Abstract
The phospholipid scramblases (PLSCR1 to PLSCR4) are a structurally and functionally unique class of proteins, which are products of a tetrad of genes conserved from Caenorhabditis elegans to humans. The best characterized member of this family, PLSCR1, is implicated in the remodeling of the transbilayer distribution of plasma membrane phospholipids but is also required for normal signaling through select growth factor receptors. Mice with targeted deletion of PLSCR1 display perinatal granulocytopenia due to defective response of hematopoietic precursors to granulocyte colony-stimulating factor and stem cell factor. To gain insight into the biologic function of another member of the PLSCR family, we investigated mice with targeted deletion of PLSCR3, a protein that like PLSCR1 is expressed in many blood cells but which, by contrast to PLSCR1, is also highly expressed in fat and muscle. PLSCR3-/- mice at 2 months of age displayed aberrant accumulation of abdominal fat when maintained on standard rodent chow, which was accompanied by insulin resistance, glucose intolerance, and dyslipidemia. Primary adipocytes and cultured bone-marrow-derived macrophages from PLSCR3-/- mice were engorged with neutral lipid, and adipocytes displayed defective responses to exogenous insulin. Plasma of PLSCR3-/- mice was elevated in non-high-density lipoproteins, cholesterol, triglycerides, nonesterified fatty acids, and leptin, whereas adiponectin was low. These data suggest that the expression of PLSCR3 may be required for normal adipocyte and/or macrophage maturation or function and raise the possibility that deletions or mutations affecting the PLSCR3-/- gene locus may contribute to the risk for lipid-related disorders in humans.
Keywords: obesity, adipocytes, lipids, macrophage, gene knockout
The phospholipid (PL) scramblase family (PLSCR1-4) represents a structurally unique class of plasma membrane proteins that were originally cloned and characterized in our laboratory (1). The first PLSCR identified (PLSCR1) was proposed to mediate the accelerated transbilayer migration of plasma membrane PL that is observed in platelets, erythrocytes, and many other cells under circumstances of elevated intracellular [Ca2+], as can occur upon platelet aggregation, cell injury by complement, and during apoptosis (1-10). PLSCR is conserved as a multigene family of four from Caenorhabditis elegans to humans; a single apparent orthologue (YJR100C) of unknown function has been identified in yeast (3, 11).
Recent data suggest a considerably more complex biology for PLSCR1 than its putative role in mediating transbilayer lipid movement (3, 5, 10, 12-19). PLSCR1 is transcriptionally up-regulated through IFN receptors and multiple growth factor receptors, and cells deficient in PLSCR1 exhibit defects in response to stimulation of the same receptors (13, 18, 20-22). When transcriptionally induced by IFN, a portion of the newly synthesized PLSCR1 localizes within the nucleus, in addition to its usual location at the plasma membrane (23). Nuclear import of PLSCR1 also occurs whenever its palmitoylation is inhibited and is mediated through its high-affinity interaction with importin-α, a cofactor of the importin/Ras/Ran nucleopore transport system (23, 24). Once imported into the nucleus, PLSCR1 tightly binds to genomic DNA, suggesting a possible role for this protein in nuclear transcription (24). Consistent with a role in growth factor receptor signaling, mice with targeted deletion of PLSCR1 exhibit defects in cell maturation and proliferation, most notably affecting granulocyte production from hematopoietic precursors in response to select growth factors, and cells deficient in PLSCR1 show defects in antiviral response to IFN (13, 14, 16, 19, 22). Although PLSCR1-/- mice exhibit perinatal granulocytopenia, they show no evidence of a defect in plasma membrane PL scramblase activity, suggesting that either the presumed role of this protein in transbilayer movement of plasma membrane PL is not correct, or, that this activity is maintained by multiple proteins, including other members of the PLSCR gene family (3, 10, 25).
We now examined the biologic role of another member of the PLSCR gene family, PLSCR3, a gene that like PLSCR1 is expressed in many blood cells, but which, by contrast to PLSCR1, is highly expressed in muscle, fat, and certain other cells and tissues (25). Whereas granulocyte production and platelet PL scramblase activity in PLSCR3-/- mice appeared normal, these animals showed a marked increase in depot fat, including that surrounding abdominal viscera, when fed a standard chow diet. They subsequently exhibited elevated plasma glucose, cholesterol (CHOL), triglycerides (TGs), and nonesterified fatty acids (NEFAs) relative to WT controls. These mice had abnormal glucose tolerance with evidence of insulin resistance, as seen in human Syndrome X (Metabolic Syndrome) (26). Adipocytes from PLSCR3-/- mice showed defects in insulin-dependent responses, including insulin-stimulated glucose uptake. Moreover, plasma levels of the adipocyte-derived cytokines adiponectin and leptin were abnormal.
Methods
Antibodies and Reagents. Anti-PLSCR3 mAb 3B4 was developed in our laboratory and does not cross-react with other members of the PLSCR gene family (data not shown). Anti-PLSCR1 mAb 1A8 has been described (23). Rabbit anti-GLUT4 antibody was from Biogenesis (Brentwood, NH); rabbit anti-insulin receptor substrate (IRS)-1, anti-IRS-2, and anti-insulin receptor (IR) antibodies were from Upstate (Charlottesville, VA); and rabbit anti-β-actin from Sigma. Horseradish peroxidase-conjugated secondary antibodies were from Jackson ImmunoResearch. Murine stem cell factor (SCF), granulocyte colony-stimulating factor (G-CSF), monocyte colony-stimulating factor, and IL-3 were from R & D Systems; Methocult 3234 was from StemCell Technologies (Vancouver); Wright-Giemsa was from Shandon (Pittsburgh); and Oil red O was from Sigma. Insulin (Humalog, Eli Lilly) was a gift from J. McCallum (The Scripps Clinic, La Jolla, CA).
PLSCR1-/- and PLSCR3-/- Mice. PLSCR1-/- mice have been described (16). In certain experiments, PLSCR1-/- mice of the inbred parental backgrounds of C57BL6J and 129SvEvBrd were each compared with the same gene deletion in the hybrid strain C57BL6J×129SvEvBRD. Targeting of the PLSCR3 gene locus in 129SvEvBRD murine embryonic stem cells was performed by Lexicon Genetics (The Woodlands, TX) by using pKOS-lacZ/MC1neo vector. The lacZ/MC1neo cassette was inserted to replace 1,127 bp of nucleotide sequence, deleting the first four exons. Clones confirming deletion by Southern blotting were implanted into pseudopregnant C57BL6J females and germ-line transmission was confirmed. No PLSCR3 antigen or mRNA was detected in homozygous PLSCR3-/- mice, by using cells and tissues (e.g., lung, fat, and platelets) that were known (in WT) to abundantly express PLSCR3. Mating of PLSCR1-/- with PLSCR3-/- mice was performed to generate a colony deficient in both genes [PLSCR(1&3)-/-]. In all experiments comparing PLSCR3-/- mice with WT, PLSCR1-/-, or PLSCR(1&3)-/-, mice were of identical genetic background (C57BL6J×129SvEvBRD). All mice were fed standard (≈5% fat) rodent chow (Harlan Teklad, Madison, WI).
Platelet coagulant activity and tail bleeding times were performed as described (16, 27).
Hematopoietic Cell Culture. Isolation and culture of bone marrow and spleen cells to induce myeloid expansion and differentiation into granulocytes or macrophages was performed essentially as described (16). To assess myeloid-committed progenitors and granulocyte differentiation, bone marrow (5 × 104) or spleen (2 × 105) cells were mixed with 1.5 ml of MethoCult 3234 supplemented with 20 ng/ml SCF and 60 ng/ml G-CSF. Cells were cultured for either 7 (bone marrow) or 10 (spleen) days, respectively. Colonies were scored as clusters containing >50 cells (16). Cells from these cultures were also evaluated by light microscopy after staining with Wright-Giemsa or Oil red O. Macrophage differentiation of bone marrow cells was routinely induced by 7-day culture in MethoCult 3234 supplemented with 5 ng/ml IL-3, 5 ng/ml granulocyte-macrophage colony-stimulating factor, and 10 ng/ml monocyte colony-stimulating factor. Alternatively, macrophage differentiation was induced by 7-day liquid culture in L929 cell-conditioned medium and adherent cells were assayed for cellular content of TGs and CHOL (esterified and nonesterified) essentially as described (28).
Tissue Histology. Adipose tissue was fixed with 10% buffered formaldehyde, embedded in paraffin, sectioned, stained with hematoxylin and eosin, and evaluated by light microscopy. Neutral lipids were visualized with Oil red O.
Light microscopy of stained cells or tissue was performed by using an Axioplan 2 microscope (Zeiss) and images were recorded with a color digital charge-coupled device camera (SV Micro color 80155).
Blood Collection. Blood plasma was routinely obtained after retro-orbital bleed into heparinized capillary tubes. Except where otherwise indicated, all animals were daytime-fasted for 6 h.
Blood Glucose and Glucose Tolerance Testing. Blood and plasma glucose concentrations were routinely measured by glucometer (Bayer). Glucose tolerance testing was performed after i.p. injection of d-glucose at a dose of 1 g/kg of body weight.
Plasma insulin, adiponectin, and leptin were each measured by using RIA kits (Linco Research, St. Charles, MO).
Analysis of Plasma Lipids and Lipoproteins. Plasma CHOL and TGs were measured by using an enzymatic assay kit from Sigma; NEFAs were measured by using an enzymatic assay kit from WAKO Chemicals USA (Richmond, VA). To determine CHOL distribution among plasma lipoproteins, plasmas from five mice of each genotype were pooled and subjected to high-resolution FPLC on Superdex 200 HR 10/30. Total, nonesterified, and esterified CHOL in each fraction was measured as described (28).
Automated Plasma Chemistries. In certain experiments, confirming measurements of plasma glucose, TGs, CHOL, and NEFAs were obtained by automated colorimetric analysis (The Jackson Laboratory).
Adipocyte Isolation. Adipocytes from gonadal or inguinal fat pads from age- and sex-matched pairs of WT and PLSCR1-/-, PLSCR3-/- or PLSCR(1&3)-/- mice were isolated essentially according to published procedures (29). After floatation, the cells were suspended in Krebs-Ringer-Hepes (KRH) buffer (120 mM NaCl/4 mM KH2PO4/1 mM MgSO4/0.75 mM CaCl2/10 mM NaHCO3/30 mM Hepes, pH 7.4).
Insulin-Stimulated Glucose Uptake. Isolated adipocytes were suspended (≈30% vol/vol) at 37°C in KRH containing 1% BSA, and prestimulated for 20 min with insulin (final concentrations, 0 to 1 × 10-6 unit/ml). Tracer uptake was initiated by addition of 0.125 μCi (1 Ci = 37 GBq) of 2-[14C]deoxy-d-glucose (PerkinElmer) in KRH containing 0.5 mM 2-deoxy-d-glucose. After 20 min, uptake was terminated by addition of 20 μM cytochalasin B, cells were recovered by floatation on dinonyl phthalate, and cell-associated radioactivity was determined by scintillation counting. Correction for nonspecific uptake was made by subtraction of cell-associated radioactivity determined for identical samples continuously maintained in cytochalasin B.
Surface Insulin Receptors. 125I-Insulin binding to adipocytes was measured at 16°C during suspension (≈20% vol/vol) in KRH containing 5% BSA, 50 μg/ml d-glucose, and 200 nM adenosine. 125I-insulin (Tyr A14, PerkinElmer) was added at a final concentration of 20 nM (≈1 μCi per tube) in the absence or presence of 100-fold excess unlabeled insulin. After 1 h, cells were separated from unbound radioactivity by floatation. Cell-associated 125I-insulin was determined by scintillation counting with correction for nonspecificity uptake.
Western Blotting. Western blotting was performed after direct lysis of platelets or adipocytes in SDS sample buffer containing 10 mM DTT. Minced soleus muscle and lung were frozen for 1 h at -80°C, homogenized, and lysed for 1 h on ice in PBS, 2% Triton X-100, 5 mM EDTA containing protease inhibitors, before addition to SDS sample buffer. Cell lysates were resolved by SDS/PAGE and transferred to nitrocellulose. Immunoblotting was performed with antibodies as indicated in the figure Legends, with detection by chemiluminescence (Pierce).
Results
Normal Procoagulant Activity and Tail-Bleeding Times in PLSCR3-/- Mice. The first member of the PLSCR gene family, PLSCR1, was originally identified based on its capacity to promote Ca2+-mediated transbilayer movement PLs (“PL scramblase” activity) when reconstituted into proteoliposomes. Because transbilayer scrambling of plasma membrane PL with consequent cell surface exposure of phosphatidylserine is thought to contribute to the initiation of blood coagulation, it was postulated that PLSCR1 deficiency might manifest as a defect in cellular procoagulant activity and possibly impaired hemostasis. However, platelets from PLSCR1-/- mice exhibited normal capacity to mobilize phosphatidylserine to the cell surface, and no apparent hemostatic abnormality was observed (16). Because PLSCR1 and PLSCR3 are coexpressed in platelets (see Fig. 2a, below), redundancy in PL scramblase activity among the two PLSCR family members might obscure a defect in activity arising from deletion of PLSCR1. However, as was observed in the case of PLSCR1-/-, we detected no difference from WT in plasma membrane PL scramblase activity of platelets isolated from either PLSCR3-/- or PLSCR(1&3)-/- mice (data not shown).
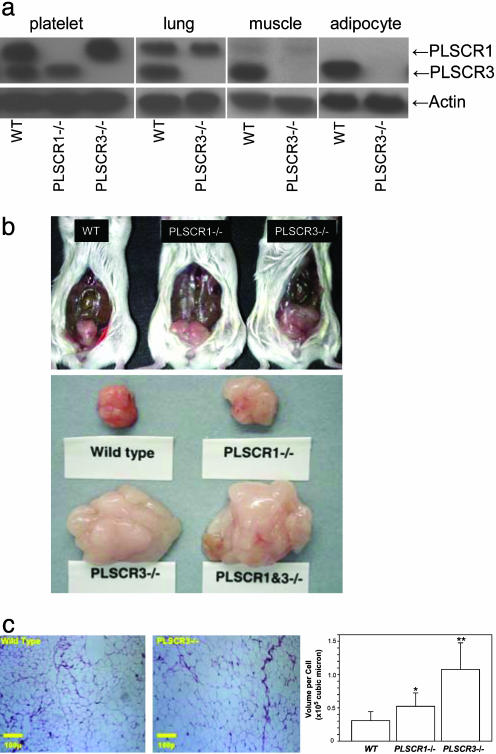
Fig. 2.
PLSCR3 is highly expressed in primary adipocytes and its deletion causes increases in fat mass with elevated adipocyte size. (a) Western blotting for PLSCR1 (35.1 kDa) and PLSCR3 (31.7 kDa) was performed on cells and tissues indicated by using monospecific mAb 1A8 and 3B4, respectively. Muscle refers to soleus skeletal muscle. Samples obtained from either WT, PLSCR1-/-, or PLSCR3-/- mice were matched for total protein. Also shown is immunoblot for β-actin (see Methods). (b) Upper photographs show uterine fat pads of WT, PLSCR1-/-, and PLSCR3-/- mice at 2 months of age. All animals were fed a standard rodent chow. This figure is representative of results obtained with >10 mice of each genotype. At ≈2 months of age, the wet weights of the gonadal fat pads from PLSCR3-/- mice (expressed as the ratio to fat pads obtained from age- and sex-matched WT controls) averaged 2.06 ± 0.1 (mean ± SD) and for PLSCR1-/-, 1.46 ± 0.3 (mean ± SD). Lower photographs show epididymal fat pads at 18 weeks of age. Wet weights were WT, 171 mg; PLSCR1-/-, 301 mg; PLSCR3-/-, 1,410 mg; PLSCR(1&3)-/-, 1,871 mg. (c) Hematoxylin and eosin staining of fat pads from WT (Left) and PLSCR3-/- (Center) epididymal fat pads. Similar results were obtained with uterine fat pads (not shown). Quantitative measurement of average cell diameter (mean ± SD) of all fields examined was as follows: WT (38.8 ± 3.3 μm); PLSCR3-/- (60.9 ± 4.0 μm). Not shown is PLSCR1-/- (44.8 ± 4.8 μm). Bar graph plots the calculated cellular volume of each, assuming spherical geometries. *, P = 0.003 (PLSCR1-/- compared with WT); **, P = 1.6E-8 (PLSCR3-/- compared with WT).
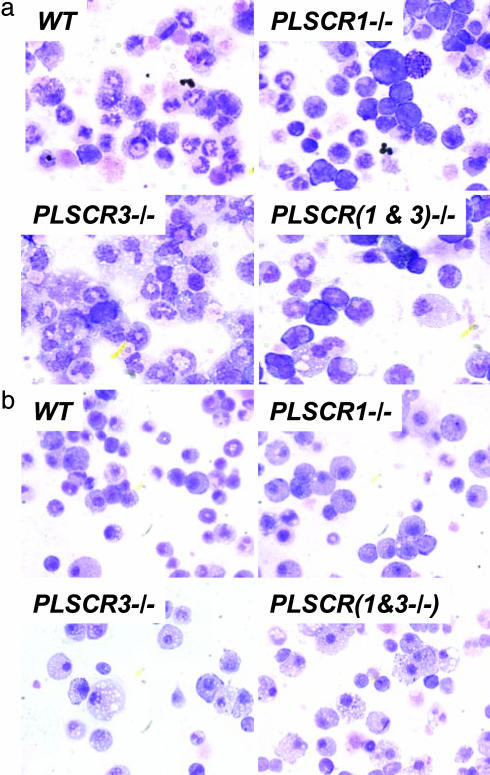
Response of PLSCR3-/- Hematopoietic Precursor Cells to Growth Factors. We previously reported that hematopoietic precursor cells from PLSCR1-/- mice exhibit an impaired proliferation and differentiation to mature granulocytes in response to SCF and G-CSF (16). Because PLSCR1 and PLSCR3 are coexpressed in a number of blood cells, we evaluated the response of PLSCR3-/- hematopoietic stem cells to these growth factors. As illustrated by Table 1, which is published as supporting information on the PNAS web site, by contrast to what was observed for PLSCR1-/-, the proliferative response of PLSCR3-/- hematopoietic cells to SCF and G-CSF was normal and indistinguishable from that of WT cells. As expected, cells from PLSCR(1&3)-/- mice reproduced the myeloid abnormalities observed for PLSCR1-/- cells. Consistent with data previously reported for PLSCR1-/-, bone marrow cells from PLSCR1-/- and PLSCR(1&3)-/- mice also exhibited impaired maturation in response to these growth factors, as evidenced by the presence of fewer cells with polylobulated or circular nuclear chromatin (characteristic of mature murine granulocytes), and a higher proportion of immature-appearing cells, distinguished by prominent round nuclei (Fig. 1a). By contrast to cells lacking PLSCR1, the nuclear morphology of the PLSCR3-/- cells was indistinguishable from that of WT, suggesting normal granulocytic maturation. However, those cells lacking PLSCR3 expression [i.e., PLSCR3-/- and PLSCR(1&3)-/-] showed a distinct increase in the number and size of lipid droplets in the cytoplasm. The neutral lipid content of these intracellular droplets was confirmed by staining with Oil Red O (data not shown). A marked increase in intracellular lipid droplets was also observed when PLSCR3-/- [and PLSCR(1&3)-/-] bone marrow-derived hematopoietic precursors were cultured under conditions to induce differentiation to macrophages (Fig. 1b). A distinct increase in cellular content of TGs (112.1 ± 14.7 vs. 40.3 ± 1.6 μg/mg cell protein) and esterified CHOL (21.8 ± 4.9 vs. 5.0 ± 3.2 μg/mg cell protein) in the PLSCR3-/- bone marrow-derived macrophages (vs. WT) was confirmed by direct assay of the cell-associated lipids (Table 2, which is published as supporting information on the PNAS web site).
Fig. 1.
In vitro induction of PLSCR3-/- granulocytes and macrophages from hematopoietic precursors. Shown are Wright-Giemsa-stained cultures of bone marrow cells after culture to induce differentiation and maturation to either granulocytes (a) or macrophages (b) as detailed in Methods (×400). Fields shown are representative of >100 surveyed from each of at least three independent experiments. (a) Note prominence of immature-appearing nuclei in association with the PLSCR1-/- genotype and the increase in frequency of cells displaying cytoplasmic lipidoid inclusions in association with the PLSCR3-/- genotype. Of 2,000 cells counted in multiple random fields, the % of cells exhibiting such lipoid inclusions was: WT, 0.8%; PLSCR1-/-, 0.9%; PLSCR3-/-, 2.2%; PLSCR(1&3)-/-, 3.4%. (b) Prevalence of cells with apparent lipoid cytoplasmic inclusions in association with the PLSCR3-/- genotype (compare Table 2).
Adiposity of PLSCR3-/- Mice. Whereas PLSCR3 is widely distributed in a variety of tissues similar to PLSCR1 (25), we noted that PLSCR3 was highly expressed in fat, where PLSCR1 was not detected by Western blotting (Fig. 2a). Our observation that myeloid-differentiated bone marrow cultures from PLSCR3-/- mice accumulated intracellular lipid prompted us to look for other abnormalities of lipid metabolism. Necropsy of PLSCR3-/- mice starting at approximately 2 months of age revealed increased size of all depot fat pads examined, relative to age, weight, and sex-matched WT mice (Fig. 2b and data not shown). A more modest increase in fat pad size was also observed in PLSCR1-/- mice, suggesting partial overlap in function of these two genes. Results for PLSCR(1&3)-/- mice closely approximated those for PLSCR3-/- mice, suggesting that expression of PLSCR3 is more significant than PLSCR1 in regulating fat accumulation. Body weights of PLSCR3-/- mice were slightly elevated by the age of 2 months, and these mice were mildly obese by age 19 weeks (33.3 ± 2.2 vs. 28.9 ± 2.8 g, PLSCR3-/- vs. WT; Table 3, which is published as supporting information on the PNAS web site). Mean cell diameters of adipocytes in fat pads from PLSCR3-/- and PLSCR1-/- mice showed increases relative to WT that were consistent with total mass (Fig. 2c). The modest but significant increase in adiposity of PLSCR1-/- mice relative to WT in the hybrid C57BL6J×129SvEvBRD strain was also observed when PLSCR1-/- and WT mice in the inbred parental genetic backgrounds (C57BL6J or 129SvEvBRD) were each compared (data not shown). This finding suggests that the increase in depot fat detected in both PLSCR1-/- and PLSCR3-/- mice (relative to matched WT) was directly related to the targeted deletions at the PLSCR1 or PLSCR3 loci (see Discussion).
Impaired Glucose Tolerance and Insulin Resistance in PLSCR3-/- Mice. As shown in Table 3, at 2 months of age fasting blood glucose levels were significantly elevated (207.0 ± 41.8 vs. 159.3 ± 32.7 mg/dl) in the PLSCR3-/- mice (compared with matched WT), and the persistence of mild hyperglycemia was confirmed at all subsequent testing points (through 6 months; data not shown). Fasting plasma insulin levels appeared somewhat higher in PLSCR3-/- mice, but these differences were not statistically significant (Table 3), suggesting that the hyperglycemia in PLSCR3-/- was secondary to insulin resistance rather than a defect in insulin secretion (see Discussion). Upon glucose tolerance testing, an ≈2-fold elevation in peak blood glucose levels was consistently obser ved in both PLSCR3-/- and PLSCR(1&3)-/- relative to WT mice (Fig. 3a). Moreover, no significant difference was observed in the glucose tolerance of PLSCR3-/- vs. PLSCR(1&3)-/- mice, suggesting a dominant influence of PLSCR3 in regulating this phenotype, consistent with the apparent dominant function of PLSCR3 in regulating fat pad size (see above).
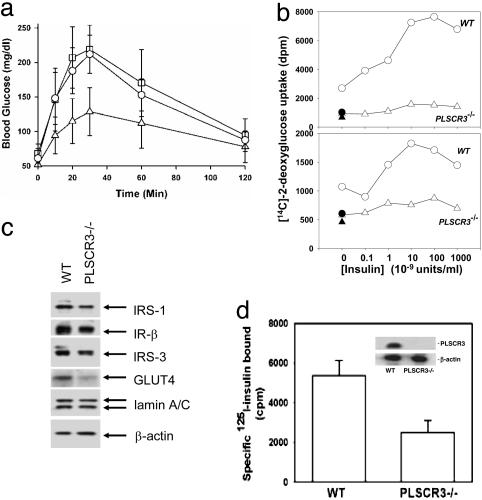
Fig. 3.
PLSCR3-/- mice show glucose intolerance and their adipocytes exhibit insulin resistance with diminished expression of IRs. (a) Glucose tolerance testing. WT (triangles), PLSCR3-/- (squares), and combined PLSCR(1&3)-/- (circles) mice were fasted for 18 h before i.p. injection of d-glucose. Blood glucose levels were measured at times indicated. Symbols represent mean ± SD (n = 8) of pooled data for four males and four females of each genotype. Animals were matched at 2 months of age (±2 d). Analysis of these data revealed no obvious or consistent difference in either fasted blood glucose or peak response to i.p. d-glucose in the male versus female animals evaluated (data not shown). These data were from a single experiment but representative of three. (b) Insulin-stimulated uptake of [14C]deoxyglucose. Experiments were performed with adipocytes isolated from epididymal (Upper) and inguinal (Lower) fat pads of 3-month-old WT and PLSCR3-/- mice. Cells were matched for total protein and prestimulated (20 min, 37°C) with insulin (concentrations on abscissa). Net influx of 2-[14C]deoxy-d-glucose was then measured 20 min after addition of tracer. ▴ and •, nonspecific uptake in WT and PLSCR3-/- adipocytes, respectively, measured in the presence of cytocholasin B. See Methods. (c) Western blotting for expression of IR, IRS-1, IRS-3, and GLUT4 in PLSCR3-/- adipocytes. Gonadal adipocytes from WT and PLSCR3-/- mice were matched on the basis of total cellular protein and Western blotted for expression of IR (β-chain), IRS-1, IRS-3, and GLUT4. The gel was also developed for β-actin and lamin (A and C). The relative band intensities by scanning densitometry (Kodak Image Station 440 CF) of these proteins detected in PLSCR3-/- adipocytes (expressed as ratios to that in WT) were: IRS-1 (0.54); IR-β (0.82); IRS-3 (0.63); GLUT4 (0.38); lamin A/C (1.21); β-actin (1.15). Data are representative of results obtained in four independent experiments with different mice. (d) Reduced expression of cell surface IRs. 125I-insulin binding to epididymal adipocytes isolated from 3-month-old WT and PLSCR3-/- mice is shown. Binding was performed at 16°C, with correction for nonspecific binding (see Methods). Cells were matched based on protein content. (Inset) Western blot of PLSCR3 and β-actin in each sample. These data, from a single experiment, are representative of two so performed.
Dyslipidemia in PLSCR3-/- Mice. Analysis of plasma lipids revealed significant increases in total CHOL (125 ± 28 vs. 85 ± 16 mg/dl), TGs (78 ± 10 vs. 56 ± 9 mg/dl), and NEFAs (1.27 ± 0.18 vs. 0.94 ± 0.26 mEq/liter) in PLSCR3-/- vs. WT mice (Table 4, which is published as supporting information on the PNAS web site). Analysis of the distribution of CHOL among the plasma lipoproteins revealed that the elevation in CHOL observed in PLSCR3-/- plasma represented a >2-fold increase in CHOL associated with apo B-containing lipoproteins (very-low-density and low-density lipoproteins), with only a small increase in high-density lipoprotein CHOL (Table 5, which is published as supporting information on the PNAS web site). Furthermore, the increase in plasma CHOL reflected a >2-fold increase in nonesterified CHOL in very-low-density and low-density lipoproteins.
Impaired Insulin-Stimulated Glucose Uptake by PLSCR3-/- Adipocytes. Adipocytes isolated from inguinal and gonadal fat pads were evaluated for insulin-stimulated uptake of 2-deoxy-d-glucose. As shown in Fig. 3b, PLSCR3-/- adipocytes exhibited a marked decrease in insulin-stimulated 2-deoxy-d-glucose uptake, compared with WT adipocytes. Western blotting revealed a decrease in cellular GLUT4 expression, which could at least partially contribute to the observed decrease in glucose transport. We also observed decreased expression of IR (detected with IR β-subunit antibody) and several of the known IR substrates (Fig. 3c). The apparent decrease in IR detected by Western blotting of primary PLSCR3-/- adipocytes was confirmed by direct measurement of 125I-insulin binding to these cells, which revealed a 50% decrease compared with controls (Fig. 3d). Consistent with this decrease in IR expression, insulin-stimulated IR autophosphorylation and Tyr-phosphorylation of substrates IRS-1 and IRS-3 were attenuated in PLSCR3-/- adipocytes (data not shown).
Abnormal Levels of Adiponectin and Leptin in PLSCR3-/- Mice. To determine whether the observed defects in PLSCR3-/- mice were associated with altered adipokine secretion, plasma concentrations of adiponectin and leptin were measured. As summarized in Table 6, which is published as supporting information on the PNAS web site, plasma adiponectin in PLSCR3-/- mice at both 8 and 19 weeks was ≈50% decreased compared with WT controls, whereas a distinct elevation of plasma leptin in the PLSCR3-/- mice was noted by 19 weeks.
Discussion
Our data suggest that the expression of PLSCR3, and to a lesser degree that of PLSCR1, is critical to the normal regulation of fat accumulation in mice. Deletion of PLSCR3 gives rise to the formation of lipid-engorged adipocytes and many of the consequences of adipocyte dysfunction that are associated in humans with obesity- and abdominal fat-related insulin resistance (26, 30-33). Cultured bone marrow-derived macrophages from these animals display distinct increases in intracellular lipid droplets, reflecting elevations in CHOL and TGs (Fig. 1b and Table 2). Although our evaluation of the effects of PLSCR gene deletion was conducted in a hybrid-strain mouse, we consider it unlikely that the phenotype we observed reflects the influence of another gene defect inadvertently selected or cosegregating with the targeted gene. We consistently observed a similar, although less pronounced, abnormality of increased adiposity in PLSCR1-/- mice (relative to WT controls) as was observed for PLSCR3-/- mice, irrespective of whether the PLSCR1 gene deletion was evaluated in the hybrid C57BL6J×129SvEvBRD background, or after backcross into either of the inbred parental strains (see Methods and Results). It is also important to note that as in humans, murine PLSCR1 and PLSCR3 are located on separate chromosomes, and that each gene is independently targeted with constructs containing unrelated sequences (25).
Obesity, in particular, the accumulation of abdominal fat with formation of enlarged, lipid-engorged adipocytes, has emerged as a key acquired risk factor for the onset of type 2 diabetes (26, 33, 34). Although originally regarded as a distinct disease, it is now recognized that type 2 diabetes (and the hyperglycemia associated with it) is often a manifestation of a broader underlying disorder referred to as Syndrome X (Metabolic Syndrome) (26). This syndrome is characterized by abdominal obesity, insulin resistance with mild glucose intolerance, dyslipidemia, mild hypertension, and increased risk for developing vascular disease. The precise causal relationship as to how increased abdominal fat leads to insulin resistance and diabetes remains to be established. Factors that have been implicated include tumor necrosis factor-α, NEFAs, and “adipokines” including leptin, adiponectin, and resistin released from adipocytes (33, 35-37). It has been suggested that the high correlation between an increase in abdominal fat with Syndrome X may relate to the fact that adipose tissue associated with abdominal viscera (in particular, in omental and mesenteric sites) drains directly into the portal venous system, thus exposing the liver to the undiluted repertoire of metabolites and secretory products of these fat depots (38, 39). Our data reveal that in mice, absence of PLSCR3 caused a phenotype that in many respects recapitulates elements of adipose-related insulin resistance in man, including formation of lipid-engorged adipocytes with increased intraabdominal fat stores, impaired glucose tolerance, dyslipidemia, and abnormal plasma levels of adiponectin and leptin. Increased fat pads, slightly elevated fasting blood glucose concentration, and impaired glucose tolerance were observed in animals as early as 8 weeks of age. Surprisingly, the abnormalities in glucose metabolism were already observed at a time when the total body weight of the PLSCR3-/- mice was only ≈10% above that of WT animals (Table 3). Because dysfunctional adipocytes (32, 36) and macrophages (40) have each been implicated in the development of systemic insulin resistance, it is of particular interest to note that, in addition to the abnormalities detected in primary adipocytes isolated from PLSCR3-/- mice, our data also suggest aberrant lipid accumulation in bone marrow-derived macrophages from these animals (Fig. 1). In bone marrow-derived macrophages, cells were examined at least 1 week after in vitro cell culture in the absence of autologous serum, implying that the observed lipid accumulation in the PLSCR3-/- cells was not the consequence of the elevated plasma TGs and CHOL of the donor mice.
Whereas our data indicate that there are major defects in the IR activation pathway of the adipocytes obtained from PLSCR3-/- animals, it is important to note that we cannot exclude that such defects arise secondarily due to the effect of PLSCR3 deletion on either (i) another cytokine or growth factor receptor pathway or metabolic process (e.g., lipid metabolism) in the adipocyte that might adversely affect its insulin-dependent responses, or (ii) an abnormality in another cell (e.g., macrophage) that might in turn adversely affect adipocyte function through a secreted systemic factor.
As previously noted, the cellular and molecular function of the PLSCR proteins remains incompletely understood. The best characterized member of this gene family, PLSCR1, was implicated to participate in the remodeling of the topology of plasma membrane PL under circumstances of elevated intracellular calcium; it also regulates cellular responses (including proliferation, maturation, and terminal differentiation) to several growth factors, including the hematopoietic growth factors G-CSF and SCF (2, 3, 16). PLSCR1 expression was not detected in mature adipocytes (by Western blotting; Fig. 2a), whereas the expression of PLSCR3 was abundant, suggesting that PLSCR1 expression is ultimately turned off in the mature adipocyte. In hematopoietic cells PLSCR1 is required for normal myeloid proliferative and maturational responses to G-CSF and SCF, its expression is markedly increased in response to these growth factors and is elevated in fully differentiated granulocytes, when compared with precursor cells (16). In contrast, the level of expression of PLSCR3 is largely unaffected (data not shown). Whether PLSCR3 plays an analogous role in signaling and the cellular response to insulin (or to other growth factors and cytokines) in the adipocyte is now unknown but is suggested by its elevated expression in WT adipocytes and by the abnormal accumulation of fat stores in the PLSCR3-/- adipocyte.
The frequency and consequence of a gene defect in humans affecting the level of expression or function of PLSCR3, or other members of the PLSCR gene family, is unknown. Deletion of the single apparent PLSCR orthologue in yeast (YJR100C) is non-lethal, as is deletion of PLSCR1, PLSCR2, PLSCR3, or, PLSCR1&3 in mice (refs. 3, 11, and 16; data not shown). The aberrant accumulation of adipocyte fat stores, with onset of insulin resistance and dyslipidemia in PLSCR3-/- and, to a lesser extent, in PLSCR1-/- mice raises the question of whether mutations affecting either the expression or function of the PLSCR proteins in humans might also influence the risk for development of similar systemic disorders.
Supplementary Material
Acknowledgments
We thank Drs. Daniel Steinberg and Joseph Witztum (University of California at San Diego) for invaluable comments and suggestions, Hongfan Peng, Daniela Junqueira, Jo-Lawrence Bigcas, Audrey Black, and David J. Bonnet for the superb technical assistance, and Drs. Luanne Peters and James McCallum for their generous assistance. This work was supported by National Institutes of Health Grants HL36946 (to P.J.S.), HL63819 (to P.J.S.), DK60484 (to A.H.), DK33651 (to J.M.O.), HL43815 (to L.K.C.), and HL35297 (to L.K.C.), and by the Stein Endowment Fund (P.J.S. and T.W.), the Hilblom Foundation (J.M.O.), and the Department of Veterans Affairs Research Service (J.M.O.). This is manuscript no. 16585-MEM from The Scripps Research Institute.
Abbreviations: PL, phospholipid(s); KRH, Krebs-Ringer-Hepes; SCF, stem cell factor; G-CSF, granulocyte colony-stimulating factor; M-CSF, monocyte colony-stimulating factor; CHOL, cholesterol; TG, triglyceride; NEFA, nonesterified fatty acid; IR, insulin receptor; IRS, insulin receptor substrate.
References
- 1.Zhou, Q. S., Zhao, J., Stout, J. G., Luhm, R. A., Wiedmer, T. & Sims, P. J. (1997) J. Biol. Chem. 272, 18240-18244. [DOI] [PubMed] [Google Scholar]
- 2.Zhao, J., Zhou, Q., Wiedmer, T. & Sims, P. J. (1998) J. Biol. Chem. 273, 6603-6606. [DOI] [PubMed] [Google Scholar]
- 3.Sims, P. J. & Wiedmer, T. (2001) Thromb. Haemostasis 86, 266-275. [PubMed] [Google Scholar]
- 4.Daleke, D. L. (2003) J. Lipid Res. 44, 233-242. [DOI] [PubMed] [Google Scholar]
- 5.Frasch, S. C., Henson, P. M., Kailey, J. M., Richter, D. A., Janes, M. S., Fadok, V. A. & Bratton, D. L. (2000) J. Biol. Chem. 275, 23065-23073. [DOI] [PubMed] [Google Scholar]
- 6.Heemskerk, J. W. M., Bevers, E. M. & Lindhout, T. (2002) Thromb. Haemostasis 88, 186-193. [PubMed] [Google Scholar]
- 7.Kagan, V. E., Fabisiak, J. P., Shvedova, A. A., Tyurina, Y. Y., Tyurin, V. A., Schor, N. F. & Kawai, K. (2000) FEBS Lett. 477, 1-7. [DOI] [PubMed] [Google Scholar]
- 8.Williamson, P., Christie, A., Kohlin, T., Schlegel, R. A., Comfurius, P., Harmsma, M., Zwaal, R. F. A. & Bevers, E. M. (2001) Biochemistry 40, 8065-8072. [DOI] [PubMed] [Google Scholar]
- 9.Williamson, P. & Schlegel, R. A. (2002) Biochim. Biophys. Acta Mol. Cell. Biol. Lipids 1585, 53-63. [Google Scholar]
- 10.Frasch, S. C., Henson, P. M., Nagosa, K., Fessler, M. B., Borregaard, N. & Bratton, D. L. (2004) J. Biol. Chem. 279, 17625-17633. [DOI] [PubMed] [Google Scholar]
- 11.Winzeler, E. A., Shoemaker, D. D., Astromoff, A., Liang, H., Anderson, K., Andre, B., Bangham, R., Benito, R., Boeke, J. D., Bussey, H., et al. (1999) Science 285, 901-906. [DOI] [PubMed] [Google Scholar]
- 12.Pastorelli, C., Veiga, J., Charles, N., Voignier, E., Moussu, H., Monteiro, R. C. & Benhamou, M. (2002) Mol. Immunol. 38, 1235-1238. [DOI] [PubMed] [Google Scholar]
- 13.Kasukabe, T., Okabe-Kado, J. & Honma, Y. (1997) Blood 89, 2975-2985. [PubMed] [Google Scholar]
- 14.Nakamaki, T., Okabe-Kado, J., Yamamoto-Yamaguchi, Y., Hino, K. I., Tomoyasu, S., Honma, Y. & Kasukabe, T. (2002) Exp. Hematol. 30, 421-429. [DOI] [PubMed] [Google Scholar]
- 15.Sun, J., Zhao, J., Schwartz, M. A., Wang, J. Y. J., Wiedmer, T. & Sims, P. J. (2001) J. Biol. Chem. 276, 28984-28990. [DOI] [PubMed] [Google Scholar]
- 16.Zhou, Q. S., Zhao, J., Wiedmer, T. & Sims, P. J. (2002) Blood 99, 4030-4038. [DOI] [PubMed] [Google Scholar]
- 17.Sun, J., Nanjundan, M., Pike, L. J., Wiedmer, T. & Sims, P. J. (2002) Biochemistry 41, 6338-6345. [DOI] [PubMed] [Google Scholar]
- 18.Nanjundan, M., Sun, J., Zhao, J., Zhou, Q., Sims, P. J. & Wiedmer, T. (2003) J. Biol. Chem. 39, 37413-37418. [DOI] [PubMed] [Google Scholar]
- 19.Kasukabe, T., Kobayashi, H., Kaneko, Y., Okabe-Kado, J. & Honma, Y. (1998) Biochem. Biophys. Res. Commun. 249, 449-455. [DOI] [PubMed] [Google Scholar]
- 20.Der, S. D., Zhou, A. M., Williams, B. R. & Silverman, R. H. (1998) Proc. Natl. Acad. Sci. USA 95, 15623-15628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhou, Q. S., Zhao, J., Al Zoghaibi, F., Zhou, A. M., Wiedmer, T., Silverman, R. H. & Sims, P. J. (2000) Blood 95, 2593-2599. [PubMed] [Google Scholar]
- 22.Dong, B., Zhou, Q., Zhao, J., Zhou, A., Harty, R. N., Bose, S., Banerjee, A., Guenther, J., Slee, R., Williams, B. R. G., et al. (2004) J. Virol. 78, 8983-8993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wiedmer, T., Zhao, J., Nanjundan, M. & Sims, P. J. (2003) Biochemistry 42, 1227-1233. [DOI] [PubMed] [Google Scholar]
- 24.Ben-Efraim, I., Zhou, Q., Wiedmer, T., Gerace, L. & Sims, P. J. (2004) Biochemistry 43, 3518-3526. [DOI] [PubMed] [Google Scholar]
- 25.Wiedmer, T., Zhou, Q. S., Kwoh, D. Y. & Sims, P. J. (2000) Biochim. Biophys. Acta Biomembr. 1467, 244-253. [DOI] [PubMed] [Google Scholar]
- 26.Zimmet, P., Alberti, K. G. & Shaw, J. (2001) Nature 414, 782-787. [DOI] [PubMed] [Google Scholar]
- 27.Tibbles, H. E., Vassilev, A., Wendorf, H., Schonhoff, D., Zhu, D., Lorenz, D., Waurzyniak, B., Liu, X. P. & Uckun, F. M. (2001) J. Biol. Chem. 276, 17815-17822. [DOI] [PubMed] [Google Scholar]
- 28.Boisvert, W. A., Black, A. S. & Curtiss, L. K. (1999) Arterioscler. Thromb. Vasc. Biol. 19, 525-530. [DOI] [PubMed] [Google Scholar]
- 29.Johnston, A. M., Pirola, L. & Van Obberghen, E. (2003) FEBS Lett. 546, 32-36. [DOI] [PubMed] [Google Scholar]
- 30.Moller, D. E. (2001) Nature 414, 821-827. [DOI] [PubMed] [Google Scholar]
- 31.Brownlee, M. (2001) Nature 414, 813-820. [DOI] [PubMed] [Google Scholar]
- 32.Weyer, C., Foley, J. E., Bogardus, C., Tataranni, P. A. & Pratley, R. E. (2000) Diabetologia 43, 1498-1506. [DOI] [PubMed] [Google Scholar]
- 33.Greenberg, A. S. & McDaniel, M. L. (2002) Eur. J. Clin. Invest. 32, Suppl. 3, 24-34. [DOI] [PubMed] [Google Scholar]
- 34.Boden, G. & Shulman, G. I. (2002) Eur. J. Clin. Invest. 32, Suppl. 3, 14-23. [DOI] [PubMed] [Google Scholar]
- 35.Saltiel, A. R. & Kahn, C. R. (2001) Nature 414, 799-806. [DOI] [PubMed] [Google Scholar]
- 36.Rajala, M. W. & Scherer, P. E. (2003) Endocrinology 144, 3765-3773. [DOI] [PubMed] [Google Scholar]
- 37.Bergman, R. N. & Ader, M. (2000) Trends Endocrinol. Metab. 11, 351-356. [DOI] [PubMed] [Google Scholar]
- 38.Montague, C. T. & O'Rahilly, S. (2000) Diabetes 49, 883-888. [DOI] [PubMed] [Google Scholar]
- 39.Masuzaki, H., Paterson, J., Shinyama, H., Morton, N. M., Mullins, J. J., Seckl, J. R. & Flier, J. S. (2001) Science 294, 2166-2170. [DOI] [PubMed] [Google Scholar]
- 40.Weisberg, S. P. M. (2003) J. Clin. Invest. 112, 1796-1808. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.