Abstract
Escherichia coli senses and signals anoxic or low redox conditions in its growth environment by the Arc two-component system. Under anaerobic conditions, the ArcB sensor kinase autophosphorylates and transphosphorylates ArcA, a global transcriptional regulator that controls the expression of numerous operons involved in respiratory or fermentative metabolism. Under aerobic conditions, the kinase activity of ArcB is inhibited by the quinone electron carriers that act as direct negative signals. Here, we show that the molecular mechanism of kinase silencing involves the oxidation of two cytosol-located redox-active cysteine residues that participate in intermolecular disulfide bond formation, a reaction in which the quinones provide the source of oxidative power. Thus, a pivotal link in the Arc signal transduction pathway connecting the redox state of the quinone pool to the transcriptional apparatus is elucidated.
Two-component signal transduction systems are widespread in prokaryotes and play extensive roles in adaptation to environmental changes (1, 2). The Arc (anoxic redox control) two-component system is an important element in the complex transcriptional regulatory network that allows facultative anaerobic bacteria, such as Escherichia coli, to sense various respiratory growth conditions and adapt their gene expression accordingly (3). This system comprises the cytoplasmic response regulator ArcA and the transmembrane sensor kinase ArcB (4, 5). ArcA is a typical response regulator possessing an N-terminal receiver domain with a conserved Asp residue at position 54 and a C-terminal helix-turn-helix DNA binding domain. In contrast, ArcB is an unorthodox sensor kinase as manifested by its unusually elaborate architecture. As a sensor, ArcB is deviant because in contrast to typical sensor kinases that have a substantial periplasmic domain for environmental sensing, ArcB has a very short periplasmic sequence of only 16 amino acid residues delimited by two canonical transmembrane segments. Interestingly, the ArcB transmembrane domain (amino acids 22-77) does not directly participate in signal sensing but rather serves as an anchor that keeps the protein close to the source of the signal (6). As a kinase, ArcB is atypical because it contains three catalytic domains: an N-terminal transmitter domain with a conserved His-292 residue, a central receiver domain with a conserved Asp-576 residue, and a C-terminal phosphotransfer domain with a conserved His-717 residue (5, 7). Moreover, in the linker that is the region connecting the catalytic domains with the transmembrane domain, there are a putative leucine zipper (8) and a Per-Arnt-Sim (PAS) domain (9).
Under reducing conditions, ArcB undergoes ATP-dependent autophosphorylation, a process shown to be enhanced by certain anaerobic metabolites such as D-lactate, acetate, and pyruvate (10, 11), and transphosphorylates ArcA via a His-292 → Asp-576 → His-717 → Asp-54 phosphorelay (12, 13). Phosphorylated ArcA (ArcA-P), in turn, represses the expression of many operons involved in respiratory metabolism and activates a few operons encoding proteins involved in fermentative metabolism (4, 14, 15). Under nonstimulating conditions, ArcB acts as a specific ArcA-P phosphatase that catalyzes the dephosphorylation of ArcA-P by a reverse Asp-54 → His-717 → Asp-576 → Pi phosphorelay (8). Recently, we reported that the quinone electron carriers act as the primary signals that silence the kinase activity of ArcB under aerobic conditions of growth (16). It was shown that the autophosphorylation of an N-terminally truncated ArcB protein (ArcB78-778) was inhibited by ubiquinone-0 (Q0, a soluble analog of ubiquinone-8) and less efficiently by menadione (a soluble analog of menaquinone-8). In agreement, in a ubi mutant blocked in the biosynthesis of ubiquinone-8, ArcB was active under aerobic growth conditions.
Here, we explore the molecular mechanism by which the oxidized forms of the quinone electron carriers inhibit the kinase activity of ArcB during aerobic growth.
Materials and Methods
Plasmids and Oligonucleotides. Construction of plasmid pQE30ArcB78-778 has been described earlier (12). To create pMX403 (ArcB78-778, C180A), the mutagenic primer 5′-CCATCGCGCGGTTAGCGCCGGAAAACTC-3′ and primer 5′-AATATCGAGCAATGCTTCTG-3′ were used in the PCR with pQE30ArcB78-778 as template. The PCR product of this reaction was purified and used as a megaprimer in combination with primer 5′-CCCGGATCCCATATGGAGCAACTGGAGGAGTCACGAC-3′ and pQE30ArcB78-778 as template. The product of the second PCR was digested with PstI and MluI, and the purified fragment was used to replace the PstI and MluI wild-type fragment of pQE30ArcB78-778. To create pMX415 (ArcB78-778, C241A) and pMX405 (ArcB78-778, C180A, C241A), a similar strategy was followed, but with primer 5′-CGGATTTCAAAGGCGGCTTTGCGCCCG-3′ and either pQE30ArcB78-778 or pMX403 as template. Sequence verification of PCR-amplified DNA was performed by the Molecular Biology Unit, Instituto de Fisiologia Celular, Universidad Nacional Autónoma de México.
Strain Construction. Strains IFC 1001, IFC1002, and IFC1003 were constructed as follows. Plasmids pMX403 (ArcB78-778, C180A), pMX415 (ArcB78-778, C241A), and pMX405 (ArcB78-778, C180A, C241A) were digested with PstI and MluI, and the mutation containing fragments were used to replace the PstI and MluI fragment of pQE30ArcB1-778 creating plasmids pMX431, pMX432, and pMX433, respectively. The BamHI fragment of plasmid pIB3 (13) was cloned into plasmid pKO3 (17), in which the NdeI site had been inactivated by digestion and subsequent fill in reaction with T4 polymerase to create plasmid pMX434. The NdeI-NruI fragment from plasmids pMX431, pMX432, and pMX433 were used to replace the NdeI-NruI fragment of plasmid pMX434 creating plasmids pMX435, pMX436, and pMX437. These pKO3 derivatives were used to replace the chromosomal arcB allele of ECL5012 as described in ref. 17.
Purification of His-6-Tagged Proteins and Phosphorylation Assays. Protein purification was performed as described in ref. 12 but under nonreducing conditions. Phosphorylation assays were carried out at room temperature in the presence of 40 μM [γ-32P]ATP [specific activity, 2 Ci/mmol (1 Ci = 37 GBq)], 50 mM Hepes (pH 7.5), 50 mM KCl, 5 mM MgCl2, 0.1 mM EDTA, and 10% glycerol. Where indicated, purified ArcB peptides (50 pmol) were preincubated with 0.25 mM Q0, 0.1 mM chloramine T, 2 mM H2O2, 5 mM DTT, or 1 mM S-methyl methanethiosulfonate (MMTS) for 30 min at 25°C. The phosphorylation reactions were initiated by the addition of [γ-32P]ATP terminated by addition of equal volume of 2× SDS sample buffer and immediately subjected to SDS/PAGE (10% polyacrylamide gels). Radioactivity of proteins resolved in the gels was determined qualitatively by autoradiography of the dried gels or quantitatively by using a PhosphorImager (Molecular Dynamics).
Tagging of ArcB with Methoxy-Polyethylene Glycol Maleimide (MAL-PEG). MAL-PEG (Mr = 5,000) was used to tag free thiols on ArcB. Purified ArcB peptides (50 pmol), untreated or pretreated with 0.5 mM Q0, were incubated in 20 mM Tris·HCl and 1 mM EDTA with 1 mM MAL-PEG at 25°C. After 1 h, the reactions were terminated by addition of equal volume of 2× SDS sample buffer and immediately subjected to SDS/PAGE. Differences in the mobility of the tagged and untagged proteins were visualized by Western blot analysis.
Modification of ArcB with 7-Chloro-4-nitrobenzo-2-oxa-1,3-diazole (NBD-Cl). Purified ArcB peptides (10 nmol) were incubated in 50 mM potassium phosphate buffer (pH 7.2) with 0.5 mM Q0 or 10 mM DTT at 25°C. After 30 min, Q0 and DTT were removed by concentration and redilution with the above buffer in a nanosep 10K (Pall). NBD-Cl (in DMSO) was added to a final concentration of 1 mM, and the modification reactions were allowed to proceed at 25°C for 1 h. Final spectra (600-300 nm) were monitored after free NBD-Cl was removed by three cycles of concentration and redilution with the above buffer.
Capturing the in Vivo Redox State of ArcB. Cells were grown aerobically in a partially filled Erlenmeyer flask that was shaking at 300 rpm or anaerobically in a completely filled screw-capped test tube. At a density of OD600 ≈ 0.4-0.5, the cells were treated with a 1/10th volume of 100% trichloroacetic acid to precipitate the proteins and stop further thiol/disulfide exchange. After a 1-h incubation on ice, the precipitated proteins were isolated by centrifugation, and the pellets were washed with acetone to remove the trichloroacetic acid. The pellet was dissolved in nonreducing 5× SDS sample buffer and split into two equal portions. One portion was treated with 100 mM DTT, and the other received an equivalent amount of buffer only. The proteins were separated by nonreducing SDS/PAGE, and ArcB was visualized by Western blot analysis as described in ref. 6.
Results
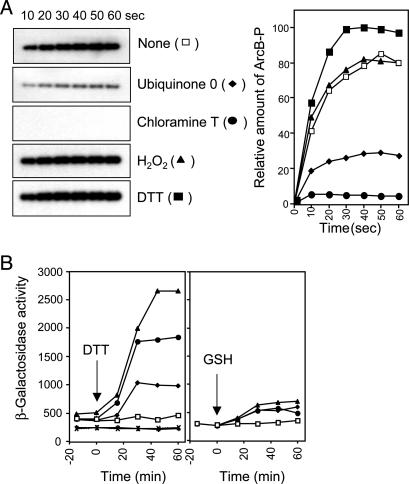
Effects of Various Modifying Agents on the Activity of ArcB. At least two alternative paths of how quinones inhibit the kinase activity of ArcB can be envisaged: the first is by stabilization of the nonautophosphorylating conformation of ArcB as a result of the allosteric binding of quinones on ArcB, whereas the alternative should be the quinone-dependent oxidation of one or several amino acids in ArcB. We favored the latter because, in an exploratory experiment, we observed that in contrast to the thiol-reducing agent DTT that slightly enhanced the in vitro phosphorylation of ArcB, Q0 and the oxidizing agent chloramine T drastically inhibited ArcB phosphorylation (Fig. 1A). Also, the facts that the chemical structures of the two inhibiting molecules are different and that chloramine T oxidizes specifically cysteine and methionine residues of proteins indicate that ArcB is sensitive to oxidation rather than to the allosteric binding of Q0.
Fig. 1.
Effects of various modifying agents on the activity of ArcB. (A) Purified ArcB78-778 (50 pmol) was incubated with [γ-32P]ATP in the presence or absence of Q0 (0.250 mM), chloramine T (0.1 mM), H2O2 (2 mM), and DTT (5 mM), and the net phosphorylation of the protein was assayed by SDS/PAGE. (Left) Autoradiograms of the gels. (Right) Net increase of ArcB-P with time in the absence (□) or presence of Q0 (♦), chloramine T (•), H2O2 (▴), or DTT (▪). (B) Effect of DTT and glutathione on the aerobic expression of λΦ(cydA′-lacZ). Four parallel cultures of strain ECL5203 (6) and its isogenic ECL5204 (ΔarcB) (6) were grown aerobically in Luria-Bertani broth containing 0.1M MOPS (pH 7.4) and 20 mM d-xylose. At OD600 of 0.3, one aliquot was withdrawn for measuring the β-galactosidase activity (depicted as -15 min). At time 0 min, DTT or glutathione was added to the cultures to final concentrations of 2.5 mM (♦), 5 mM (•), 10 mM (▴), or nothing (□), and the β-galactosidase activity was followed. × and + depict ECL5204 (ΔarcB) with no addition and 10 mM DTT, respectively. (Left) Addition of DTT. (Right) Addition of glutathione. The data represent the averages from four experiments (variations were <10% from the mean).
Further support to our hypothesis was provided by the finding that the membrane-permeating reductant DTT was able to activate ArcB during aerobic growth. It was found that addition of DTT to the aerobic culture of a wild-type strain led to an immediate increase of the expression of the ArcA-P activatable cydA′-lacZ reporter in a concentration-dependent manner (Fig. 1B). The observed effect was exerted through ArcB because DTT did not affect the expression of the reporter in a ΔarcB strain (Fig. 1B). In agreement, 2-mercaptoethanol, another membrane-permeating thiol-reductant, did also activate ArcB during aerobiosis (data not shown), whereas glutathione, a reducing agent not able to permeate the plasma membrane because of its lipophobic nature, did not (Fig. 1B). In this context, it is of relevance to mention that the membrane-permeating reductants generate an intracellular reducing environment that affects protein folding and, in particular, disulfide bond formation (18, 19). Considering that ArcB possesses a cytoplasm-located signal reception site (6), it is activated only by the membrane-permeating thiol-reductants during aerobiosis, and it has two unique cysteine residues (Cys-180 and Cys-241) in the linker region, we postulated that it might respond to changes in redox conditions through thiol oxidation and reduction. It is important to point out that both Cys-180 and Cys-241 are conserved in the ArcB homologues of different bacterial species including Salmonella typhimurium, Shigella flexneri, Yersinia pestis, Erwinia carotovora, and Photorhabdus luminescens (20-23). However, the ArcB homologue of Haemophilus influenzae represents an intriguing exception because it lacks almost the entire linker region, corresponding to amino acid residues 93-271 of E. coli ArcB (24), and therefore the two conserved cysteine residues. Nevertheless, in contrast with most other homologues that possess only two cysteine residues, ArcB of H. influenzae possesses five cysteine residues that are located at positions 47, 268, 472, 574, and 596 of the protein.
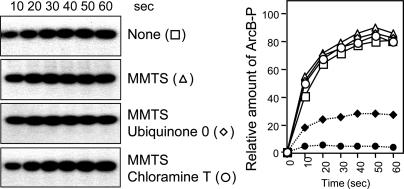
Cys-180 and Cys-241 of ArcB Are Required for Kinase Silencing. To test whether the two cysteine residues participate in the mechanism that controls the activity of ArcB, we took advantage of the fact that upon alkylation, cysteine residues are protected from further oxidation and are also unable to form disulfide bonds. Therefore, the effect of MMTS, which is a small, neutral, highly soluble molecule that reacts specifically with the free sulfhydryl groups on cysteine side chains to form Cys-S-CH3, was examined on the in vitro response of ArcB to Q0 and chloramine T (Fig. 2). MMTS per se did not affect the phosphorylation kinetics of ArcB, indicating that the ligand does not interfere with the activity of the protein. However, in contrast to the significant inhibition of the unprotected protein by Q0 and chloramine T (Fig. 1 A), the MMTS-blocked protein was unresponsive to both Q0 and chloramine T (Fig. 2). It thus appears that free cysteine thiols are required for the action of the inhibiting agents, supporting the involvement of Cys-180 and/or Cys-241 in the mechanism for ArcB regulation.
Fig. 2.
Effect of alkylation on the inhibition of ArcB by Q0 and chloramine T. Purified ArcB78-778 (50 pmol) was preincubated with MMTS (1 mM) for 30 min, and the kinetics of [γ-32P]ATP-dependent phosphorylation in the presence or absence of Q0 (0.250 mM) and chloramine T (0.1 mM) were assayed by SDS/PAGE. (Left) Autoradiograms of the gels. (Right) Time course of ArcB phosphorylation in the absence (□) or presence of Q0 (♦), chloramine T (•), MMTS (▵), MMTS and Q0 (⋄), or MMTS and chloramine T (○).
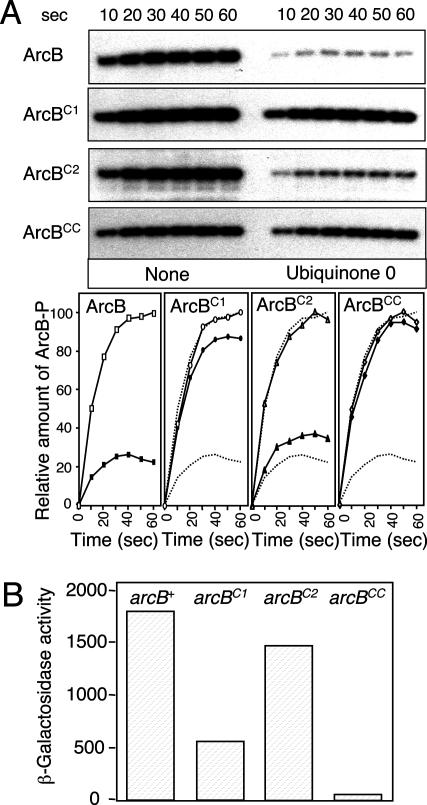
To provide definite evidence for the participation of the two cysteine residues in the above mechanism, Cys-180 and/or Cys-241 were mutated to Ala. The resulting mutant peptides ArcB78-778, C180A (ArcBC1), ArcB78-778, C241A (ArcBC2), and ArcB78-778, C180A, C241A (ArcBCC) were purified and tested for their response to Q0 by in vitro [γ32P]ATP-dependent phosphorylation (Fig. 3A). In agreement with earlier results, Q0 drastically inhibited the phosphorylating activity of ArcB, which decreased to ≈25%, compared with the one in the absence of Q0. In contrast, ArcBC1 was barely affected by Q0 as it retained >85% of its activity, confirming that Cys-180 plays a central role in the regulation of ArcB. On the other hand, ArcBC2 was significantly inhibited by Q0, retaining ≈35% of its activity. However, despite the high degree of Q0-dependent inhibition, ArcBC2 retained more of its activity than the wild-type ArcB, suggesting that Cys-241 might participate in the regulation of ArcB. Finally, no Q0-dependent inhibition was observed in the case of ArcBCC. Hence, our results confirm the requirement of at least Cys-180 for silencing the phosphorylating activity of ArcB and also indicate a probable involvement of Cys-241 in this process. They also suggest that there are no other oxidizable residues in the protein that are essential for its regulation. To verify the involvement of Cys-180 and Cys-241 in the in vivo regulation of ArcB, we replaced the chromosomal arcB+ allele by arcBCys180Ala (arcBC1), arcBCys241Ala (arcBC2), or arcBCys180Ala, Cys241A (arcBCC) in a strain bearing the ArcA-P-repressible λΦ(lldP′-lacZ) reporter, and their aerobic β-galactosidase activity levels were assayed (Fig. 3B). In agreement with the in vitro results, the expression of the reporter was slightly repressed in the arcBC2 mutant strain but strongly repressed in the arcBC1 and arcBCC mutant strains, as the β-galactosidase activity dropped, respectively, to ≈84%, ≈32%, and ≈3.5% of its level in the wild-type strain. Thus, replacement of the cysteine residues in ArcB results in a highly active kinase even under aerobic conditions of growth. Hence, our in vitro and in vivo data confirm the requirement of the two cysteine residues for ArcB regulation, although Cys-180 appears to play a more decisive role in this process than Cys-241.
Fig. 3.
Requirement of the two cysteine residues for ArcB silencing by Q0. (A) A total of 50 pmol of purified ArcB78-778 (ArcB), ArcB78-778, C180A (ArcBC1), ArcB78-778, C241A (ArcBC2), and ArcB78-778, C180A, C241A (ArcBCC) were incubated with [γ-32P]ATP in the presence or absence of Q0 (0.250 mM), and the net phosphorylation of the protein was assayed by SDS/PAGE. (Upper) Autoradiograms of the gels. (Lower) Net increase of protein-P with time in the absence (open symbols) or presence of Q0 (filled symbols). The kinetics of ArcB phosphorylation are depicted in all panels as dotted lines. (B) Aerobic expression of λΦ(lldP′-lacZ). Strains ECL5002 (λΦ[lldP′-lacZ]) (6), IFC1001 (arcBC180A Kanr λΦ[lldP′-lacZ]), IFC1002 (arcBC241A Kanr λΦ[lldP′-lacZ]), and IFC1003 (arcBC180A, C241A Kanr λΦ[lldP′-lacZ]) were grown in Luria-Bertani broth containing 0.1 M MOPS (pH 7.4) and 20 mM d-xylose supplemented with 20 mM l-lactate as an inducer. β-Galactosidase activity was assayed and expressed in Miller units. The data are averages from four experiments (variations were <10% from the mean).
Both Cysteine Residues of ArcB Undergo Q0-Dependent Oxidation. The oxidation status of the purified ArcB before and after treatment with Q0 was then determined. The large molecular mass compound MAL-PEG (Mr = 5,000), which forms covalent adducts with free thiol groups, was used to covalently tag ArcB that was reduced at cysteine residues. It is noteworthy that, although maleimides react also with amino groups, the reaction with SH groups is ≈1,000 times more rapid at neutral pH. The wild type and the three mutant ArcB peptides (ArcB, ArcBC1, ArcBC2, and ArcBCC) were treated with MAL-PEG and fractionated by SDS/PAGE, and the protein was detected by Western blot analysis (Fig. 4A). It was found that treatment of ArcB with MAL-PEG resulted in three shifted complexes (i, ii, and iii), indicating pegylation of ArcB at more than one site. However, treatment of ArcBC1 with MAL-PEG yielded only one complex with the same apparent mobility as complex ii. Likewise, treatment of ArcBC2 with MAL-PEG yielded a complex migrating at the same position as complex i. In contrast, treatment of ArcBCC with MAL-PEG did not yield any complex, confirming the specificity of the reaction. Thus, ArcB pegylated at both cysteine residues corresponds to complex iii, in accord with the expectation to exhibit the slowest mobility. The anomalous mobility of ArcBC1-PEG and ArcBC2-PEG is most likely due to changes in the fold of the two adducts resulted by the different positioning of MAL-PEG on the mutant peptides. Finally, when the proteins were first treated with Q0 and then incubated with MAL-PEG, no complex formation was attained, verifying that both cysteine residues undergo Q0-dependent oxidation that readily blocks formation of covalent adducts with MAL-PEG.
Fig. 4.
Redox state of ArcB. (A) Tagging ArcB with MAL-PEG. Purified ArcB peptides (50 pmol), untreated or pretreated with 0.5 mM Q0, were incubated with 1 mM MAL-PEG at 25°C. After 1 h, the reactions were terminated and differences in the mobility of the tagged and the untagged proteins were visualized by Western blot analysis with specific ArcB polyclonal antibodies. (B) Spectroscopic characteristics of NBD-modified ArcB. Purified ArcB peptides (10 nmol) were pretreated with either 0.5 mM Q0 or 10 mM DTT and incubated with NBD-Cl (1 mM). Final spectra (600 to 300 nm) were monitored after free NBD-Cl was removed by three cycles of concentration and redilution in a nanosep 10K. Solid line, pretreatment with DTT; dotted line, pretreatment with Q0. (C) Immunoblot analysis. Aerobically or anaerobically grown E. coli cells were harvested at an OD600 ≈0.4-0.5 and treated with trichloroacetic acid to precipitate the proteins and stop further thiol/disulfide exchange. Precipitated proteins were dissolved in nonreducing SDS sample buffer and divided into two equal portions, one of which was treated with 100 mM DTT. Proteins were separated on nonreducing SDS/PAGE, followed by Western blot analysis with specific ArcB polyclonal antibodies. The 88-kDa band corresponds to the size of full-length ArcB monomer, and the 170-kDa band corresponds to an ArcB dimer.
ArcB Dimerizes Through Intermolecular Disulfide Bond Formation. We next attempted to identify the chemical nature of the oxidized cysteine intermediates. The pathway for thiol oxidation begins with the formation of sulfenic acid (-SOH), which rapidly condenses to form disulfide bonds (-S-S-). In proteins, however, steric hindrance may prevent an oxidized thiol from forming a disulfide, and a stable sulfenic acid intermediate is sustained. Cysteine sulfenic acids have been proposed as intermediates in a number of catalytic and regulatory proteins (25) but have been difficult in identifying because of their inherent instability (26). Nevertheless, reaction between the electrophilic reagent NBD-Cl with reduced thiols and sulfenic acids in proteins have been shown to lead to the covalent incorporation of spectroscopically detectable conjugates with absorption maxima at 420 and 347 nm, respectively (27). Therefore, to examine whether a stable sulfenic acid is formed in ArcB, DTT-reduced or Q0-treated ArcB was reacted with NBD-Cl, and the absorbance characteristics of the resulting NBD adducts were determined (Fig. 4B). As expected, the reaction of NBD-Cl with the DTT-treated protein formed a conjugate with maximal absorbance at 420 nm, characteristic for Cys-S-NBD. On the other hand, when NBD-Cl was incubated with the Q0-treated ArcB, the spectra showed neither the Cys-S-NBD peak at 420 nm nor the Cys-S(O)-NBD species at 347 nm. This is consistent with the formation of either an intramolecular or two intermolecular disulfide bonds that result in the blockage of the thiols of both cysteine residues from reacting with NBD-Cl. In agreement, the DTT-treated ArcBC1 and ArcBC2 both formed the Cys-S-NBD adduct with maximal absorbance at 420 nm, but the Q0-treated proteins failed to form a detectable adduct with NBD-Cl (Fig. 4B). In contrast, neither the DTT- nor the Q0-treated ArcBCC formed a detectable adduct with NBD-Cl, confirming the specificity of the reaction. Thus, it appears that the two cysteine residues participate in intermolecular disulfide-bond formation.
We therefore examined whether disulfide-bond formation in ArcB occurs in vivo. To this end, anaerobic or aerobic grown E. coli cells were treated with 10% trichloroacetic acid to denature the cellular proteins, a process that effectively fixes ArcB sulfhydryls into their oxidized or reduced states. Subsequently, the cellular proteins were subjected to nonreducing SDS electrophoresis followed by Western blot analysis with ArcB-specific antibodies. The extracts from anaerobically grown cells revealed a single ≈88-kDa band, corresponding to the size of full-length ArcB monomer. However, in the aerobic extracts, the predominant portion of ArcB migrated as an ≈170-kDa dimer, the mobility of which was shifted to that of the monomer upon treatment with DTT (Fig. 4C). Hence, the two cysteine residues in ArcB participate in intermolecular disulfide bond formation, leading to dimerization and thereby silencing of its kinase activity.
ArcB Is Oxidized Specifically by Quinones. Because the activity of many redox-regulated proteins appears to be controlled by H2O2-induced disulfide bond formation, we examined the effect of this oxidant on the activity of ArcB. Curiously, H2O2 did not affect the in vitro phosphorylation of ArcB (Fig. 1 A). Also, addition of elevated H2O2 concentrations (0.1-2 mM) in an anaerobic culture did not affect the expression of the ArcA-P repressible λΦ(lldP′-lacZ) reporter, verifying the in vitro result (data not shown). On the other hand, shifting the anaerobic culture to aerobiosis led to an instantaneous increase in reporter expression (data not shown). In this respect, it has to be mentioned that although molecular oxygen per se does not have a direct effect on the activity of ArcB (28), its presence leads to the oxidation of the quinone electron carriers that in turn inhibit ArcB phosphorylation. In fact, quinones have been shown to efficiently oxidize the periplasmic located cysteine residues of DsbB, a membrane-bound protein that is a central component in thiol oxidation reactions (29). Hence, ArcB is not only insensitive to molecular oxygen but also to the aerobic byproduct H2O2 and therefore seems to have evolved to specifically sense the oxidized forms of the quinone electron carriers.
Discussion
We have provided evidence that the molecular event of redox signaling by ArcB is intermolecular disulfide bond formation and reduction, a reaction in which quinones act as direct oxidants. Despite that the cytosol of both prokaryotic and eukaryotic cells provides a highly reducing environment, which is almost untenable for disulfide bond formation (30, 31), a number of cytoplasmic proteins that are regulated by reversible disulfide bond formation are known. Some examples include the transcriptional factors Yap1p in Saccharomyces cerevisiae, OxyR and the chaperone Hsp33 in E. coli, and the CtrJ and the membrane-bound kinase RegB in Rhodobacter capsulatus (31-36). However, our in vivo and in vitro analyses indicate that disulfide bond formation in ArcB may differ from that of the other redox regulators in that it appears to respond specifically to the oxidized forms of quinones rather than to molecular oxygen or to reactive-oxygen species such as H2O2.
E. coli expresses branched electron-transfer chains that can deal with various electron donors and several alternative electron acceptors. A central component in these networks is the quinone pool that acts as a redox mediator. That is to act both as collector of electrons from dehydrogenases and as donor to reductases and oxidases, thereby serving as the crossroad of electron transfer. As such, its redox state will change rapidly in response to variations in external conditions that affect electron flow in the electron-transfer chains, for example a shift from anaerobiosis to aerobiosis. In this case, the electrons will rapidly flow toward O2, and the quinone pool will shift to its oxidizing state. This will enable the electron transfer from the cysteine residues of ArcB to quinones, which will result in disulfide bond formation and immediate silencing of the kinase activity of the protein (Fig. 5). This apparently simple mechanism will allow a rapid response of bacterial cells to their environment, as the redox signals will be instantly transduced into the transcriptional apparatus.
Fig. 5.
The ArcB sensor kinase and a model for its redox regulation. (A) Schematic representation of ArcB. The linker region contains a putative leucine zipper (8) and a Per-Arnt-Sim (PAS) domain (9). Depicted, in the linker region, are also the cysteine residues 180 and 241. The primary transmitter domain (H1) contains the conserved His-292 and the catalytic determinants N, G1, and G2. The G1 and G2 sequences typify nucleotide-binding motifs. The receiver domain (D1) contains the conserved Asp-576, and the phosphotransfer domain (H2) contains the conserved His-717. (B) A simplified model for ArcB inactivation. Upon a shift from anaerobic to aerobic conditions of growth, the quinone pool shifts to its oxidized state. This allows the electron transfer from the Cys-180 of ArcB to quinones that leads to the formation of an intermolecular disulfide bond between the Cys-180 of two monomers, and results in a significant reduction of the kinase activity of ArcB. As the electrons rapidly flow toward O2 via either cytochrome bd or bo oxidase, the quinone pool maintains its oxidized state and induces the formation of a second disulfide bond between the two Cys-241, resulting in the complete silencing of the ArcB kinase activity.
Thus, thiol oxidation and reduction in the cytosolic portion of ArcB provides a pivotal link in the Arc signal transduction pathway that connects the redox state of the quinone pool to the transcriptional apparatus.
Acknowledgments
We thank E. C. C. Lin, F. Åslund, D. González-Halphen, R. Pérez-Montfort, A. Gómez-Puyou, N. Sánchez, A. Peña, and B. Michel for advice and/or for critically reading the manuscript. This work was supported by Consejo Nacional de Ciencia y Technología Grant 37342-N, National Institutes of Health Research Grant R03 TW06003, and Korean Ministry of Science and Technology Grant M1-0311-00-0081.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: Q0, ubiquinone-0; MMTS, S-methyl methanethiosulfonate; MAL-PEG, methoxy-polyethylene glycol maleimide; NBD-Cl, 7-chloro-4-nitrobenzo-2-oxa-1,3-diazole.
References
- 1.Parkinson, J. S. & Kofoid, E. C. (1992) Annu. Rev. Genet. 26, 71-112. [DOI] [PubMed] [Google Scholar]
- 2.Hoch, J. A. & Silhavy, T. J. (1995) Two-Component Signal Transduction (Am. Soc. Microbiol., Washington, DC).
- 3.Lynch, A. S. & Lin, E. C. (1996) in Escherichia coli and Salmonella: Cellular and Molecular Biology, eds. Neidhardt, F. C., Curtis, R., III, Ingraham, A. L., Lin, E. C. C., Low, K. B., Magasanik, B., Reznikoff, W. S., Riley, M., Schaechter, M. & Umbarger, H. E. (Am. Soc. Microbiol., Washington, DC), pp. 1526-1538.
- 4.Iuchi, S. & Lin, E. C. (1988) Proc. Natl. Acad. Sci. USA 85, 1888-1892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Iuchi, S., Matzuda, Z., Fujiwara, T. & Lin, E. C. (1990) Mol. Microbiol. 4, 715-727. [DOI] [PubMed] [Google Scholar]
- 6.Kwon, O., Georgellis, D., Lynch, A. S., Boyd, D. & Lin, E. C. (2000) J. Bacteriol. 182, 2960-2966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ishige, K., Nagasawa, S., Tokishita, S. & Mizuno, T. (1994) EMBO J. 13, 5195-5202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Georgellis, D., Kwon, O., De Wulf, P. & Lin, E. C. (1998) J. Biol. Chem. 273, 32864-32869. [DOI] [PubMed] [Google Scholar]
- 9.Taylor, B. L. & Zhulin, I. B. (1999) Microbiol. Mol. Biol. Rev. 63, 479-506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Georgellis, D., Kwon, O. & Lin, E. C. (1999) J. Biol. Chem. 274, 35950-35954. [DOI] [PubMed] [Google Scholar]
- 11.Rodriguez, C., Kwon, O. & Georgellis, D. (2004) J. Bacteriol. 186, 2085-2090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Georgellis, D., Lynch, A. S. & Lin, E. C. (1997) J. Bacteriol. 179, 5429-5435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kwon, O., Georgellis, D. & Lin, E. C. (2000) J. Bacteriol. 182, 3858-3862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brondsted, L. & Atlung, T. (1994) J. Bacteriol. 176, 5423-5428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lynch, A. S. & Lin, E. C. (1996) J. Bacteriol. 178, 6238-6249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Georgellis, D., Kwon, O. & Lin, E. C. (2001) Science 292, 2314-2316. [DOI] [PubMed] [Google Scholar]
- 17.Link, A. J., Phillips, D. & Church, G. M. (1997) J. Bacteriol. 179, 6228-6237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gelman, M. S. & Prives, J. M. (1996) J. Biol. Chem. 271, 10709-10714. [DOI] [PubMed] [Google Scholar]
- 19.Lodish, H. F. & Kong, N. (1993) J. Biol. Chem. 268, 20598-20605. [PubMed] [Google Scholar]
- 20.McClelland, M., Sanderson, K. E., Spieth, J., Clifton, S. W., Latreille, P., Courtney, L., Porwollik, S., Ali, J., Dante, M., Du, F., et al. (2001) Nature 413, 852-856. [DOI] [PubMed] [Google Scholar]
- 21.Jin, Q., Yuan, Z., Xu, J., Wang, Y., Shen, Y., Lu, W., Wang, J., Liu, H., Yang, J., Yang, F., et al. (2002) Nucleic Acids Res. 30, 4432-4441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Parkhill, J., Wren, B. W., Thomson, N. R., Titball, R. W., Holden, M. T., Prentice, M. B., Sebaihia, M., James, K. D., Churcher, C., Mungall, K. L., et al. (2001) Nature 467, 523-527. [DOI] [PubMed] [Google Scholar]
- 23.Duchaud, E., Rusniok, C., Frangeul, L., Buchrieser, C., Givaudan, A., Taourit, S., Bocs, S., Boursaux-Eude, C., Chandler, M., Charles, J. F., et al. (2003) Nat. Biotechnol. 11, 1307-1313. [DOI] [PubMed] [Google Scholar]
- 24.Manukhov, I. V., Bertsova, Y. V., Trofimov, D. Y., Bogachev, A. V. & Skulachev, V. P. (2000) Biochemistry (Moscow) 65, 1321-1326. [PubMed] [Google Scholar]
- 25.Claiborne, A., Miller, H., Parsonage, D. & Ross, R. P. (1993) FASEB J. 7, 1483-1490. [DOI] [PubMed] [Google Scholar]
- 26.Kice, J. L. (1980) Adv. Phys. Org. Chem. 17, 65-181. [Google Scholar]
- 27.Ellis, H. R. & Poole, L. B. (1997) Biochemistry 36, 15013-15018. [DOI] [PubMed] [Google Scholar]
- 28.Iuchi, S., Chepuri, V., Fu, H. A., Gennis, R. B. & Lin, E. C. (1990) J. Bacteriol. 172, 6020-6025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bader, M., Muse, W., Ballou, D. P., Gassner, C. & Bardwell, J. C. (1999) Cell 98, 217-227. [DOI] [PubMed] [Google Scholar]
- 30.Gilbert, H. F. (1990) in Advances in Enzymology and Related Areas of Molecular Biology, ed. Meister, A. (Wiley, New York), pp. 69-172. [DOI] [PubMed]
- 31.Hwang, C., Sinskey, A. J. & Lodish, H. F. (1992) Science 257, 1496-1502. [DOI] [PubMed] [Google Scholar]
- 32.Kuge, S., Arita, M., Murayama, A., Maeta, K., Izawa, S., Inoue, Y. & Nomoto, A. (2001) Mol. Cell. Biol. 21, 6139-6150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zheng, M., Aslund, F. & Storz, G. (1998) Science 279, 1718-1721. [DOI] [PubMed] [Google Scholar]
- 34.Jakob, U., Muse, W. & Bardwell, J. C. A. (1999) Cell 96, 341-352. [DOI] [PubMed] [Google Scholar]
- 35.Masuda, S., Dong, C., Swem, D., Setterdahl, A. T., Knaff, D. B. & Bauer, C. E. (2002) Proc. Natl. Acad. Sci. USA 99, 7078-7083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Swem, L. R., Kraft, B. J., Swem, D. L., Setterdahl, A. T., Masuda, C., Knaff, D. B., Zaleski, J. M. & Bauer, C. E. (2003) EMBO J. 22, 4699-4708. [DOI] [PMC free article] [PubMed] [Google Scholar]