Abstract
T cells generally recognize peptide antigens bound to MHC proteins through contacts with residues found within or immediately flanking the seven- to nine-residue sequence accommodated in the MHC peptide-binding groove. However, some T cells require peptide residues outside this region for activation, the structural basis for which is unknown. Here, we have investigated a HIV Gag-specific T cell clone that requires an unusually long peptide antigen for activation. The crystal structure of a minimally antigenic 16-mer bound to HLA-DR1 shows that the peptide C-terminal region bends sharply into a hairpin turn as it exits the binding site, orienting peptide residues outside the MHC-binding region in position to interact with a T cell receptor. Peptide truncation and substitution studies show that both the hairpin turn and the extreme C-terminal residues are required for T cell activation. These results demonstrate a previously unrecognized mode of MHC-peptide-T cell receptor interaction.
Keywords: antigen presentation, receptors, antigen, protein conformation
Class II MHC proteins are cell-surface glycoproteins that bind antigens in the form of short peptides and present them for recognition to T cell receptors (TCRs) on the cell surface of CD4+ T cells (1). Naturally processed peptides isolated from class II MHC proteins found in antigen-presenting cells are usually 15-25 residues long (2, 3). The central region of these peptides interacts directly with class II MHC proteins, typically with specific recognition of an approximate nine-residue stretch (4). X-ray crystallography of human and murine class II MHC proteins has revealed that peptides bind to the protein in an extended polyproline type II conformation, with several peptide side chains bound into polymorphic pockets that line the peptide-binding groove (5-12). A hydrogen-bond network between the conserved residues on the class II MHC and the peptide main-chain carbonyl and amide groups, independent of the sequence of the peptide, stabilizes the MHC-peptide complex (13), and enforces the polyproline conformation that directs some of the side chains into the MHC pockets and leaves the others accessible for TCR interactions (14). Generally, pockets accommodate the side chains of peptide residues at the P1, P4, P6, and P9 positions, with smaller pockets or shelves in the binding site accommodating the P3 and P7 residues. Minor variations on this theme have been observed, for example, in some complexes the P9 interactions are weak or absent (5, 10). In the canonical conformation, the side chains of residues at positions P -1, P2, P5, and P8 are solvent-accessible and point toward the TCR, with portions of other side chains and the peptide main chain also exposed for potential TCR interaction in the central region of the complex.
The interaction of TCR with MHC-peptide complexes also is expected to be relatively stereotyped, with complementarity-determining regions (CDRs) from the Vα and Vβ domain lying across the peptide-MHC complex, typically with CDR3 loops of both variable domains extending down over the center of the peptide and the CDR1 and CDR2 loops contacting the α-helices (4, 15). Detailed structural information on the MHC-TCR interaction is available only for two class II MHC-TCR complexes (16-18), in which TCR CDR3 loops contact the MHC-bound peptides within the region circumscribed by MHC-peptide contacts, at the P -1, P2, P3, P5, and P8 positions. For both these systems, earlier mapping experiments had identified these positions as sites where amino acid substitution abrogated T cell activation without disturbing MHC-peptide interaction (16, 17). Mutagenesis studies have suggested that many other MHC-TCR pairs interact in this manner (ref. 4 and references therein). The structures provide no evidence for TCR contacts with peptide residues outside the peptide-binding groove, although peptide-mapping experiments have suggested that for some peptides the flanking P11 residue also can be an important T cell contact (18-21).
Here, we investigate a complex that uses a different mode of class II MHC peptide-TCR interaction. The crystal structure of an HIV Gag(p24)-derived peptide bound to HLA-DR1 reveals an unusual bent conformation for the bound peptide. A hairpin turn involving residues P9-P12 folds the peptide back over itself after it exits the binding groove, orienting the P13 side chain for interaction with TCR. A Gag(p24)-restricted CD4+ T cell clone requires both the hairpin turn and the P13 residue for activation.
Methods
Peptide Synthesis. Peptides were synthesized by using solid-phase fluorenylmethoxycarbonyl chemistry, and were cleaved, deprotected, and purified by using standard methods (22). The identity and homogeneity of each peptide was verified by using reverse-phase HLPC and matrix-assisted laser desorption ionization/time-of-flight MS. Peptide concentration was determined by using amino acid analysis.
Protein Expression and Purification. For peptide binding experiments, the extracellular portion of HLA-DR1 (DRA*0101, DRB1*0101) was produced in insect cells as soluble empty αβ heterodimers, as described (23). For x-ray crystallography, the extracellular portion of HLA-DR1 was produced by expression of isolated subunits in Escherichia coli inclusion bodies, followed by refolding in vitro and immunoaffinity purification as described (24). SEC-3B2 superantigen was expressed as a soluble protein in E. coli and isolated from the periplasmic fraction as described (25).
T Cell Activation Assay. The CD4+ T cell clone AC-25 and the corresponding autologous B cell line were derived as described (26). Briefly, the clone was derived after stimulating peripheral blood mononuclear cells with whole p24 antigen, resting for 2 weeks, then restimulating and plating at limiting dilution on a round-bottom 96-well plate. The AC-25 T cell clone was maintained by restimulation every 2 weeks with IL-2 (100 units/ml), 12F6 [anti-CD3 antibody obtained from J. Wong (Massachusetts General Hospital; 0.1 μg/ml)], and 106 feeder cells were irradiated at 30 Gy (26) in R-10-RPMI medium 1640 (Sigma), with penicillin-streptomycin (50 units/ml and 50 μg/ml, Mediatech, Herndon, VA), Hepes (2.38 mg/ml, Mediatech), l-glutamine (Mediatech), plus 10% heat-inactivated human AB serum (Sigma). Proliferation and enzyme-linked immunospot assays were performed in the same medium. For proliferation assays, autologous B lymphoblastoid cell lines were irradiated (120 Gy) and resuspended with the appropriate peptide antigen, and plated in triplicate with AC-25 cells (50,000 cells per well). After 48 h, 1 μCi (1 Ci = 37 GBq) of 3H-thymidine was added, and, after an additional 17 h, plates were harvested onto glass fiber filters. Results were expressed as the difference between the counts in the presence and absence of antigen, with differences >1,000 considered to be significant (26). For IFN-γ enzyme-linked immunospot assays, B lymphoblastoid cell lines (5 × 104), peptide antigen (1 μg/ml), and AC-25 T cells (50 cells per well) were added to 96-well plates coated with anti-IFN-γ antibody (Mabtech, Mariemont, OH). After overnight incubation, cells were discarded and the plates were washed, and captured IFN-γ was detected by using biotinylated anti-IFN-γ (Endogen, Cambridge, MA), streptavidin-alkaline phosphatase (Mabtech), and nitroblue tetrazolium/5-bromo-4-chloro-3-indolyl phosphate (Bio-Rad) was added. Background responses to wells with no antigen or irrelevant antigen ranged from 0 to 2.5 spots per well. Responses greater than five spots per well and more than five times the maximum background were considered significant.
Peptide-Binding Assay. A competition assay was used to determine binding affinities of peptides. HLA-DR1 (25 nM) was mixed together with biotinylated Ha[306-318] peptide (25 nM) and various concentrations of unlabeled competitor peptide (10-12 to 10-5 M). The mixtures were incubated for 3 days at 37°C in 100 mM sodium phosphate buffer, pH 5.5, containing 50 mM NaCl, 1 mg/ml PMSF, 37 μg/ml iodoacetamide, 10 mM EDTA, 0.02% NaN3, and 0.5 mg/ml octyl glucoside, followed by detection of bound biotinylated peptide using an immunoassay that used anti-DR1 capture antibody LB3.1 and alkaline phosphate-labeled streptavidin.
Crystallization. HLA-DR1-peptide complexes, purified by gel filtration, were mixed with equimolar SEC3-3B2, and crystals were grown at 4°C by vapor diffusion in hanging drops, by using 1 μl of protein complex (10 mg/ml) mixed with 1 μl of precipitant solution (5-10% ethylene glycol/100 mM sodium acetate, pH 5.2-5.6/2-6% polyethylene glycol 4000 for Gag[PG13] or 2-6% polyethylene glycol/5000 monomethyl ether for Gag[PP16]). For x-ray diffraction experiments, the crystals were soaked for ≈1 min in a cryoprotectant solution consisting of 25% ethylene glycol in the mother liquor and were then flashed-cooled in liquid nitrogen.
Data Collection and Processing. For the Gag[PG13] complex, a 2.25-Å data set was collected from a single crystal (300 × 200 × 200 μm) on an R-AXIS IV image plate detector by using CuKα radiation from a rotating anode source. For the Gag[PP16] complex, a 2.45-Å data set was collected on a single crystal (80 × 60 × 60 μm) on a Mar180 image plate detector, also by using CuKα radiation. Collected data were processed and scaled by using the programs denzo, scalepack, and the CCP4 package (27, 28).
Structure Determination. The structure of each complex was determined by molecular replacement. Coordinates for another complex of E. coli-derived HLA-DR1 and SEC3-3B2, [Protein Data Bank (PDB) ID code 1PYW; ref. 22] were used as the search model, after removal of waters and the peptide. Refinement was carried out by using the program CNS (29) and manual inspection and rebuilding were carried out by using the program xtalview (30). The final model was verified for distortions on the secondary structure features by using the program procheck (31). Coordinates and structure factors for HLA-DR1/Gag[PP16]/SEC3-3B2 and HLA-DR1/Gag[PG13]/SEC3-3B2 have been deposited in the PDB database (ID codes 1SJE and 1SJH, respectively).
Results
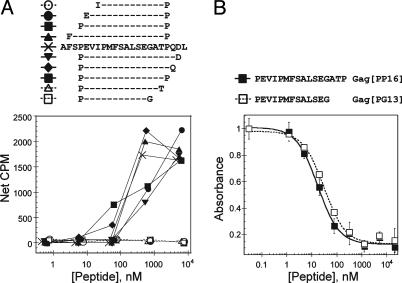
Residues at the C Terminus of an HIV-Gag-Derived Peptide Are Required for T Cell Activation but Not for MHC-Peptide Interaction. A CD4+ T cell clone (AC-25) isolated from an individual acutely infected with HIV-1 recognizes an antigen derived from the HIV Gag (p24) protein bound to the human class II MHC protein HLA-DR1. As reported (32), AC-25 requires an unusually long peptide for activation. Only three residues can be removed from either end of a Gag(p24)-derived 22-mer without affecting T cell activation (Fig. 1A and ref. 32). Removal of additional residues from either the N or C terminus of the minimal gag-derived 16-mer PEVIPMFSALSEGATP (Gag[PP16]) results in abrogation of T cell activation, as shown by a cellular proliferation assay (Fig. 1A), and also in abrogation of cytotoxicity and cytokine secretion functions (32).
Fig. 1.
A Gag(p24)-specific T cell clone requires an unusually long peptide for activation. (A) T cell activation, measured by using a proliferation assay and peptide-pulsed autologous antigen-presenting cells. Open symbols and dashed lines indicate peptides that do not activate AC-25 T cell proliferation at any concentration tested. (B) MHC-peptide binding, measured by using a competition binding assay. Curves fits reflect IC50 values of 29 ± 1 nM (Gag[PP16]) and 28 ± 3 nM (Gag[PG13]), which correspond to Kd values of ≈6 and 9 nM, assuming simple two-state binding. The Gag[PG13] peptide, which is not able to activate the AC-25 T cell clone, exhibits apparent peptide-binding affinity indistinguishable from that of the activating Gag[PP16] peptide. CPM, counts per minute.
For HLA-DR1, the most important determinants of peptide binding are a hydrophobic residue at position P1 and a small residue at position P6, with additional but weaker preferences at P4, P7, and P9 (33, 34). Within the Gag[PP16] peptide, the preferred binding frame is expected to be VIPMFSALS (key MHC-peptide contacts are underlined) (35, 36). A shorter Gag-derived peptide containing this sequence, PEVIPMFSALSEG (Gag[PG13]), binds to HLA-DR1 with an apparent dissociation constant of <10 nM, which is essentially identical to that of the longer Gag[PP16] peptide (Fig. 1B) However, this peptide is inactive (Fig. 1A, open squares). The inability of the Gag[PG13] peptide to induce activation responses in a T cell clone that recognizes the longer peptide is unexpected, and could indicate an unusual binding frame or mode of T cell interaction.
Crystal Structure of HLA-DR1 in Complex with Gag[PG13] and Gag[PP16] Peptides. To determine the actual peptide-binding frame of these peptides and to evaluate the importance of the C-terminal residues of Gag[PP16] not present in Gag[PG13], the crystal structures of HLA-DR1 bound to Gag[PP16] and Gag[PG13] were determined (Table 1). The complexes were crystallized in the presence of SEC3-3B2, an affinity-matured variant of a staphylococcal enterotoxin that interacts with HLA-DR1 outside the binding groove, and which has been used to promote crystallization (37). For both complexes, electron density for HLA-DR1, peptide, and superantigen was clear and continuous except for HLA-DR1 βl09-111, a disordered loop away from the peptide-binding site, for SEC3-3B2 residues 99-105, a region located away from the interaction site with HLA-DR1, and for the Gag[PP16] terminal proline residue, which was not observed. Electron density for the Gag[PP16] penultimate threonine was observed but was not sufficiently clear to allow definitive placement of the side chain.
Table 1. Data collection and refinement statistics.
| Gag[PP16]* | Gag[PG13]* | |||
|---|---|---|---|---|
| Crystal parameters | ||||
| Space group | R3 | R3 | ||
| Cell dimensions a, c, Å | 172.75, 121.40 | 172.46, 121.46 | ||
| Data collection | Overall | Highest-resolution shell | Overall | Highest-resolution shell |
| Resolution limits, Å | 20.0-2.45 | 2.54-2.45 | 50.0-2.25 | 2.33-2.25 |
| Unique reflections | 49,701 | 4,962 | 64,060 | 6,424 |
| Total reflections | 286,608 | 26,633 | 321,541 | 28,138 |
| Completeness, % | 100.0 | 100.0 | 99.8 | 99.8 |
| Mean I/σ (I) | 11.3 | 3.4 | 9.6 | 1.9 |
| Rsym, intensities, % | 8.4 | 41.3 | 13.8 | 48.9 |
| Refinement | ||||
| Rfree/(Rcryst)† | 22.3/(19.8) | 31.2/(27.8) | 24.5/(22.0) | 31.2/(28.4) |
| Model | Average B factor, Å2 | No. of residues (atoms) | Average B factor, Å2 | No. of residues (atoms) |
| HLA-DR1 | 39.9 | 369 (3,035) | 41.5 | 368 (3,026) |
| Peptide | 34.7 | 15 (109) | 38.3 | 13 (97) |
| SEC3-3B2 | 38.8 | 231 (1,900) | 40.9 | 229 (1,883) |
| Waters | 42.8 | 264 | 42.5 | 275 |
| Ramachandran plot | ||||
| Favored/allowed/ | 87.0/12.3/0.6/0.2 | 87.3/12.2/0.5/0.0 | ||
| generous/disallowed, % | ||||
Crystals contained HLA-DR1 bound to the indicated Gag(p24)-derived peptide and the superantigen SEC3-3B2.
R factor on structure factors for reflections omitted from the refinement and used as a test set (10% of total, Rfree) or reflections included in the refinement (90% of total, Rcryst).
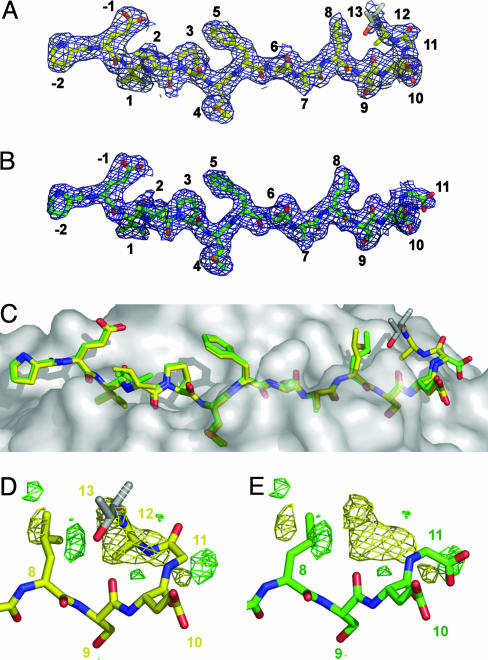
Both Gag[PP16] and Gag[PG13] peptides bind in the expected frame, with Val (P1), Met (P4), Ser (P6), and Ser (P9) accommodated in pockets in the peptide-binding groove (Fig. 2 A and B, and Fig. 6, which is published as supporting information on the PNAS web site). The Gag[PG13] peptide adopts the usual polyproline conformation. The Gag[PP16] peptide adopts that same conformation within the MHC peptide-binding groove, but, just outside the groove, the peptide bends sharply at P10 and doubles back over itself (Fig. 2C). These differences are highlighted in the Gag[PP16] - Gag[PG13] difference Fourier map (Fig. 2 D and E). The largest features in this map are a positive peak above the peptide P9-P11 residues (8.4σ) together with a negative peak beyond the Gag[PG13] C terminus (7.5σ), which indicate the deviation of the Gag[PP16] peptide from the usual extended conformation, and positive and negative peaks (± 6.5σ) surrounding the side chain of Leu (P8), which indicate an ≈90° rotation of the side chain (Fig. 2 D and E).
Fig. 2.
Crystal structures of DR1/Gag[PP16]/SEC3 and DR1/Gag[PG13]/SEC3. (A) A |2 Fo - Fc| electron density map calculated by using model phases and contoured at 1σ for the Gag[PP16] complex in the vicinity of the peptide. Carbon atoms are yellow, nitrogen atoms are blue, and oxygen atoms are red. (B) Corresponding map of the Gag[PG13] complex, but with carbon atoms being green. (C) Surface of HLA-DR1 showing superposition of bound Gag[PP16] (yellow) and Gag[PG13] (green) peptides. (D and E) Difference Fourier map|FGag[PP16] - FGag[PG13]| calculated by using Gag[PP16] model phases, contoured at 4σ, with positive (Gag[PP16]) density in yellow and negative (Gag[PG13]) density in green, overlaid with the Gag[PP16] peptide (D) or the Gag[PG13] peptide (E). No significant positive or negative difference peaks were observed outside this region.
Overall, the Gag[PG13] and Gag[PP16] peptides bind to HLA-DR1 in essentially identical conformations from the N terminus through P7, with the Gag[PG13] peptide P8-P11 region extending conventionally out of the site, and the Gag[PP16] peptide P9-P12 region forming a hairpin turn. The turn places the Gag[PP16] peptide P13 (Thr) side chain near the P8 (Leu) side chain, and changes the orientation of Leu (P8) relative to that found in the Gag[PG13] peptide (Fig. 2C).
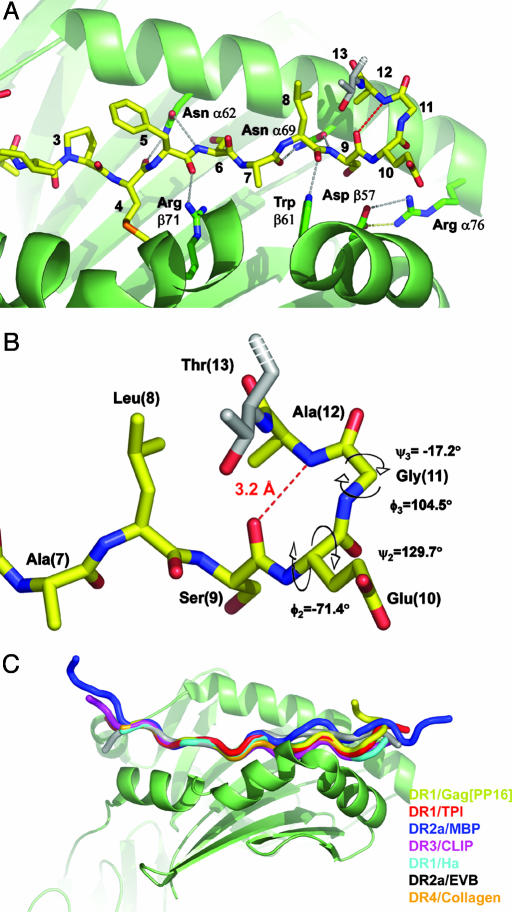
The C-Terminal Region of Gag[PP16] Adopts a Type II β-Turn. Both Gag[PG13] and Gag[PP16] peptides bind with the usual main-chain hydrogen-bonding scheme involving HLA-DR1 residues Glnα9, Asnα62, Asnα69, Argα76, Aspβ57, Trpβ61, Argβ71, Hisβ81, and Asnβ82 (although with intervening waters for the Gag[PP16] interaction with Argα76 and Aspβ57; Fig. 3A). The Gag[PP16] peptide makes an additional intramolecular main-chain hydrogen bond, involving the carbonyl oxygen of the P9 residue (Ser) and the amide nitrogen of P12 (Ala; Fig. 3B). This interaction is characteristic of a β-turn, a four-residue structural motif found frequently in proteins (38). In Gag[PP16], the SEGA (P9-P12) sequence forms a β-turn, with main-chain backbone φ and ψ angles characteristic of the type II β-turn (Fig. 3B). In type II β-turns, the oxygen atom of the carbonyl of residue 2 (P10) crowds the Cβ atom of residue 3, which is therefore usually glycine (39). The Gag[PP16] peptide has glycine at this position (P11).
Fig. 3.
A type II β-turn at the C-terminal region of the Gag[PP16] peptide. (A) C-terminal region of the DR1 peptide-binding site with bound Gag[PP16] peptide showing conserved MHC peptide hydrogen bonds. Conserved MHC peptide hydrogen bonds in this region are shown as gray dashed lines, with the Gag[PP16] intraturn hydrogen bond between the nitrogen of Ala (P12) and the carbonyl oxygen of Ser (P9) shown as a red dashed line. (B) Closeup view of the Gag[PP16] hairpin, showing the intraturn hydrogen bond and φ-ψ values characteristic of a type II β-turn. (C) Alignment of HLA-DR crystal structures exhibiting ordered peptide density beyond P10. Complexes were aligned by least-squares fitting of α1 and β1 domains. Peptides are shown as Cα traces. DR1/TP1 is red (PDB ID code 1KLU), DR2a/MBP is blue (PDB ID code 1FV1), DR3/CLIP is magenta (PDB ID code 1A6A), DR1/Ha is cyan (PDB ID code 1DLH), DR2a/EBV is gray (PDB ID code 1H15), and DR4/collagen is orange (PDB ID code 2SEB).
This bent conformation has not been observed previously among complexes of peptides bound to class II MHC molecules. An alignment of Gag[PP16] with all other crystal structures of HLA-DR1 complexes exhibiting ordered density beyond P10 is shown in Fig. 3C. In each of these structures, the peptide extends straight out of the binding site, without any pronounced bends or turns.
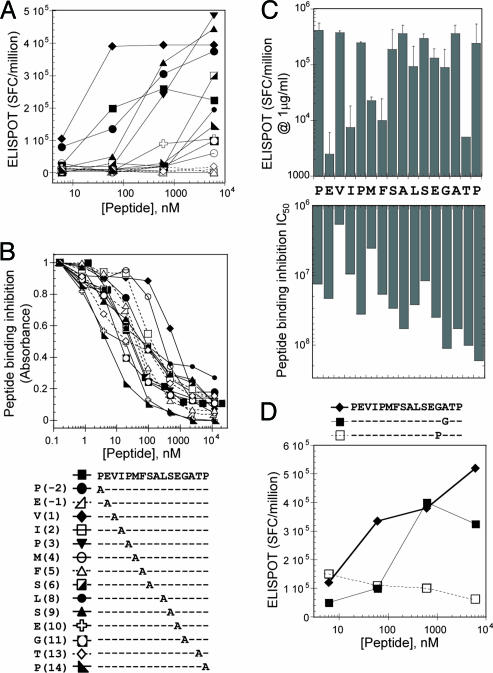
The C-Terminal Hairpin Is Required for T Cell Activation. Alanine-scanning mutagenesis of the Gag[PP16] peptide was used to determine the importance of the C-terminal hairpin in AC-25 TCR interaction, and to confirm the physiological relevance of the peptide conformation observed in the crystal structure. Each Gag[PP16] peptide residue except for Ala (P7) and Ala (P12) was changed independently to alanine, and standard MHC peptide binding and T cell proliferation assays were performed (Fig. 4 A and B).
Fig. 4.
Alanine-scanning mutagenesis studies of Gag[PP16]. (A) AC-25 T cell activation, measured by using an enzyme-linked immunospot ELISPOT assay SFC, spot-forming centers. (B) HLA-DR1 peptide binding, measured by using a competition binding assay. The key for A and B is at the bottom. (C) Effect of alanine substitution on T cell activation and MHC peptide-binding affinity shown together on a logarithmic scale. For T cell activation, values represent the response induced by 1 μg/ml peptide concentration (average of two independent experiments). For MHC peptide binding, IC50 values in nM are shown (average of two independent experiments). In both assays, shorter bars represent decreased activity. Values for alanine positions reflect the unmodified Gag[PP16] peptide. (D) T cell activation assay of additional substitutions at Gly(P11) and Ala(P12).
In the MHC peptide-binding assay (Fig. 4 B and C), no single-alanine substitution abolished MHC-peptide interaction, but significant reductions in binding affinity were observed upon alanine substitution of Val (P1) and Met (P4), and, to a lesser extent, Ser (P9). These effects are consistent with the binding frame observed in the crystal structures. [Both Ala and Ser are preferred residues at P6 (34), and no significant effect is expected or observed for this substitution].
In the T cell activation assay, alanine substitutions of Glu (P -1), Ile (P2), Phe (P5), and Thr (P13) had dramatic effects, abolishing activation at all concentrations tested (Fig. 4 A and C). In the Gag[PP16] crystal structure, the side chains of residues at the P -1, P2, and P5 positions were observed to be oriented away from the peptide-binding site as in other class II MHC-peptide complexes, and the effects of alanine substitutions at these positions are consistent with conventional MHC-TCR interaction. In the Gag[PP16] crystal structure, the hairpin turn orients Thr (P13) above the bulk of the peptide, in the vicinity of the P8 side chain. The large effect of Thr (P13)-to-alanine substitution at this position on T cell activation but not MHC-peptide interactions suggests that this residue and the hairpin turn play an important role in TCR interaction.
To further establish the contribution of the residues at the Gag[PP16] C terminus to T cell activation, additional substitutions were analyzed at Gly(P11) and Ala(P12) (Fig. 4D). Ala(P12) was substituted by glycine, to evaluate a possible role of the alanine side chain in TCR recognition, which would not have been apparent in the initial alanine scan. This substitution was tolerated well by the TCR (Fig. 4D, ▪). To evaluate the role of the reverse turn, Gly(P11) was substituted by proline, which (unlike alanine) cannot be accommodated in a type II turn. Gly(P11)-to-Pro substitution abrogated T cell activation (Fig. 4D, □), again highlighting the importance of the hairpin turn in T cell activation.
Discussion
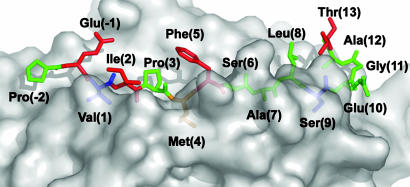
The crystal structure of the HIV Gag-derived peptide Gag[PP16] bound to HLA-DR1 shows that it binds in an unexpected bent conformation, with a hairpin turn at one end of the binding groove that orients its C-terminal region above the remainder of the peptide and in position to interact with TCR. Truncation analysis shows that this C-terminal region is important for recognition by a CD4+ T cell clone derived from an HIV-infected person. The effects of alanine scanning and site-specific substitutions introduced into the peptide show that the Thr residue at position P13 has no effect on binding to the class II MHC protein, but is crucial for TCR interaction, along with conventional T cell contacts at P -1, P2, and P5 (Fig. 5). The hairpin turn would appear to be important in aligning the threonine for recognition by TCR because introduction of a proline residue that interferes with turn formation blocked T cell activation.
Fig. 5.
The Gag[PP16] hairpin turn orients important C-terminal residues in position to interact with TCR. Side view of surface of HLA-DR1 with the Gag[PP16] peptide shown as a stick model. Peptide side chains essential for T cell activation are red and those side chains important for peptide binding to HLA-DR1 are blue. Met (P4) affects both MHC peptide binding and T cell activation and is orange.
The Gag[PP16] C-terminal hairpin would appear to represent a physiologically relevant conformation, rather than one induced only by crystallization or recombinant MHC peptide production. First, several β-turn prediction algorithms (39-42) identify the SEGA (P9-P12) sequence as having strong type II β-turn potential, with contributions from the Glu at position 2, in addition to the strongly preferred Gly at position 3. The Thr (P13) side chain is in van der Waals contact with Leu (P8), which also would tend to stabilize the turn and would be able to form an intrachain hydrogen bond between the Thr hydroxyl and the main-chain carbonyl oxygen at position 1 of the type II β-turn. Moreover, no crystal contacts are found in the vicinity of the hairpin. Second, AC-25 T cell recognition of the Gag[PP16] peptide appears to depend on the C-terminal β-hairpin turn, as shown by the Gly(P11)-to-Pro substitution, which abrogates T cell activation, as well as by the pattern of important T cell contact residues (Fig. 5). The AC-25 T cell clone specific for this conformation was isolated from an HIV-infected subject who was treated with antiretroviral therapy during primary HIV infection. The clonotype presumably was elicited in response to viral infection, and was selected in vitro by using peripheral blood mononuclear cells stimulated with intact Gag protein. This finding suggests that both naturally infected and Gag protein-treated cells present on their surface an activating Gag-derived epitope that presumably includes the hairpin conformation. However, we note that the hairpin is not obligatory for T cell recognition of this region of Gag: the same Gag (24) epitope as studied here has been identified also as the target of an HLA-DR4-restricted T cell clone (32) and another HLA-DR1-restricted T cell clone (43). Both of these clones appear to recognize a conventional 10-residue sequence, EVIPMFSALS, contained within the Gag[PG13] sequence. Finally, whereas P13 is well outside the range of peptide residues generally considered to be seen by the TCR, comparison of the Gag[PP16] structure with previously determined class II MHC-TCR complexes (44-46) suggests that a TCR in a normal orientation could contact Thr directly by interacting with the CDR3 and/or CDR2 loops on Vβ.
How frequently might we expect such unconventional recognition of bent peptides? Except for the HLA-DR1-Gag[PP16] complex reported here, hairpin turns or other bent or bulged conformations have not been observed in the class II MHC crystal structures reported to date. However, several of the complexes crystallized do not contain residues outside of the P1-P10 region, and many complexes carry their peptides as covalent attachments to the MHC β-subunit, where the linker between the peptide C terminus and the MHC β N terminus could interfere with potential turn formation (5, 7, 47-50). Thus, at present we may not have a complete picture of the degree of potential conformational variability for peptides bound to class II MHC proteins. The large number of main-chain hydrogen bonds in the central region of the peptide constrain the conformation of a peptide bound to class II MHC proteins, but, outside of the P1-P10 region, the peptide would appear to be unconstrained by the MHC. A peptide turn or bend would therefore have to be stabilized by interactions provided by the peptide, as observed here for Gag[PP16]. In general, the conformational stability of structures formed by short peptides is thought to be low, but any particular sequence able to form a stable conformation potentially could be recognized by TCR.
TCR recognition of residues outside the MHC peptide-binding groove has been observed previously in cellular studies. A flanking tryptophan at the P11 position of a lysozyme peptide bound to IAk has been shown to be important in T cell activation (18-20). Such flanking interactions have been proposed to be a common feature of MHC-TCR interaction (20), although this is controversial (51). Note that the P11 side chain is oriented toward the TCR for a MHC-bound peptide canonical polyproline repeat, and does not require a peptide bend, bulge, or turn for recognition by TCR. There have been fewer reports of T cells that are sensitive to peptide residues lying further outside a reliably predicted MHC-binding region, or with patterns of T cell contact residues that suggest deviations from the canonical polyproline extended structure. In one study, Met was found at P12 (48, 52), and is located at the end of a sequence with high propensity for β-turn formation (41). In another, critical T cell contacts were mapped to positions several residues N-terminal to the expected MHC-binding regions (19).
In conclusion, these studies show that peptides can bind to class II MHC proteins in a previously unobserved conformation in which residues outside the binding groove can bend back over the peptide and become accessible for T cell recognition. These results provide evidence for a mode of MHC peptide-TCR interaction that should be considered more widely in prediction, testing, and design of putative T cell antigens.
Supplementary Material
Acknowledgments
We thank Jennifer D. Stone for helpful discussions. This work was supported by National Institutes of Health Grants R01-AI38996 (to L.J.S.), R01-AI01698 (to P.J.N.), F31-GM64859 (to Z.Z.-R.), and the Doris Duke Charitable Foundation (to P.J.N.). Research was carried out (in part) at the National Synchrotron Light Source, Brookhaven National Laboratory (Upton, NY), which is supported by the U.S. Department of Energy, Division of Material Sciences, and Division of Chemical Sciences, under Contract DE-AC02-98CH10886. Structural figures were generated by using the program pymol (53).
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: TCR, T cell receptor; PDB, Protein Data Bank.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID codes 1SJE and 1SJH).
References
- 1.Watts, C. (1997) Annu. Rev. Immunol. 15, 821-850. [DOI] [PubMed] [Google Scholar]
- 2.Chicz, R. M., Urban, R. G., Lane, W. S., Gorga, J. C., Stern, L. J., Vignali, D. A. & Strominger, J. L. (1992) Nature 358, 764-768. [DOI] [PubMed] [Google Scholar]
- 3.Rudensky, A., Preston-Hurlburt, P., Hong, S. C., Barlow, A. & Janeway, C. A., Jr. (1991) Nature 353, 622-627. [DOI] [PubMed] [Google Scholar]
- 4.Rudolph, M. G. & Wilson, I. A. (2002) Curr. Opin. Immunol. 14, 52-65. [DOI] [PubMed] [Google Scholar]
- 5.Corper, A. L., Stratmann, T., Apostolopoulos, V., Scott, C. A., Garcia, K. C., Kang, A. S., Wilson, I. A. & Teyton, L. (2000) Science 288, 505-511. [DOI] [PubMed] [Google Scholar]
- 6.Smith, K. J., Pyrdol, J., Gauthier, L., Wiley, D. C. & Wucherpfennig, K. W. (1998) J. Exp. Med. 188, 1511-1520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fremont, D. H., Dai, S., Chiang, H., Crawford, F., Marrack, P. & Kappler, J. (2002) J. Exp. Med. 195, 1043-1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stern, L. J., Brown, J. H., Jardetzky, T. S., Gorga, J. C., Urban, R. G., Strominger, J. L. & Wiley, D. C. (1994) Nature 368, 215-221. [DOI] [PubMed] [Google Scholar]
- 9.Ghosh, P., Amaya, M., Mellins, E. & Wiley, D. C. (1995) Nature 378, 457-462. [DOI] [PubMed] [Google Scholar]
- 10.Li, Y., Li, H., Martin, R. & Mariuzza, R. A. (2000) J. Mol. Biol. 304, 177-188. [DOI] [PubMed] [Google Scholar]
- 11.Lee, K. H., Wucherpfennig, K. W. & Wiley, D. C. (2001) Nat. Immunol. 2, 501-507. [DOI] [PubMed] [Google Scholar]
- 12.Liu, X., Dai, S., Crawford, F., Fruge, R., Marrack, P. & Kappler, J. (2002) Proc. Natl. Acad. Sci. USA 99, 8820-8825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McFarland, B. J. & Beeson, C. (2002) Med. Res. Rev. 22, 168-203. [DOI] [PubMed] [Google Scholar]
- 14.Jardetzky, T. S., Brown, J. H., Gorga, J. C., Stern, L. J., Urban, R. G., Strominger, J. L. & Wiley, D. C. (1996) Proc. Natl. Acad. Sci. USA 93, 734-738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hennecke, J. & Wiley, D. C. (2001) Cell 104, 1-4. [DOI] [PubMed] [Google Scholar]
- 16.Sant'Angelo, D. B., Waterbury, G., Preston-Hurlburt, P., Yoon, S. T., Medzhitov, R., Hong, S. C. & Janeway, C. A., Jr. (1996) Immunity 4, 367-376. [DOI] [PubMed] [Google Scholar]
- 17.Krieger, J. I., Karr, R. W., Grey, H. M., Yu, W. Y., O'Sullivan, D., Batovsky, L., Zheng, Z. L., Colon, S. M., Gaeta, F. C., Sidney, J., et al. (1991) J. Immunol. 146, 2331-2340. [PubMed] [Google Scholar]
- 18.Vignali, D. A. & Strominger, J. L. (1994) J. Exp. Med. 179, 1945-1956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Muller, C. P., Ammerlaan, W., Fleckenstein, B., Krauss, S., Kalbacher, H., Schneider, F., Jung, G. & Wiesmuller, K. H. (1996) Int. Immunol. 8, 445-456. [DOI] [PubMed] [Google Scholar]
- 20.Arnold, P. Y., La Gruta, N. L., Miller, T., Vignali, K. M., Adams, P. S., Woodland, D. L. & Vignali, D. A. (2002) J. Immunol. 169, 739-749. [DOI] [PubMed] [Google Scholar]
- 21.Srinivasan, M., Marsh, E. W. & Pierce, S. K. (1991) Proc. Natl. Acad. Sci. USA 88, 7928-7932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zavala-Ruiz, Z., Sundberg, E. J., Stone, J. D., DeOliveira, D. B., Chan, I. C., Svendsen, J., Mariuzza, R. A. & Stern, L. J. (2003) J. Biol. Chem. 278, 44904-44912. [DOI] [PubMed] [Google Scholar]
- 23.Stern, L. J. & Wiley, D. C. (1992) Cell 68, 465-477. [DOI] [PubMed] [Google Scholar]
- 24.Frayser, M., Sato, A. K., Xu, L. & Stern, L. J. (1999) Protein Expression Purif. 15, 105-114. [DOI] [PubMed] [Google Scholar]
- 25.Andersen, P. S., Lavoie, P. M., Sekaly, R. P., Churchill, H., Kranz, D. M., Schlievert, P. M., Karjalainen, K. & Mariuzza, R. A. (1999) Immunity 10, 473-483. [DOI] [PubMed] [Google Scholar]
- 26.Norris, P. J., Sumaroka, M., Brander, C., Moffett, H. F., Boswell, S. L., Nguyen, T., Sykulev, Y., Walker, B. D. & Rosenberg, E. S. (2001) J. Virol. 75, 9771-9779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Collaborative Computational Project 4 (1994) Acta Crystallogr. D 50, 760-763.15299374 [Google Scholar]
- 28.Otwinowski, Z. & Minor, W. (1997) Methods Enzymol. 276, 307-326. [DOI] [PubMed] [Google Scholar]
- 29.Brunger, A. T., Adams, P. D., Clore, G. M., DeLano, W. L., Gros, P., Grosse-Kunstleve, R. W., Jiang, J. S., Kuszewski, J., Nilges, M., Pannu, N. S., et al. (1998) Acta Crystallogr. D 54, 905-921. [DOI] [PubMed] [Google Scholar]
- 30.McRee, D. E. (1999) J. Struct. Biol. 125, 156-165. [DOI] [PubMed] [Google Scholar]
- 31.Laskowski, R. A., Moss, D. S. & Thornton, J. M. (1993) J. Mol. Biol. 231, 1049-1067. [DOI] [PubMed] [Google Scholar]
- 32.Norris, P. J., Moffett, H. F., Brander, C., Allen, T. M., O'Sullivan, K. M., Cosimi, L. A., Kaufmann, D. E., Walker, B. D. & Rosenberg, E. S. (2004) AIDS Res. Hum. Retroviruses 20, 315-326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.O'Sullivan, D., Sidney, J., Del Guercio, M. F., Colon, S. M. & Sette, A. (1991) J. Immunol. 146, 1240-1246. [PubMed] [Google Scholar]
- 34.Hammer, J., Takacs, B. & Sinigaglia, F. (1992) J. Exp. Med. 176, 1007-1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rammensee, H., Bachmann, J., Emmerich, N. P., Bachor, O. A. & Stevanovic, S. (1999) Immunogenetics 50, 213-219. [DOI] [PubMed] [Google Scholar]
- 36.Sturniolo, T., Bono, E., Ding, J., Raddrizzani, L., Tuereci, O., Sahin, U., Braxenthaler, M., Gallazzi, F., Protti, M. P., Sinigaglia, F. & Hammer, J. (1999) Nat. Biotechnol. 17, 555-561. [DOI] [PubMed] [Google Scholar]
- 37.Redpath, S., Alam, S. M., Lin, C. M., O'Rourke, A. M. & Gascoigne, N. R. (1999) J. Immunol. 163, 6-10. [PubMed] [Google Scholar]
- 38.Richardson, J. S. (1981) Adv. Protein Chem. 34, 167-339. [DOI] [PubMed] [Google Scholar]
- 39.Hutchinson, E. G. & Thornton, J. M. (1994) Protein Sci. 3, 2207-2216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chou, P. Y. & Fasman, G. D. (1974) Biochemistry 13, 211-222. [DOI] [PubMed] [Google Scholar]
- 41.Chou, K. C. & Blinn, J. R. (1997) J. Protein Chem. 16, 575-595. [DOI] [PubMed] [Google Scholar]
- 42.Wilmot, C. M. & Thornton, J. M. (1988) J. Mol. Biol. 203, 221-232. [DOI] [PubMed] [Google Scholar]
- 43.Boritz, E., Palmer, B. E., Livingston, B., Sette, A. & Wilson, C. C. (2003) J. Immunol. 170, 1106-1116. [DOI] [PubMed] [Google Scholar]
- 44.Reinherz, E. L., Tan, K., Tang, L., Kern, P., Liu, J., Xiong, Y., Hussey, R. E., Smolyar, A., Hare, B., Zhang, R., et al. (1999) Science 286, 1913-1921. [DOI] [PubMed] [Google Scholar]
- 45.Hennecke, J., Carfi, A. & Wiley, D. C. (2000) EMBO J. 19, 5611-5624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hennecke, J. & Wiley, D. C. (2002) J. Exp. Med. 195, 571-581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Siebold, C., Hansen, B. E., Wyer, J. R., Harlos, K., Esnouf, R. E., Svejgaard, A., Bell, J. I., Strominger, J. L., Jones, E. Y. & Fugger, L. (2004) Proc. Natl. Acad. Sci. USA 101, 1999-2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhu, Y., Rudensky, A. Y., Corper, A. L., Teyton, L. & Wilson, I. A. (2003) J. Mol. Biol. 326, 1157-1174. [DOI] [PubMed] [Google Scholar]
- 49.Kersh, G. J., Miley, M. J., Nelson, C. A., Grakoui, A., Horvath, S., Donermeyer, D. L., Kappler, J., Allen, P. M. & Fremont, D. H. (2001) J. Immunol. 166, 3345-3354. [DOI] [PubMed] [Google Scholar]
- 50.Fremont, D. H., Monnaie, D., Nelson, C. A., Hendrickson, W. A. & Unanue, E. R. (1998) Immunity 8, 305-317. [DOI] [PubMed] [Google Scholar]
- 51.Sant'Angelo, D. B., Robinson, E., Janeway, C. A., Jr., & Denzin, L. K. (2002) Eur. J. Immunol. 32, 2510-2520. [DOI] [PubMed] [Google Scholar]
- 52.Naujokas, M. F., Southwood, S., Mathies, S. J., Appella, E., Sette, A. & Miller, J. (1998) Cell. Immunol. 188, 49-54. [DOI] [PubMed] [Google Scholar]
- 53.Delano, W. (2002) The pymol Molecular Graphics System (Delano Scientific, San Carlos, CA).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.