores exceeded the highest voting score such that more medium binders were found above it than high binders below it. For comparison with the method of Parker et al. (14), we used the matrices made available by U.S. National Institutes for Health BioInformatics and Molecular Analysis Section (BIMAS; http://bimas.dcrt.nih.gov/molbio/hla_bind) followed by the same procedure as described above for locating high-binding peptides.
To obtain randomized proteomes of the various organisms, the program shuffleseq, publicly available through the European Molecular Biology Open Software Suite (http://www.hgmp.mrc.ac.uk/Software/EMBOSS), was used to randomly permute the order of amino acids in each protein while preserving the length and amino acid composition of the original sequence. Last, to test statistical significance of differences between shuffled and unshuffled human genomes for each allele, we pooled the two shuffled genomes and performed a χ2 test of independence on the two-by-two table of peptides predicted to be ligands versus peptides predicted not to be ligands for shuffled and unshuffled genomes.
Results
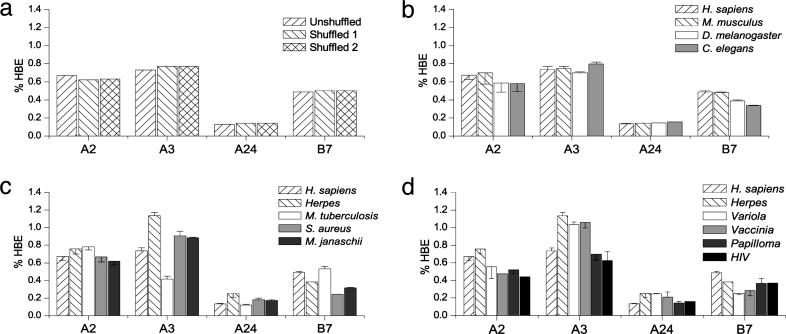
We predicted class I peptide ligands for four of the nine HLA super types by using newly devised algorithms that outperform published algorithms in predicting known ligands (13). These four super types encompass the specificities of a significant fraction of class I gene products encoded by humans (15). Fig. 1 shows the fraction of peptides encoded by the predicted human transcripts in the Celera annotation of the human genome assembly Release 26 (44,448 ORFs), which is predicted to bind with high affinity to the indicated class I superfamily members. To determine whether this represents positive or negative selection for peptide binding, we predicted the frequency of ligands in two artificial proteomes created by randomly permuting amino acids in each human ORF. These shuffled proteomes consist of the same sets of proteins but with random arrangements of the same amino acids in each protein. The two shuffled proteomes varied noticeably less from one another than either did from the unshuffled proteome. Although the relative values were similar in all cases, statistically significant differences existed according to a χ2 comparison of predicted ligand counts in shuffled versus unshuffled genomes (P value of <0.005 for all alleles), particularly in A2-binding peptides, which were overrepresented by 7.2%, and A3-binding peptides, which were underrepresented by 5.2%. To preserve structural characteristics of the protein, we also tried permuting only hydrophilic residues among each other and hydrophobic residues among each other. On the whole, the simulated human Refseq proteins had a very similar A2-binding profile as the shuffled proteins. We also examined whether a bias in class I binding peptides would appear in peptides known to be encoded by polymorphic regions relative to those encoded by nonpolymorphic ones as a means of testing whether sequences containing epitopes might be under greater evolutionary pressure. The analysis found no significant bias in either direction (data not shown). We extended our analysis to the immunopeptidome of another mammal, Mus musculus. The frequency of predicted human class I ligands was highly similar to the frequency in the human immunopeptidome, although the frequency of A2 ligands was even more positively biased, fully 18% more than predicted based simply on a random distribution of amino acids in individual proteins. Human and murine class I molecules show different, although slightly overlapping, peptide specificities, which has been interpreted as the result of convergent evolution (16). Hence, whereas the comparison of the human and murine immunopeptidomes supports the interpretation that evolutionary pressure has not influenced the human immunopeptidome, it could also be explained by convergent evolution of class I specificity.
Fig. 1.
Immunopeptidomics of humans and their pathogens. (a) Comparison of the percentage of peptides predicted to bind MHC (percentage of high-binding epitopes, % HBE) in the predicted human proteome and two artificial proteomes generated by randomly permuting the amino acids in each predicted ORF of the human proteome. The figure displays results for the MHC class I alleles A2, A3, A24, and B7. (b) Comparison of the percentage of peptides predicted to bind MHC in the predicted proteomes of H. sapiens, M. musculus, D. melanogaster, and C. elegans. The figure displays results for the MHC class I alleles A2, A3, A24, and B7. Error bars show the deviation between measured results and the results derived from artificial proteomes created by randomly permuting all residues in each ORF. (c) Comparison of the percentage of peptides predicted to bind MHC in the predicted proteomes of H. sapiens, herpesviruses, M. tuberculosis, S. aureus, and M. janaschii. The figure displays results for the MHC class I alleles A2, A3, A24, and B7. Error bars show the deviation between measured results and the results derived from artificial proteomes created by randomly permuting all residues in each ORF. (d) Comparison of the percentage of peptides predicted to bind MHC in the predicted proteomes of H. sapiens, herpesviruses, human papillomaviruses, HIV, vaccinia, and variola. The figure displays results for the MHC class I alleles A2, A3, A24, and B7. Error bars show the deviation between measured results and the results derived from artificial proteomes created by randomly permuting all residues in each ORF.
To further understand the potential significance of these biases in the human immunopeptidome, we turned to two multicellular organisms, D. melanogaster and C. elegans. As invertebrates, these organisms lack an adaptive immune system and therefore do not express class I molecules or functional homologs. Because these organisms do not parasitize vertebrates, they have evolved independently of the cellular immune system. As seen in Fig. 1, their immunopeptidomes showed nearly the identical biases to those of human and mouse in the frequencies of predicted class I ligands. This finding indicates that the biases we observed in humans and mice do not reflect an influence of the MHC on protein evolution to favor or disfavor class I binding peptides. Rather, the excess of A2-binding peptides must be due to the nonrandom distribution of binding residues in protein domains.
Relative to the other three super types tested, the A2 super type favors the binding of hydrophobic peptides, which are overrepresented in the signal sequences used to target proteins to the secretory compartment and transmembrane regions. To examine the effect of these domains on the frequency of A2-binding peptides, we identified likely endoplasmic reticulum-targeted proteins by the presence of a predicted NH2-terminal signal sequence (24.7% of the proteome) and calculated the frequencies of peptides in signal sequences, nonsignal portions of signal sequence-containing proteins, and proteins lacking signal sequences altogether. This revealed that the frequency of A2-binding peptides in nonsignal sequence proteins matched the randomized sequence. Most of the bias in signal peptide-containing proteins occurred within the signal sequence itself (6.5-fold increase), with the remainder of the A2 protein sequences exhibiting only a slight bias (1.3-fold). Extending this analysis to the other three super types revealed a surprisingly strong bias (2.9-fold) in B7 ligands for location in signal sequences and a slight bias (1.5- to 1.7-fold) for A3 and A24 super-type ligands.
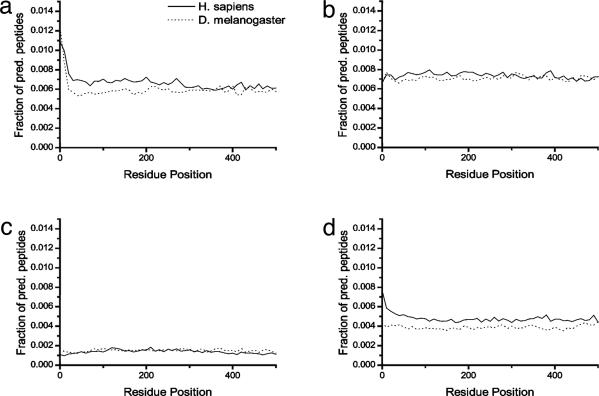
We further analyzed position-related differences in class I ligand frequencies by plotting the frequency of predicted peptides versus position in their respective proteins (Fig. 2). This confirmed the bias toward A2 and B7 peptides in signal sequences, which otherwise were distributed fairly evenly throughout the rest of the proteins, although A2 peptides were slightly less frequent at the COOH terminus. A3 peptides were evenly distributed within proteins, whereas A24 peptides were slightly less likely (≈1.5-fold) to reside at either terminus. Similar biases were found in D. melanogaster proteins (Fig. 2), demonstrating that they reflect predilections for combinations of amino acids in certain regions of proteins that have not been influenced by coevolution with class I genes.
Fig. 2.
Peptides as a function of position in protein. Fraction of predicted high-binding peptides as a function of the position of the first residue of the peptide within its respective protein. Each data point represents the sum of 10 consecutive residue positions to reduce noise in the plots. (a) A2. (b) A3. (c) A24. (d) B7.
Our findings indicate that human class I binding has exerted little if any effect on the human proteome. But what of pathogens? Given the role of CD8+ T cells in controlling viruses and other intracellular pathogens, avoidance of T cell recognition in combination with high mutation rates may result in a decrease in the frequency of binding peptides. We therefore examined the immunopeptidomes of a number of important human viruses, including the eight human herpesviruses, variola virus, and vaccinia virus (smallpox agent and vaccine), HIV, and human papilloma virus. In response to TCD8+ selection, herpesviruses have evolved numerous proteins that interfere with TCD8+ activation and function. Despite the evidence that they have evolved to evade human immune surveillance, herpesviruses analyzed either as a group (Fig. 1) or individually (data not shown) failed to demonstrate a significant depletion in class I binding peptides as compared with their shuffled proteome. The same was found for the poxviruses, papilloma virus, and HIV (Fig. 1). The latter result is particularly revealing because TCD8+ has been documented to influence the evolution of selected simian immunodeficiency virus and HIV determinants in some infected individual monkeys and patients (17). Our findings suggest that such selection has little effect on the presence of class I binding peptides in HIV.
We extended this analysis to two pathogenic bacteria, M. tuberculosis and S. aureus, and to the thermophilic archeobacterium M. janaschii, which has evolved to live at extreme temperatures and pressures and is therefore incapable of being pathogenic in humans. Although considerable variation occurred in the number of predicted determinants between the three microorganisms, there was no consistent pattern across the various HLA super types (Fig. 1). Furthermore, the differences in ligand frequencies were consistent with differences in amino acid content, as shown by comparison of predicted ligand frequencies with that predicted for the randomized sequences.
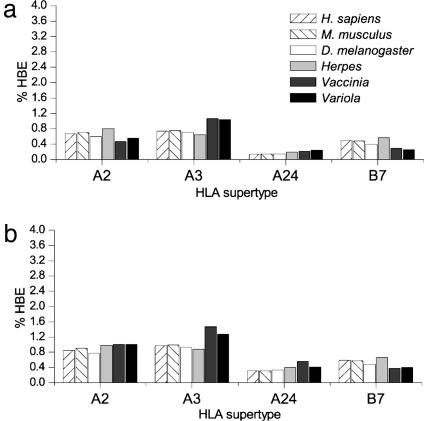
Finally, we asked whether our findings reflect some limitation in our ligand-predicting algorithms. We therefore repeated many of these analyses using the Parker et al. (14) predictive algorithms. Fig. 3 shows a comparison of a few reference proteomes: Homo sapiens, M. musculus, D. melanogaster, herpesvirus, vaccinia virus, and variola virus for both methods. The absolute value of any given fraction of peptides predicted to bind varied considerably, depending on the method, reflecting the differing sensitivities and specificities of the methods. Qualitatively, our voting methodology and that of Parker et al. (14) showed almost identical relative distributions of ligand frequencies.
Fig. 3.
Comparison of predictions derived from different ligand prediction methods. (a) Our voting methods (13). (b) The matrix-based method of Parker et al. (14).
Discussion
Although we find statistically significant differences in predicted ligand frequencies between distinct organisms and between normal and shuffled proteomes of single organisms, no meaningful overall trend is evident. Proteomes predicted to be overrepresented in some ligands relative to their shuffled versions or to other proteomes tend to be underrepresented in others. Most significantly, no clear association exists between ligand frequency and the presence of an adaptive cellular immune system. From this, we conclude that differing relative frequencies of particular ligands in particular proteomes are unlikely to have anything to do with evolutionary selection exerted by the peptide-binding specificity of class I molecules.
With few exceptions, the ligand profile of a given proteome is much closer to that of its shuffled proteome than to any other unshuffled proteome. This finding suggests that differences in ligand frequencies between organisms largely reflect different relative abundances of amino acids in individual proteins. This observation does not, however, rule out the possibility that a small absolute change in ligand frequencies is induced by changes in amino acid order so as to evade immune surveillance. A case in point is HIV, where solid evidence for immune selection of escape mutants exists. We note, however, that immune escape from one T cell population can lead to recognition by another, and this would score as neutral in a global analysis of ligand frequencies.
The absence of a general positive or negative ligand bias in the human proteome, particularly in polymorphic regions, suggests that selection against ligands is not a significant factor in protecting against autoimmunity. This lack of bias does not necessarily mean that autoimmunity does not exert selective pressure on antigen recognition, which could be controlled at the level of T cell antigen receptor repertoire evolution. On the other hand, the lack of such a bias in pathogens, particularly viral pathogens with a limited number of gene products to tend to, is remarkable as they would seem to derive an obvious benefit in evolutionary fitness by minimizing class I ligands. Why do they not bother? The likely answer is that polymorphism in class I binding specificity has made it impossible to create proteins that lack class I ligands: mutations that abrogate binding to one class I allomorph create ligands for other allomorphs.
Our analysis is based solely on predicted ligand binding. Whereas the predictive algorithm we use is demonstrably better than others in retrospective analysis (13), we do not pretend that it is perfect. Conservatively, it probably correctly identifies approximately half of the class I binding peptides. Another limitation of our analysis is the fact that only a fraction of class I binding peptides will ever be provided to class I molecules in biologically significant quantities because of limitations in antigen processing (18). This limitation is less of a problem with viral proteins, which are generally expressed at much higher levels than most cellular proteins, and therefore will express a higher percentage of predicted ligands at biologically significant levels. Even for cellular proteins, however, to the extent the biologically significant fraction of predicted peptides is not too small, any bias should still be evident in analyzing the total predicted ligand pool. Ultimately, sorting out these issues will require the careful characterization of the immunopeptidome as determined by quantitative mass spectroscopy of peptides recovered from class I molecules.
These limitations do not apply to a major question addressed by our analysis, which is the extent to which class I molecules use common motifs in amino acid sequences as a basis for their specificity. Here, the answer is clear. NH2-terminal leader sequences are a preferred source of peptides, particularly for HLA-A2-super-type allomorphs. Outside of this, however, class I binding preferences occur independently of specific sequence preferences present in the proteome.
Abbreviation: SNP, single nucleotide polymorphism.
References
- 1.Schubert, U., Anton, L. C., Gibbs, J., Norbury, C. C., Yewdell, J. W. & Bennink, J. R. (2000) Nature 404, 770-774. [DOI] [PubMed] [Google Scholar]
- 2.Reits, E. A., Vos, J. C., Gromme, M. & Neefjes, J. (2000) Nature 404, 774-778. [DOI] [PubMed] [Google Scholar]
- 3.Venter, J. C., Adams, M. D., Myers, E. W., Li, P. W., Mural, R. J., Sutton, G. G., Smith, H. O., Yandell, M., Evans, C. A., Holt, R. A., et al. (2001) Science 291, 1304-1351. [DOI] [PubMed] [Google Scholar]
- 4.Mural, R. J., Adams, M. D., Myers, E. W., Smith, H. O., Miklos, G. L., Wides, R., Halpern, A., Li, P. W., Sutton, G. G., Nadeau, J., et al. (2002) Science 296, 1661-1671. [DOI] [PubMed] [Google Scholar]
- 5.Schuler, G. D., Epstein, J. A., Ohkawa, H. & Kans, J. A. (1996) Methods Enzymol. 266, 141-162. [DOI] [PubMed] [Google Scholar]
- 6.Peterson, J. D., Umayam, L. A., Dickinson, T. M., Hickey, E. K. & White, O. (2001) Nucleic Acids Res. 29, 123-125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cole, S. T., Brosch, R., Parkhill, J., Garnier, T., Churcher, C., Harris, D., Gordon, S. V., Eiglmeier, K., Gas, S. & Barry, C. E., III, et al. (1998) Nature 393, 537-544. [DOI] [PubMed] [Google Scholar]
- 8.Bult, C. J., White, O., Olsen, G. J., Zhou, L., Fleischmann, R. D., Sutton, G. G., Blake, J. A., FitzGerald, L. M., Clayton, R. A., Gocayne, J. D., et al. (1996) Science 273, 1058-1073. [DOI] [PubMed] [Google Scholar]
- 9.The C. elegans Sequencing Consortium (1998) Science 282, 2012-2018. [DOI] [PubMed] [Google Scholar]
- 10.Celniker, S. E., Wheeler, D. A., Kronmiller, B., Carlson, J. W., Halpern, A., Patel, S., Adams, M., Champe, M., Dugan, S. P., Frise, E., et al. (2002) Genome Biol. 3, RESEARCH0079. Epub Dec 23. [DOI] [PMC free article] [PubMed]
- 11.Stenson, P. D., Ball, E. V., Mort, M., Phillips, A. D., Shiel, J. A., Thomas, N. S., Abeysinghe, S., Krawczak, M. & Cooper, D. N. (2003) Hum. Mutat. 21, 577-581. [DOI] [PubMed] [Google Scholar]
- 12.Sherry, S. T., Ward, M. H., Kholodov, M., Baker, J., Phan, L., Smigielski, E. M. & Sirotkin, K. (2001) Nucleic Acids Res. 29, 308-311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Florea, L., Halldorsson, B. V., Kohlbacher, O., Schwartz, R., Hoffman, S. & Istrail, S. (2003) in Proceedings of the 2nd Computational Systems Bioinformatics Conference (IEEE Computer Society, Los Alamitos, CA), pp. 17-26. [PubMed]
- 14.Parker, K. C., Bednarek, M. A. & Coligan, J. E. (1994) J. Immunol. 152, 163-175. [PubMed] [Google Scholar]
- 15.Marsh, S. G. E., Parham, P. & Barber, L. D. (2000) The HLA Facts Book (Academic, San Diego).
- 16.Sette, A., Sidney, J., Livingston, B. D., Dzuris, J. L., Crimi, C., Walker, C. M., Southwood, S., Collins, E. J. & Hughes, A. L. (2003) Immunogenetics 54, 830-841. [DOI] [PubMed] [Google Scholar]
- 17.McMichael, A. (1998) Cell 93, 673-676. [DOI] [PubMed] [Google Scholar]
- 18.Yewdell, J. W. & Bennink, J. R. (1999) Annu. Rev. Immunol. 17, 51-88. [DOI] [PubMed] [Google Scholar]