Abstract
Clinical applications of mesenchymal stromal cells (MSCs) require the manufacture of large cell lots, which involves multiple passages for cell expansion and sometimes genetic modification. MSCs from various sources, including bone marrow (BM), exhibit high donor-to-donor variability in their growth characteristics. This can lead to unpredictable manufacturing outcomes with respect to success or failure of individual lots. Early determination of lot success has the potential to reduce the cost and improve the efficiency of the MSC manufacturing process. However, methods that effectively predict lot growth potential early in the manufacturing process are currently lacking. Here we report that the growth potential of an MSC lot can be predicted a few days after BM plating based on α-smooth muscle actin (αSMA) protein expression in large colony-forming unit-fibroblast (CFU-f) colonies. The proposed prediction method could be a useful tool to prospectively determine MSC lot success or failure.
Key words: Mesenchymal stromal cells (MSCs), Colony-forming units-fibroblast (CFU-f), α-Smooth muscle actin (αSMA), Assay
INTRODUCTION
Mesenchymal stromal cells (MSCs) were first detected in bone marrow (BM) cultures as cells forming adherent clonal fibroblastic colonies and capable of undergoing more than 20 population doublings, hence their name: colony-forming units-fibroblast (CFU-f)1,2. The ability of these culture-expanded cells to differentiate into bone, cartilage, reticular, and adipose tissue and to transfer hematopoietic microenvironment3,4, along with their potent secretome5 and immunomodulatory properties6, brought them to the frontier of cell therapeutic efforts. More than 350 clinical trials currently use expanded MSCs from BM or other sources as a potential treatment for various skeletal, degenerative, and immune disorders. Moreover, large cell lots of allogeneic MSCs can be manufactured for “off-the-shelf” use on a number of patients. The manufacturing of large BM-MSC lots, however, can be sometimes challenging due to the high variability in the growth potential of MSCs derived from different BM donors7–9. It would therefore be advantageous to have tools to predict MSC culturing outcome and terminate unsuccessful lots early in the manufacturing process.
CFU-f assay enumerates clonogenic adherent cells (CFU-f) within the BM preparation. At BM plating densities ensuring linearity between numbers of plated cells and resulting colonies, colony number/105 plated cells [colony-forming efficiency (CFE)] is dependent on the donor, the BM specimen harvesting method, the cell isolation steps (i.e., washing), and the culturing protocol10,11. In 7–10 days, the cultures contain colonies that vary greatly in size. CFU-fs represent a mixture of committed and intermediate progenitors and stem cells; they are typically distinguished on the basis of the potency of their colonies to undergo single, bilineage, or trilineage mesenchymal differentiation. It is generally believed that in culture, stem cells are more prolific than both committed and intermediate progenitors, but have longer lag period after BM explantation than committed progenitors12. In light of this, the following assays have been proposed as predictors of MSC growth potential: trilineage differentiation potential of colonies13,14, CFE after the first passage9, monitoring of proliferation and viability at early passages15, and using motility as a marker of multipotent (stem) cells16. However, these approaches require MSC passaging and are laborious, which makes them impractical as growth predictors for cell manufacturing.
α-Smooth muscle actin (αSMA) is a contractile actin isoform, a hallmark of vascular smooth muscle cells and myofibroblasts that differentiate from many mesenchymal cell types in response to tissue injury and repair; in the wound, it is stimulated by the increased stiffness of the substrate17. In cultured MSCs, αSMA colocalizes with stress fibers developing during culturing18. Sorting passaged MSCs by cell size (small and large cell populations) concomitantly sorts them by expression of αSMA (low and high, correspondingly) and by clonal and differentiation potential (high and low, correspondingly). Moreover, knockdown of αSMA or plating cells onto a soft surface restores clonogenicity and differentiation potential19. Thus, to a certain degree, αSMA expression in early MSCs could be regarded as an artifact of culturing these mechanosensitive cells on a stiff plastic surface20. While there is widespread use of plastic surfaces in manufacturing of MSCs, the variability among BM donors with respect to their MSC response to the stiff plastic culture surface has not been studied yet.
Here we describe a novel colorimetric colony-based assay that can be used to predict the growth rates of MSC lots later in the manufacturing culture process based on parameters measured only few days after BM plating. The αSMA/lactate dehydrogenase (LDH)-based colony (ALC) assay quantifies both αSMA expression and intracellular LDH activity in CFU-f colonies. We show that the output of the ALC assay on day 10 of BM culturing correlates well with the subsequent MSC lot growth up to passage 3.
MATERIALS AND METHODS
BM Cell Preparation, Plating, and Culturing
Undiluted BM aspirates from healthy individual human donors were purchased from Lonza (Walkersville, MD, USA) or AllCells (Alameda, CA, USA) and delivered overnight with cold packs. Lonza donors (samples D121–D128 and L1) were 21- to 25-year-old males, while AllCells donors (AC12–14) were 32- to 33-year-old males. The aspirate sample (1–3 ml) was diluted three times in MSC growth medium consisting of α minimum essential medium (αMEM; Mediatech, Manassas, VA, USA) supplemented with 10% fetal bovine serum (lot selected for MSC growth; HyClone, Logan, UT, USA), GlutaMAX (Invitrogen, Thermo Fisher Scientific; www.thermofisher.com), and penicillin/streptomycin (Mediatech) and centrifuged at 200 × g for 8 min. The supernatant was carefully removed, and the pellet was resuspended to the prior volume of the diluted sample. White blood cell (WBC) counts were determined in aliquots after diluting them in Erythrocyte Lysis Reagent (Sigma-Aldrich, St. Louis, MO, USA). For the analysis of CFU-f colonies, washed BM cells were resuspended at 6.6 × 104 cells/ml and plated into 96-well black microplates with a clear bottom (100 μl/well; Costar, Corning, NY, USA); two rows were left empty. This cell concentration resulted in growth of colonies in less than 30% of wells, thus ensuring that there would be a low probability of having more than one colony per well. For mass culture plating, BM cells were resuspended and plated at about 2–4 × 105 WBC/cm2 into a T75 flask. On day 3, the majority of the nonadherent cells were removed during medium change. Thereafter, medium was changed every 2–3 days. On day 10, microplates were used in the ALC assay, while mass cultures were cultured an additional 0–4 days before passaging. For passaging, cells were lifted with 0.25% trypsin/EDTA (Mediatech), counted using trypan blue (Sigma-Aldrich), and replated at about 0.5–1 × 104 cells/cm2. Subsequent passages (up to 3) were performed when cultures reached 70%–80%, which took 4–7 days. MSC mass culture growth rate at passage n (GRn) was calculated as follows:
where dHn is the cell density at harvest of passage n, dPn is the cell density at plating of passage n, and dWBC is the cell density at initial BM plating.
ALC Assay
In preliminary experiments, an assay that can simultaneously measure αSMA in a colony and the colony’s cell number was developed using passaged MSCs. Intracellular LDH activity, a surrogate for cell number, was colorimetrically measured in cells processed for immunodetection of αSMA, that is, after light formalin fixation21 [4% paraformaldehyde (PFA) for 20 min; Electron Microscopy Sciences, Hartfield, PA, USA], permeabilization [0.2% Triton X-100; 20 min; Sigma-Aldrich], and incubation with anti-αSMA antibody conjugated to horseradish peroxidase (HRP; Abcam, Cambridge, MA, USA). This residual LDH activity was found to be proportional to plated MSC numbers and/or numbers of cell nuclei quantified using a fluorescent nuclear dye Hoechst 33342 (Molecular Probes, Thermo Fisher Scientific) and image analysis by Cytation 5 multimode plate reader, objective 4×, and Gen5 software (BioTek, Winooski, VT, USA).
Therefore, for the colonies’ measurements, two colorimetric assay protocols—residual LDH activity and αSMA protein expression detection—were incorporated into a single protocol performed on the plate with cultured adherent BM cells; for each assay, empty wells were designated for standards and controls. Wells designated for LDH standards and HRP controls were kept unused until indicated in the protocol below. On day 10 after BM plating, plates were washed with phosphate-buffered saline (PBS), fixed with 4% PFA for 20 min, and then the fixative was replaced with PBS. Meanwhile, surrogate αSMA standards were prepared by serially diluting AffiniPure Donkey Anti-Mouse IgG (Jackson Immunoresearch Laboratories, West Grove, PA, USA) in PBS and adsorbing the solutions in the designated wells for 1 h; these wells were then blocked with the MSC growth medium. All wells (except for those designated for LDH and αSMA standards and HRP controls) were then washed once and incubated with 0.2% Triton X-100 in PBS for 20 min, followed by blocking in 0.5% normal donkey serum (Jackson Immunoresearch Laboratories) for 30 min. The wells (except for those designated for LDH standards and HRP controls) were then incubated with monoclonal anti-αSMA antibody conjugated to HRP (1:1,000) for 1 h and washed three times with PBS. The LDH assay was then performed. LDH standards were prepared in designated wells by serially diluting bovine LDH (Sigma-Aldrich), and then catalyst/dye (0.25 ml:11 ml) mixture from LDH Cytotoxicity Detection kit (Clontech Laboratories, Mountain View, CA, USA) was added to all wells with cultures and LDH standards. The signals were read at 490 nm with correction at 650 nm. After LDH detection, plates were washed once and prepared for HRP detection. To ensure that readings on all plates would be comparable across different experiments, an HRP control prepared from highly stabilized HRP (Sigma-Aldrich) was used on each plate. Wells containing cultures, surrogate αSMA standards, and HRP controls were filled with the HRP substrate 3,3′,5,5′-tetramethylbenzidine (TMB; eBioscience, San Diego, CA, USA). The absorbance was measured at 370 nm with the correction at 492 nm. Both LDH and HRP signals were quantified using standard curves generated in SoftMax Pro (Molecular Devices, Sunnyvale, CA, USA); thus, LDH activity units were expressed in mU/ml, and αSMA expression units represented the corresponding concentration of anti-mouse antibody in ng/ml.
Data Processing for ALC Assay
The processing of ALC assay data was conducted using a programmed Excel template (Microsoft, Redmond, WA, USA). HRP control data were used to make adjustments between different plates and experiments if needed. Then for each plate the background values for both measured parameters were calculated: for LDH, as the average of all wells without a colony + 1 standard deviation (SD), and for HRP, as the average of all wells without colonies. The background was subtracted from the corresponding dataset. This method enabled the elimination of wells with less than 50 cells from further calculations; at the same time, less stringent conditions for HRP signals prevented the exclusion of colonies with very low levels of αSMA expression. The αSMA expression was normalized to the corresponding LDH signal from the same well (a surrogate for αSMA/cell). All colony data were sorted from high to low LDH levels, and the percentage of colonies with LDH ≥0.4 mU/ml (“large colonies”) was determined. This threshold typically categorized more than 15% colonies as large in the BM lots. In these large colonies, normalized αSMA expression was averaged. Each BM lot was thus characterized by the average αSMA level in cells of large colonies [termed here as Av(αSMA/LDH)LC].
Cell Proliferation Assay and Immunofluorescence
Proliferation assay was conducted on colonies using Click-iT Plus 5-ethynyl-2′-deoxyuridine (EDU) Alexa Fluor 594 Imaging Kit (Life Technologies, Frederick, MD, USA) according to the manufacturer’s protocol. Cells were labeled with EDU for 5 h. After performing EDU detection, the colonies were blocked with 0.3% normal donkey serum and probed with fluorescein isothiocyanate (FITC)-conjugated anti-αSMA monoclonal antibody (Sigma-Aldrich), followed by washing. Nuclei were counterstained using Hoechst 33342. To quantify the percentage of EDU+ nuclei, images of colonies were acquired using Cytation 5 under a 1.25× objective and analyzed using Gen5 software. The colony data were then sorted in Excel. Colonies exhibiting highest and lowest % EDU (three to four colonies/group) were selected and acquired under 4× magnification to visualize and quantify αSMA mean fluorescence intensity using ImageJ as mean gray value with the subtraction of the corresponding background. For the analysis of αSMA and EDU distribution in one colony, the image of a colony was digitally enlarged and αSMA+ areas were selected; then equal areas were selected in the αSMA− region of a colony. Total and EDU+ nuclei were manually counted in these areas.
Statistics
Statistical analysis (paired or unpaired t-test and linear regression analysis) and graphing were done using GraphPad Prism 6 (San Diego, CA, USA). A value of p < 0.05 was considered statistically significant.
RESULTS
Correlation Between Cell Proliferation and αSMA Expression in Day 10 Colonies
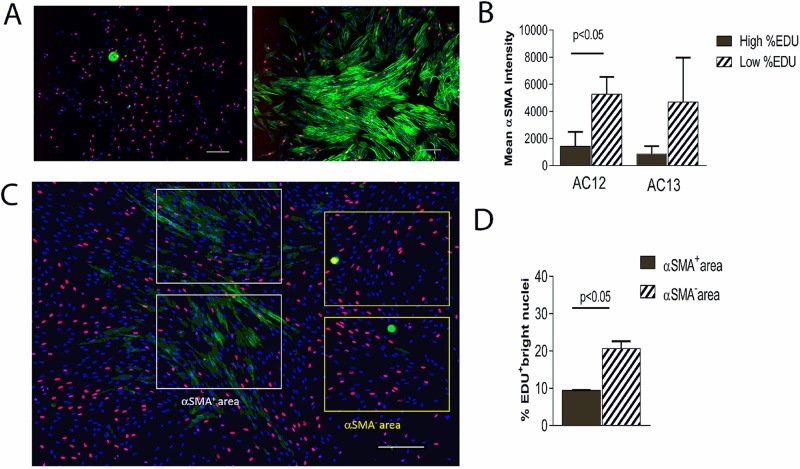
In an MSC lot, highly proliferative colonies (high % EDU incorporation) exhibited lower αSMA expression than did colonies with low % EDU (Fig. 1A). In two lots, colonies with the highest % EDU and the lowest % EDU exhibited correspondingly low and high mean fluorescence intensity for αSMA, and the effect was either statistically significant or a strong trend (Fig. 1B). There was a negative correlation between the level of αSMA expression and the percent of nuclei with bright EDU staining (Fig. 1C and D). αSMA+, but not αSMA−, areas also contained nuclei with very low levels of EDU in addition to brightly stained ones. Taken together, these observations indicated that αSMA expression appeared negatively correlated with both the colony’s overall proliferation rate and the proliferation status within the colony.
Figure 1.
Correlation between α-smooth muscle actin (αSMA) expression and cell proliferation in mesenchymal stromal cell (MSC) colonies. Colonies were labeled with 5-ethynyl-2′-deoxyuridine (EDU) for 5 h prior to fixation, and EDU (red) and αSMA (green) were detected; nuclei were stained with Hoechst dye (blue). (A) αSMA expression in representative large colonies with high % EDU (left) and low % EDU (right). (B) In two lots, four colonies with highest % EDU (AC12: EDU >39% and AC13: EDU >28%) and four colonies with lowest % EDU (AC12: EDU <18% and AC13: EDU <15%) were labeled using fluorescein isothiocyanate (FITC)-conjugated anti-αSMA; the mean FITC intensities of colonies in the high and low % EDU groups were compared. (C) A highly proliferative colony expressing some αSMA is shown; in areas designated as αSMA+ and αSMA− (white and yellow rectangles, respectively), percent bright EDU+ nuclei was compared (D). Scale bars: 200 μm. Error bars represent SD between two rectangles.
Correlating ALC Assay Data on Day 10 With the Subsequent Mass Culture Growth
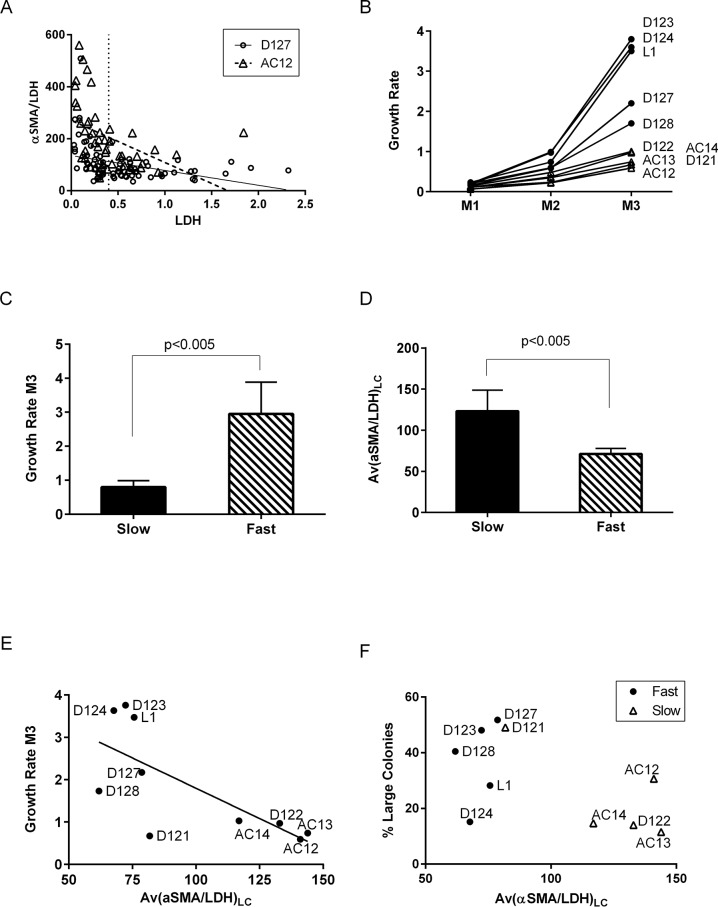
On day 10, LDH values (colony size) negatively correlated with αSMA/LDH values (surrogate for αSMA/cell) in 9 out of 10 tested BM-MSC lots (34–89 colonies/lot; p < 0.05), and the distribution pattern of colonies on the αSMA/LDH versus LDH plot was found to be characteristic for individual BM lots (Fig. 2A). Because of the exponential character of cell growth, the contribution of large colonies in mass culture population is substantially greater than that of small colonies. Therefore, we arbitrarily defined large colonies as those with LDH ≥0.4 mU/ml (i.e., colonies with approximately 2,000 cells, based on nuclei quantification). ALC assay output allowed determining percentage of large colonies, their averaged αSMA/LDH, and CFE and subsequently testing the correlation of these parameters with the corresponding mass culture growth.
Figure 2.
Correlation of ALC assay (day 10) and mass culture growth (passage 3) data in 10 BM lots. (A) Normalized αSMA [αSMA/lactate dehydrogenase (LDH)] plotted against colony size (LDH) for each colony (dot) for two representative lots: AC12 (triangles) and D127 (circles). Linear regression lines are shown. Vertical dotted line shows the arbitrary threshold for large colonies. (B) Mass culture growth rate curves from three passages (M1–M3) for 10 lots. MSC lots with growth rate above 1 were categorized as fast (black circles), and the others as slow (clear triangles). (C, D) Comparison of fast and slow MSC groups: (C) mass culture growth rates on passage 3 and (D) average αSMA/LDH in large colonies (LDH ≥0.4). (E) Correlation between average αSMA/LDH in large colonies and growth rate on passage 3; the line shows linear regression for 10 lots; nonzero slope, p < 0.02 and R 2 = 0.5138. Note that D121 is an outlier; its exclusion gives p < 0.01 and R 2 = 0.6846. (F) ALC data plot: percentage of large colonies versus average αSMA/LDH in them. Note clear separation of lots with the subsequent fast and slow growth.
The outcome of mass culturing the set of 10 MSC lots was studied during three passages (Fig. 2B). The lots could be categorized as either slow or fast growing based on their cumulative growth rates (GRs) at passage 3 (GR ≤1 or >1, respectively); the difference in GRs between two groups was statistically significant (Fig. 2C).
The three lots that were produced from BM aspirates obtained at AllCells and, coincidentally, from older donors (see Materials and Methods) were all in the slow-growing group; however, this group also contained two lots produced from younger donors from BM aspirates purchased at Lonza, indicating that the donor’s age is not a good predictor of MSC growth.
MSC growth data were compared to CFE, percent of large colonies, and Av(αSMA/LDH)LC. Slow-growing lots had significantly higher Av(αSMA/LDH)LC than did fast-growing lots (Fig. 2D). Interestingly, this was true irrespective of how large colonies were defined; however, if defined as those with LDH ≥0.4, the statistical difference between Av(αSMA/LDH)LC of the two groups was the greatest (p < 0.005 and R 2 = 0.71). There was no correlation between CFE or percentage of large colonies and mass culture growth.
There was a significant linear correlation between mass culture growth rate and Av(αSMA/LDH)LC (slope nonzero with p < 0.02, R 2 = 0.514) (Fig. 2E); removing one outlier (D121) made the correlation stronger. Plotting percentage of large colonies against Av(αSMA/LDH)LC (Fig. 2F) separated all data points into two groups, one consisted of four lots that all exhibited slow growth and another consisted of all five fast-growing lots plus D121, a slow-growing lot. The culturing data on this lot suggested that it was unintentionally overgrown at passage 2, which could explain why this lot grew slower than was predicted by the ALC assay. Acknowledging the small sample size, the existing data suggested that an Av(αSMA/LDH)LC >100 predicted subsequent slow growth of mass culture with 100% positive and 80% negative predictive values and 100% specificity and 83% sensitivity.
DISCUSSION
The goal of this study was to develop a method to predict MSC lot failure early in the BM culturing process, preferably before the first passage. Commercial manufacture defines failure as production of an insufficient number of cells after a defined number of passages using a standardized culture process. We hypothesized that the growth properties of an MSC lot can be predicted on the basis of the characteristics of CFU-f colonies, specifically on αSMA expression levels in large colonies. By using a simple colorimetric assay, we determined the average αSMA level in cells of large colonies on day 10 after BM plating and showed that this parameter is negatively correlated with the subsequent growth rate of MSC lot up to passage 3.
Scoring MSC lots in part based on large colonies can be justified because of the major contribution of large, but not small, colonies to the resulting cell population. Although we defined large colonies arbitrarily, by intracellular LDH levels corresponding to more than approximately 2,000 cells/colony, the percentage of large colonies likely reflects the proportion of stem cells in the adherent cell population of the donor BM lot. Indeed, several studies have shown that large colonies of CFU-f are formed by MSCs (i.e., tripotent), in contrast to smaller colonies that are believed to be formed by more differentiated progenitors13,22,23.
Our MSC growth-predictive model is based on αSMA expression in large colonies. Strong αSMA expression is known to be a hallmark of large, flat, stress fiber-abundant, nonproliferating senescent MSCs appearing with passaging24. A recent study using passaged MSCs demonstrated a direct correlation of αSMA expression with the substrate stiffness and reduced clonogenicity and showed αSMA regulation by yes-associated protein/transcriptional coactivator with PDZ-binding motif (YAP/TAZ) pathway19. Our study shows that αSMA is detectable before the first passage, at the CFU-f colony formation stage, that is, on day 10 or even earlier after BM plating. We find a donor-to-donor variability in αSMA expression levels in colonies and establish a correlation of αSMA levels in large colonies with future growth rates of MSC lots.
The fact that the percentage of large colonies was not a predictor of mass culture growth relates to the fact that the proliferation of large colonies at a given time does not predict lot growth, though it generally inversely correlates with the colony’s level of αSMA. In addition to the culture substrate stiffness, αSMA expression responds to specific culture conditions, such as medium change frequency (i.e., the removal of factors secreted by growing MSC population and contaminating nonadherent BM cells). Indeed, expression of αSMA can be potently induced by transforming growth factor-β (TGF-β) and inhibited by platelet-derived growth factor-BB (PDGF-BB)25, both factors known to be secreted by MSCs. The influence of the microenvironment on MSCs’ ability to proliferate is also evident from a correlation between expression of αSMA and proliferation in different areas of the same colonies. This observation is in agreement with the notion of a reciprocity between the inner and the outer region of a colony in the expression of extracellular matrix (ECM) proteins and cell cycle-related proteins26. Thus, it is reasonable to suggest that the adaptation of MSCs from an individual donor to given culture conditions is a complex process determined by the functioning of various pathways including those regulating αSMA expression, the production of certain growth factors (by MSCs and contaminating cells), and the deposition of ECM. It appears that the average αSMA expression level in cells of large colonies could be a reflection of various genetic and external factors shaping MSC growth potential in rigidly defined manufacturing conditions. Identification of pathways and specific factors regulating the adaptation of MSCs to in vitro culture conditions is a matter of future studies. The assay described here can help to exclude lots with low growth potential and could be especially effective if lot exclusion criteria are adjusted for specific manufacturing conditions that may differ from those described here. In conclusion, the ALC assay assesses αSMA in large CFU-f colonies a few days after plating of BM. It provides a simple and easily standardized measurement that predicts the subsequent growth of MSCs. It could be readily incorporated as a screen to increase efficiency and reduce cost in manufacturing large-scale batches of MSCs for clinical use.
ACKNOWLEDGMENT
All the authors are employees of SanBio.
REFERENCES
- 1. Friedenstein AJ, Chailakhjan RK, Lalykina KS. The development of fibroblast colonies in monolayer cultures of guinea-pig bone marrow and spleen cells. Cell Tissue Kinet. 1970;3(4):393–403. [DOI] [PubMed] [Google Scholar]
- 2. Friedenstein AJ, Gorskaja JF, Kulagina NN. Fibroblast precursors in normal and irradiated mouse hematopoietic organs. Exp Hematol. 1976;4(5):267–74. [PubMed] [Google Scholar]
- 3. Friedenstein AJ, Deriglasova UF, Kulagina NN, Panasuk AF, Rudakowa SF, Luria EA, Ruadkow IA. Precursors for fibroblasts in different populations of hematopoietic cells as detected by the in vitro colony assay method. Exp Hematol. 1974;2(2):83–92. [PubMed] [Google Scholar]
- 4. Owen M, Friedenstein AJ. Stromal stem cells: Marrow-derived osteogenic precursors. Ciba Found Symp. 1988;136:42–60. [DOI] [PubMed] [Google Scholar]
- 5. Paul G, Anisimov SV. The secretome of mesenchymal stem cells: Potential implications for neuroregeneration. Biochimie 2013;95(12):2246–56. [DOI] [PubMed] [Google Scholar]
- 6. Menard C, Tarte K. Immunoregulatory properties of clinical grade mesenchymal stromal cells: Evidence, uncertainties, and clinical application. Stem Cell Res Ther. 2013;4(3):64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Zhukareva V, Obrocka M, Houle JD, Fischer I, Neuhuber B. Secretion profile of human bone marrow stromal cells: Donor variability and response to inflammatory stimuli. Cytokine 2010;50(3):317–21. [DOI] [PubMed] [Google Scholar]
- 8. Wegmeyer H, Broske AM, Leddin M, Kuentzer K, Nisslbeck AK, Hupfeld J, Wiechmann K, Kuhlen J, von Schwerin C, Stein C, Knothe S, Funk J, Huss R, Neubauer M. Mesenchymal stromal cell characteristics vary depending on their origin. Stem Cells Dev. 2013;22(19):2606–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Digirolamo CM, Stokes D, Colter D, Phinney DG, Class R, Prockop DJ. Propagation and senescence of human marrow stromal cells in culture: A simple colony-forming assay identifies samples with the greatest potential to propagate and differentiate. Br J Haematol. 1999;107(2):275–81. [DOI] [PubMed] [Google Scholar]
- 10. Kuznetsov SA, Friedenstein AJ, Robey PG. Factors required for bone marrow stromal fibroblast colony formation in vitro. Br J Haematol. 1997;97(3):561–70. [DOI] [PubMed] [Google Scholar]
- 11. Mannello F, Tonti GA. Concise review: No breakthroughs for human mesenchymal and embryonic stem cell culture: Conditioned medium, feeder layer, or feeder-free; medium with fetal calf serum, human serum, or enriched plasma; serum-free, serum replacement nonconditioned medium, or ad hoc formula? All that glitters is not gold! Stem Cells 2007;25(7):1603–9. [DOI] [PubMed] [Google Scholar]
- 12. Cordeiro-Spinetti E, de Mello W, Trindade LS, Taub DD, Taichman RS, Balduino A. Human bone marrow mesenchymal progenitors: Perspectives on an optimized in vitro manipulation. Front Cell Dev Biol. 2014;2:7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Russell KC, Lacey MR, Gilliam JK, Tucker HA, Phinney DG, O’Connor KC. Clonal analysis of the proliferation potential of human bone marrow mesenchymal stem cells as a function of potency. Biotechnol Bioeng. 2011;108(11):2716–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Bertolo A, Mehr M, Janner-Jametti T, Graumann U, Aebli N, Baur M, Ferguson SJ, Stoyanov JV. An in vitro expansion score for tissue-engineering applications with human bone marrow-derived mesenchymal stem cells. J Tissue Eng Regen Med. 2016;10(2):149–61. [DOI] [PubMed] [Google Scholar]
- 15. Deskins DL, Bastakoty D, Saraswati S, Shinar A, Holt GE, Young PP. Human mesenchymal stromal cells: Identifying assays to predict potency for therapeutic selection. Stem Cells Transl Med. 2013;2(2):151–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Gothard D, Dawson JI, Oreffo RO. Assessing the potential of colony morphology for dissecting the CFU-F population from human bone marrow stromal cells. Cell Tissue Res. 2013;352(2):237–47. [DOI] [PubMed] [Google Scholar]
- 17. Hinz B. Formation and function of the myofibroblast during tissue repair. J Invest Dermatol. 2007;127(3):526–37. [DOI] [PubMed] [Google Scholar]
- 18. Charbord P, Lerat H, Newton I, Tamayo E, Gown AM, Singer JW, Herve P. The cytoskeleton of stromal cells from human bone marrow cultures resembles that of cultured smooth muscle cells. Exp Hematol. 1990;18(4):276–82. [PubMed] [Google Scholar]
- 19. Talele NP, Fradette J, Davies JE, Kapus A, Hinz B. Expression of alpha-smooth muscle actin determines the fate of mesenchymal stromal cells. Stem Cell Reports 2015;4(6):1016–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hinz B. The myofibroblast: Paradigm for a mechanically active cell. J Biomech. 2010;43(1):146–55. [DOI] [PubMed] [Google Scholar]
- 21. Baba N, Sharma HM. Histochemistry of lactic dehydrogenase in heart and pectoralis muscles of rat. J Cell Biol. 1971;51(3):621–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Russell KC, Phinney DG, Lacey MR, Barrilleaux BL, Meyertholen KE, O’Connor KC. In vitro high-capacity assay to quantify the clonal heterogeneity in trilineage potential of mesenchymal stem cells reveals a complex hierarchy of lineage commitment. Stem Cells 2010;28(4):788–98. [DOI] [PubMed] [Google Scholar]
- 23. Mareddy S, Dhaliwal N, Crawford R, Xiao Y. Stem cell-related gene expression in clonal populations of mesenchymal stromal cells from bone marrow. Tissue Eng Part A 2010;16(2):749–58. [DOI] [PubMed] [Google Scholar]
- 24. Galmiche MC, Koteliansky VE, Briere J, Herve P, Charbord P. Stromal cells from human long-term marrow cultures are mesenchymal cells that differentiate following a vascular smooth muscle differentiation pathway. Blood 1993;82(1):66–76. [PubMed] [Google Scholar]
- 25. Kinner B, Zaleskas JM, Spector M. Regulation of smooth muscle actin expression and contraction in adult human mesenchymal stem cells. Exp Cell Res. 2002;278(1):72–83. [DOI] [PubMed] [Google Scholar]
- 26. Ylostalo J, Bazhanov N, Prockop DJ. Reversible commitment to differentiation by human multipotent stromal cells in single-cell-derived colonies. Exp Hematol. 2008;36(10):1390–402. [DOI] [PMC free article] [PubMed] [Google Scholar]