Abstract
Studies reporting term pregnancy and the production of genetically identical offspring from isolated blastomeres of early stage embryos have been carried out in small and large animals. However, very little is known about the effects of embryo splitting on the development and reproductive competency of human embryos. In this study, we investigated the effects of embryo splitting on profile of microRNAs (miRNAs) detected in their spent blastocyst medium (SBM) by comparative analysis of miRNA profiles in SBM of human twin embryos created by blastomere biopsy and SBM of blastocysts that resulted in a healthy pregnancy and live birth following embryo transfer. The profile of miRNA secretion in in vitro culture media consistently distinguishes twin from control embryos. We found that six miRNAs are significantly more abundant in SBM from twin embryos, while nine are significantly more abundant in SBM from euploid implanted blastocysts. These nine include miRNA-30c, a previously reported marker of blastocyst implantation potential. Furthermore, 22.9% of miRNAs secreted by twin embryos were never detected in SBM from normal reproductively competent blastocysts, or from trophectoderm (TE) samples from normal blastocysts donated for the research. The miRNA profile, unique to twin blastocysts, might be a result of differential lineage commitment in these embryos.
Keywords: : embryo splitting, human preimplantation embryos, miR-30c, miRNA, spent blastocyst medium
Introduction
MicroRNAs (miRNAs) are evolutionarily conserved, single-stranded noncoding RNA molecules of ∼22 nucleotides in length and are considered to be major transcriptional/posttranscriptional regulators of gene expression. In the human genome, more than 1,000 miRNAs have been identified (miRBase, www.mirbase.org). Human blastocysts express large number of miRNAs [1]. They are secreted and can be found in embryo culture media. Attempts have been made to link specific miRNAs detected in the medium with embryo ploidy status and reproductive competence to predict in vitro fertilization (IVF) outcome, and several candidates have been identified [2–5]. Although further studies are warranted, a comprehensive profile of miRNAs secreted by human preimplantation embryos in spent culture media is taking shape [6].
Several studies have suggested that the splitting of human embryos might result in morphologically adequate viable blastocysts [7,8]. However, qualitative analyses of the embryos created in such a way have been relatively limited. We recently analyzed 176 twin embryos created by blastomere separation of 88 human embryos from either early (2–5 blastomeres, n = 43) or late (6–10 blastomeres, n = 45) cleavage stage. Half of the blastomeres in each embryo were biopsied and placed into an empty zona pellucida (ZP) prepared in advance by removing the cellular content from immature oocytes or degraded, clinically unsuitable embryos. The blastomere donor embryos were dubbed twin A and the blastomere recipients twin B [9]. We found that they have distinctive features in comparison with normal blastocysts obtained through fertilization. Twin split embryo blastocysts were smaller and less cellular, the first fate decision was somewhat delayed, inner cell mass (ICM) was either absent or small and poor quality, and the majority of cells were expressing ICM and trophectoderm (TE) markers simultaneously (Fig. 1).
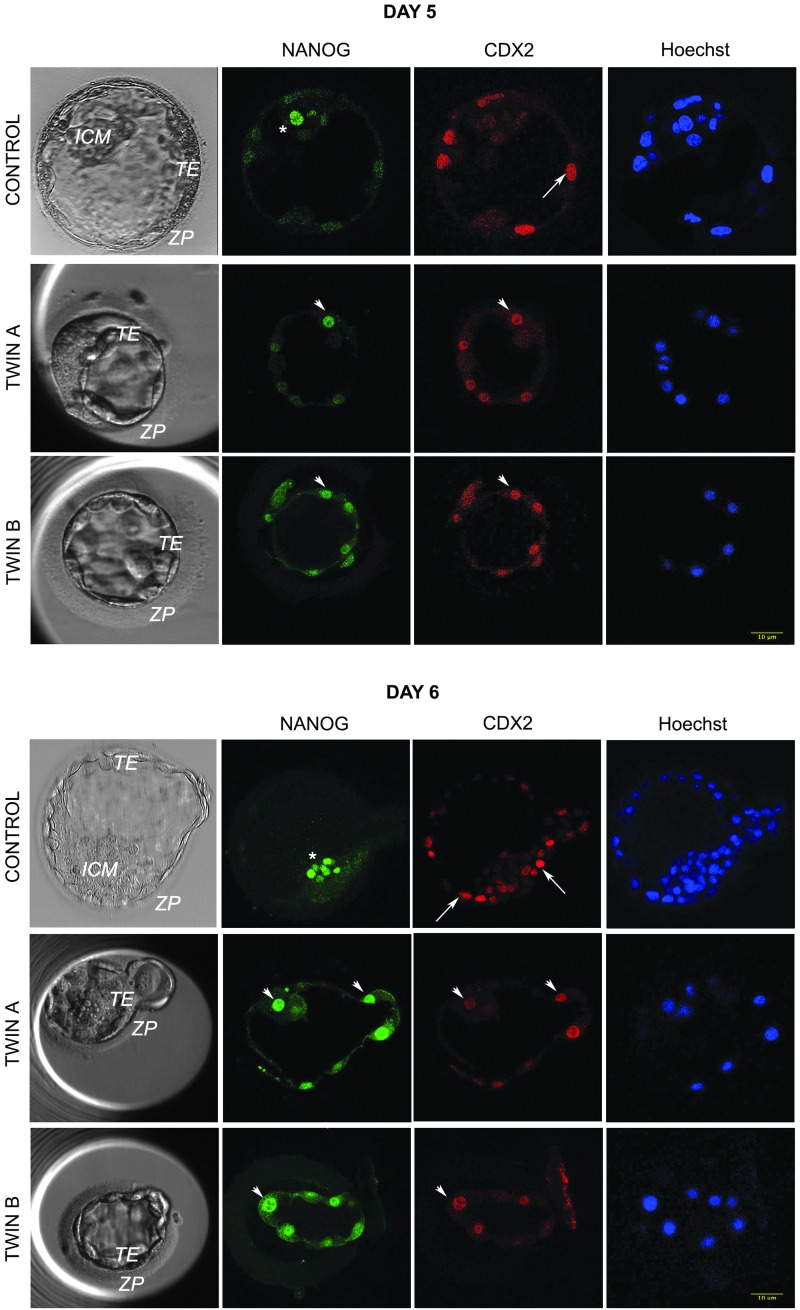
FIG. 1.
Expression of the lineage markers in twin and control embryos at day 5 postfertilization. Twin embryos generated by embryo splitting are smaller, often with no distinctive ICM. They are less cellular in comparison with nonmanipulated control embryos and most of the cells have a dual expression of ICM and TE markers. Green: ICM marker NANOG. Red: TE marker CDX2. Blue: DNA dye Hoechst 33342. In control embryo, asterisk marks ICM cell expressing NANOG and arrow TE cell expressing CDX2. In twins A and B, the arrowheads point to cells with dual expression of NANOG and CDX2. ICM, inner cell mass; TE, trophectoderm.
To address whether such changes in the phenotype of embryos generated by embryo splitting affect miRNA secretion, we compared miRNA profile in spent blastocyst medium (SBM) of seven pairs of twins and seven blastocysts generated by IVF. All control blastocysts resulted in live births upon single embryo transfer.
Materials and Methods
The work described in this study is done under license from the UK Human Fertilisation and Embryology Authority (research license Nos.: R0075 and R0133) and also has local ethical approval (UK National Health Service Research Ethics Committee Reference: 06/Q0702/90). Informed consent was obtained from all embryo donors and the experiments conformed to the principles set out in the WMA Declaration of Helsinki and the NIH Belmont Report. No financial inducements are offered for donation. All the embryos used in this project were cryopreserved with slow freezing method.
Embryo culture
Embryo culture and splitting using the blastomere biopsy technique has been described in detail previously [9]. All the embryos were split at the late cleavage stage (6–9 cells). Half of the blastomeres were aspirated and removed from a donor embryo and placed in an empty human ZP, previously prepared from clinically unsuitable oocytes or embryos. After blastomere transfer, the donor (Twin A, n = 7) and recipient embryos (Twin B, n = 7) were cultured up to the blastocyst stage (day 5/6). SBM was collected as described [2]. Briefly, the embryos were cultured from day 3 postfertilization in a 35-μL drop of Quinn's Advantage Blastocyst Medium (Cooper Surgical) with 5% Quinn's Advantage Human Serum Albumin (Cooper Surgical) in a humidified atmosphere containing 5% O2 and 6% CO2. At the blastocyst stage, before transfer, 25 μL of SBM was taken for analyses. As a control, we used SBM of embryos that resulted in a healthy pregnancy and live birth following embryo transfer (n = 7; Fig. 2). “No Template” controls and unconditioned blank culture media were also run in parallel to exclude any possible false positive amplification from the subsequent analyses.
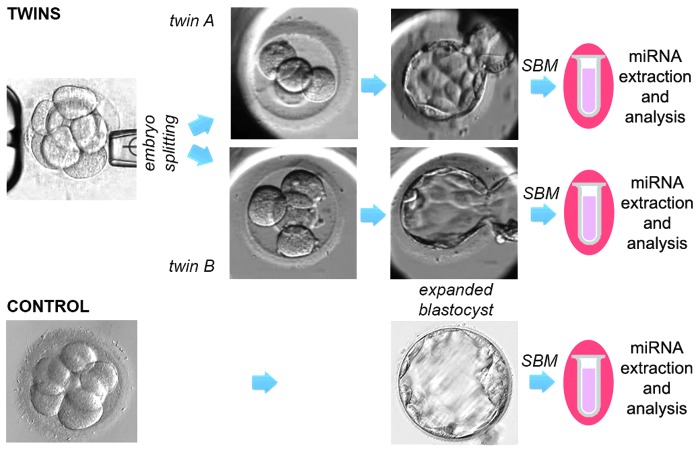
FIG. 2.
Study design. Control embryos were cultured undisturbed up to fully expanded blastocyst stage, and SBM was collected before thawing. All seven blastocysts were diagnosed as euploid and healthy pregnancies ensued after transfer. Twin embryos instead were produced after embryo splitting at the cleavage stage. Twin A were cultured up to the blastocyst stage in their own zona pellucida and twin B were cultured in host zona pellucida. SBM was collected at the blastocyst stage. miRNAs were extracted and analyzed from seven controls and seven pairs of twins. miRNAs, microRNAs; SBM, spent blastocyst medium.
miRNA isolation, profiling, and screening
miRNA isolation and profiling were described in detail previously [2]. Briefly, miRNAs were isolated through a magnetic beads-based extraction (Anti-miRNA Bead Capture Purification Kit human panel A; Life Technologies), and after targeted retrotranscription and preamplification steps using the Megaplex RT and Preamp human panel A primers pool (Life Technologies), samples were analyzed in quantitative polymerase chain reaction (qPCR) through the TaqMan Low Density Array (TLDA) cards on a ViiA7 instrument (Life Technologies). Data processing and statistical analyses were performed through the RealTime StatMiner software (Integromics) and the SPSS software (IBM). Data were compared also with miRNAs profiled from five ICM-free TE samples obtained from normal blastocysts donated for the research, as previously described [2]. The primer sequences for targets within the human panel A + B are listed on the Thermo Fisher's website http://tools.thermofisher.com/content/sfs/manuals/4473439C.pdf
GenoSplice Technology performed quality control (QC), processing, and further analyses of the data using endogenous control for normalization. For each miRNA X of each sample S, the samples were normalized according to the formula 2−ΔCtX,S = 2−[CtX,S − mean(CtU6,S)]; samples 6A and 6B were omitted.
Single assays
Eluted leftover SBM samples from twin A, twin B, and randomly selected historical controls of euploid implanted blastocysts, respectively, were processed for single assays' analyses through qPCR. Blank (molecular biology grade water) and media samples never exposed to embryos were processed in parallel as negative controls. We tested single assays for the U6-snRNA, miR-30c, and miR-203. The first was chosen to confirm its weak reliability as detected through the miRNome panel, the second to confirm the differential expression in the comparison of twin A versus twin B, while the last miRNA was chosen as potential normalizer since it showed a stable trend in the panels' analysis among all these samples (Twin A Ct 26.3 ± 1.1, 24.1–27.4; Twin B Ct 25.8 ± 1.2, 23.5–27.3; euploid implanted blastocysts 24.1 ± 1.1, 22.9–25.5). The minimum variance of the median method performed through the StatMiner software (Integromics) confirmed its eligibility to this aim.
The primers for retrotranscription reaction of the three assays were pooled together (5 μL each) and diluted through 485 μL of nuclease-free water. The retrotranscription mix was composed as follows: 6 μL of primers pool, 0.3 μL of 100 mM dNTPs (with dTTP), 3 μL of MultiScribe™Reverse Transcriptase 50 U/μL, 1.5 μL of 10× Reverse Transcription Buffer, 0.2 μL of RNase Inhibitor 20 U/μL, 1.5 μL of nuclease-free water (TaqMan® Small RNA Assays protocol; Life Technologies), and 2.5 μL of eluted leftover sample. The thermal protocol was conducted on a 2720 Thermal Cycler (Life Technologies) as follows: 16°C for 30′, 42°C for 30′, 85°C for 5′, and 4°C ∞. No preamplification step is entailed by this protocol, and thus, we raised the threshold Ct level for detection to 37 cycles. qPCR mix was composed as follows: 0.5 μL of TaqMan Small RNA Assay (20 ×), 5 μL of TaqMan Universal PCR Master Mix II (2 ×) no UNG, 2 μL of nuclease-free water, and 2.5 μL of sample. qPCR was performed on a ViiA7 machine (Life Technologies) according to manufacturer's protocol. Each assay was run in triplicate to exclude the technical variability due to pipetting error.
The primer sequences for single assays are: hsa-miR-30c-5p (cat No. 000419) target sequence UGUAAACAUCCUACACUCUCAGC, u6 snRNA (cat No. 001973) target sequence GUGCUCGCUUCGGCAGCACAUAUACUAAAAUUGGAACGATACAGAGAAGAUUAGCAUGGCCCCUGCGCAAGGAUGACACGCAAAUUCGUGAAGCGUUCCAUAUUUU, and hsa-miR-203 (cat No. 000507) target sequence GUGAAAUGUUUAGGACCACUAG.
The Ct values for miR-30c were normalized on miR-203 ones and the differential expression analyses were conducted through the ΔΔCt method.
Results
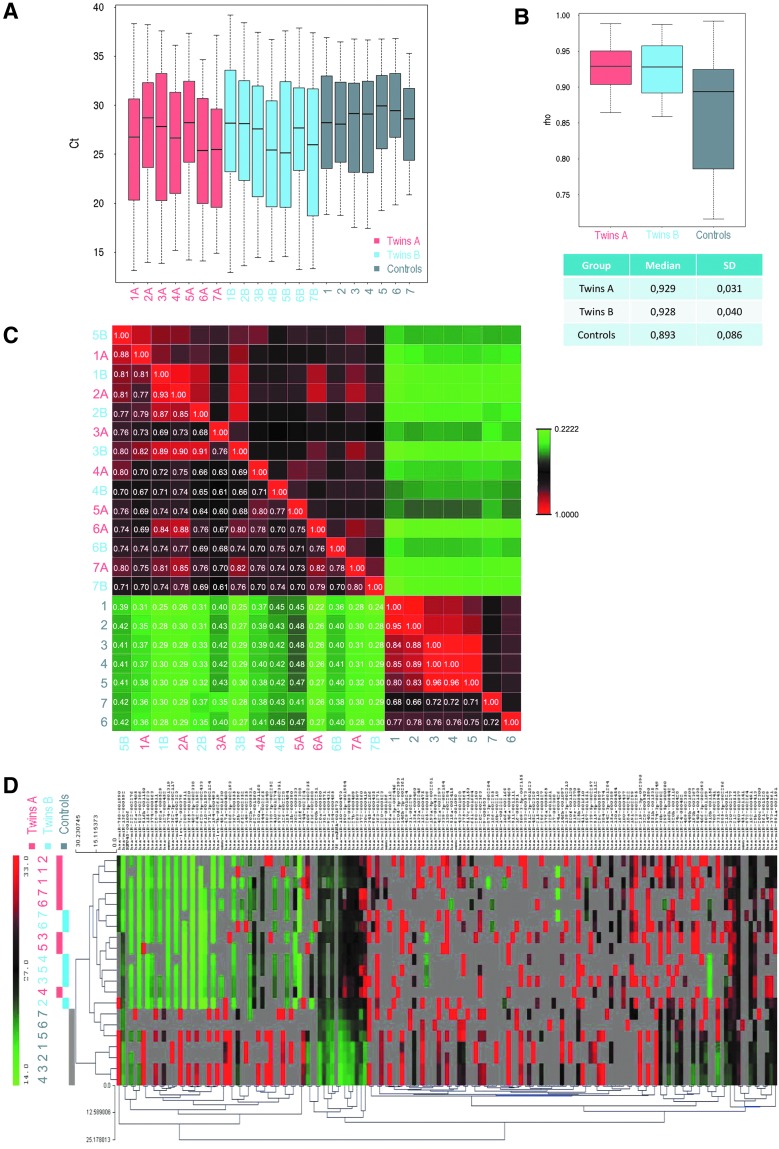
Using a medium throughput analysis, we compared the profile of 377 miRNA sequences from the SBM of twin embryos (n = 7 pairs; Supplementary Figs. S1 and S2; Supplementary Data are available online at www.liebertpub.com/scd) with those from control embryos that resulted in live birth after single embryo transfer (n = 7). Correlation between samples of each group was high for all conditions (Fig. 3A). Correlation between samples in the control group was somewhat lower than between the samples in Twin A and Twin B groups (Fig. 3B). Pearson's correlation matrix of the raw Ct values consistently distinguished control embryo from twin embryos (Fig. 3C). Heat map revealed that there were no significant clusters by experimental group; however, there were high numbers of missing Ct values (Fig. 3D).
FIG. 3.
Estimation of the correlation between samples within each group. (A) Distribution of Ct values for twin A, twin B, and control embryos. (B) Distribution of Spearman test rho values (Ct) indicates that median correlations are very high for all conditions. Correlation between twin A (0.929 ± 0.031) and twin B (0.928 ± 0.040) samples is similar, whereas between control samples is lower (0.893 ± 0.086). (C) Pearson's correlation matrix comparing raw Ct data among all analyzed samples. A clear discrimination between twin embryos and controls can be deduced from the low correlation of miRNA profiles from SBM after in vitro culture. The numbers show the actual correlation value among all analyzed samples. (D) Heat map of Ct values from twin A, twin B, and controls indicates no significant cluster by experimental group and high number of missing values.
To minimize experimental error, we analyzed only miRNA with at least 60% of Ct values above the threshold that was set as 35 in each compared experimental group (namely 5/7 valid values for control SBMs and 9/14 valid values for twin SBM).
Data analysis using common intracellular normalizers
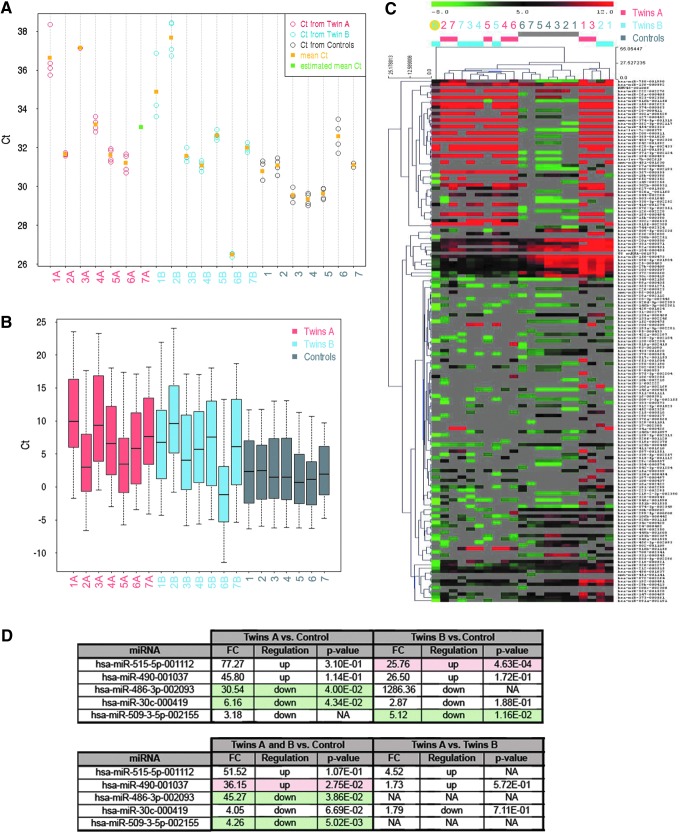
Commonly used intracellular normalizers, such as small nuclear RNA U6, often show large fluctuations in samples measuring secreted miRNAs [10]. In our case, after QC analyses, only one out of three endogenous controls was available (U6). There were no Ct values for endogenous control small nucleolar RNA RNU44 and only eight values for RNU48. Despite Ct of U6 endogenous control varying across the samples, Ct values of U6 were homogenous within each sample (Fig. 4A). Since the four different U6 reporters provided approximately the same Ct values for the same sample, U6 can be used for computing −ΔCt normalization of the other miRNAs. However, even though reproducibility across the samples was good, due to very low U6 Ct values in the sample 6B, we excluded the twins 6A and 6B from the analyses (Fig. 4B, C).
FIG. 4.
Quality of the endogenous control and effect of the normalization with the endogenous control. (A) Ct values of the endogenous control U6 are heterogeneous between the samples and homogenous within each sample. The sample 6B has very low Ct value. (B) While distribution of −ΔCt values in samples from control embryo group is relatively homogenous, the values are much more heterogeneous in samples from both twin groups indicating that expressing or missing miRNA can be different for each sample. (C) Heat map of −ΔCt values from twin A, twin B, and controls indicates no significant cluster by experimental group, high number of missing values, and that 6B sample (yellow dot) does not cluster with others. (D) miRNAs were regulated in at least one comparison (FC ≥1.5 and P value ≤0.05), where U6 was used for normalization. FC, fold-change.
We found that miR-515-5p and miR-490 were detected at significantly higher levels, whereas miR-486-3p, miR-30c, and miR-509-3-5p had significantly lower concentrations in twins in comparison with control embryos [fold-change (FC) ≥1.5, P value ≤0.05; Fig. 4D]. However, a lack of two internal reference genes (RNU44 and RNU48) and large fluctuations in the third one (U6) for secreted miRNAs prompted us to approach data analysis using a different type of normalization: the global normalization strategy [11].
Data analysis using global geometric mean of expression of all detected miRNAs
Global normalization strategy consisted of three successive steps. Any Ct value above the threshold set at 35 was considered as a noise and was discarded from further analysis. The arithmetic average Ct value for all detected miRNAs was then calculated for each individual sample and, subsequently, subtracted from each individual Ct value for that sample. The procedure results in normalized expression values in logE scale (E being the base of the exponential amplification function, with 2 being a good estimate); for individual miRNAs, values are inversely correlated with expression levels. Whole-genome quantitative real-time polymerase chain reaction based miRNA profiling in combination with a global mean normalization strategy has proven to be the most sensitive and accurate approach for high throughput miRNA profiling, whose effectiveness when applied to low input samples, as well as SBMs, has been validated and reported previously [2].
MicroRNA profile in SBMs is similar among twins A and B, but significantly different from miRNA profile of control embryos
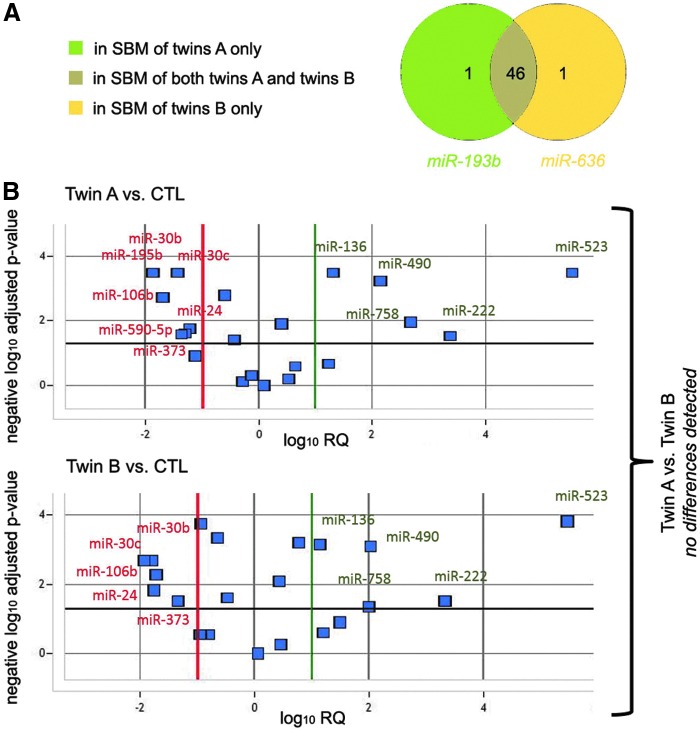
Seven pairs of SBMs from twin embryos were compared to SBMs from reproductively competent blastocysts. After all QC steps, 48 miRNAs were found to be consistently secreted in IVF culture media by twin A and B blastocysts, while 59 by control ones. The twins A and B shared 46 out of 48 miRNA detected in their SBM. miRNA detected only in SBM of twin A was miR-193b, whereas miR-636 was detected only in SBM of twin B (Fig. 5A). Differences between miRNA profiles in SBM of twin A versus control blastocyst were overlapping with differences of profiles in SBM of twin B versus control blastocysts (Fig. 5B).
FIG. 5.
Comparison between miRNAs in SBM from twins A and B. (A) More than 95% (46/48) of miRNAs detected are shared by SBM from both twins A and B. (B) The volcano plots comparing twin A or twin B versus controls are almost overlapping, in fact no significant difference can be detected when comparing instead twin A versus twin B.
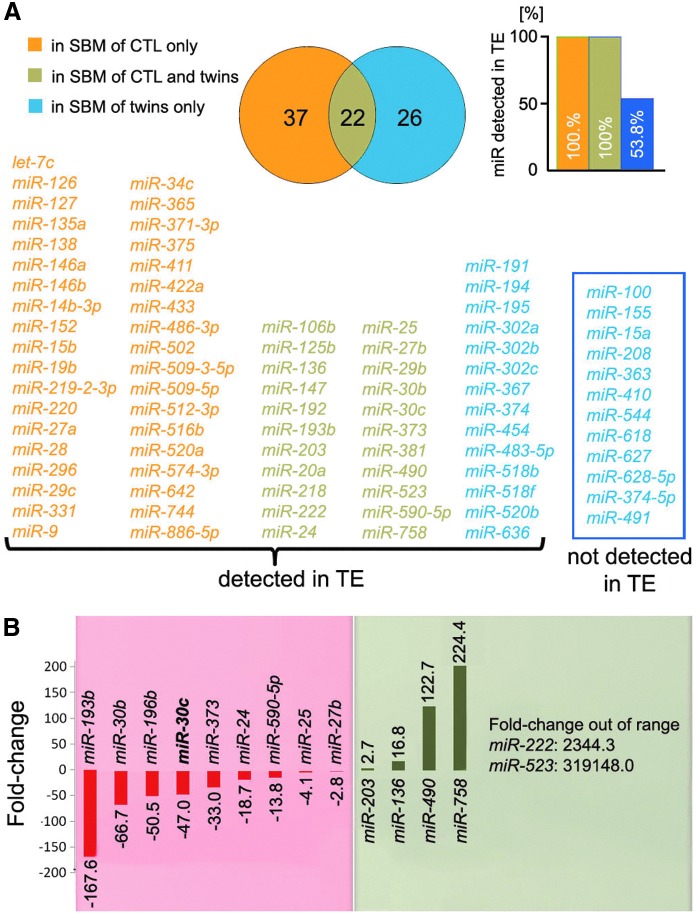
Twenty-two miRNAs were shared between SBM of control and twin embryos (Fig. 6A). Thirty-seven miRNAs were detected solely in SBM of controls and 26 only from twin and never secreted by nonmanipulated human blastocysts generated by IVF. All 59 miRNAs detected in SBM of control blastocyst were also detected in ICM-free TE cells [2]. However, only 53.8% (14/26) of the twin-specific secreted miRNAs were coexpressed from normal blastocyst-derived TE cells, whereas 12 were not detected in TE cells previously. Among these 12, only 1, miR-374-5p, was expressed in ICM (A. Capalbo, unpublished data). The detection of those novel 12 miRNAs may suggest abnormal blastocyst development and/or be an indicator of different lineage commitment stage in the split embryos.
FIG. 6.
Comparison between twin and control SBM miRNA expression profile. (A) Thirty-seven miRNAs (orange) were found solely in SBM from control blastocysts and are all coexpressed from TE cells; 22 miRNAs (olive) were found in SBMs from both control and twin blastocysts and are all coexpressed from TE cells; 26 miRNAs (blue) were found exclusively in SBM from twin blastocysts, but only 53.8% (14/26, blue, framed) are coexpressed from TE cells. (B) The volcano plot shows nine miRNAs significantly less abundant in SBM from twins versus control reproductively competent blastocysts, with a FC variable between −167.6 and −2.8, and six miRNAs instead significantly more abundant, with a FC between 2.7 and 319,148. Interestingly, miR-30c, putative biomarker of reproductive potential, was found in the former cluster with a FC −47.0.
A relative quantification analysis was conducted, and a volcano plot was generated by comparing the detection levels in SBM from twin versus controls of the 22 common miRNAs (Fig. 6B). Six miRNAs were found to be significantly more abundant in the twin samples (miR-203, miR-136, miR-490, miR-758, miR-222, and miR-523), whereas nine miRNAs were more abundant in the control samples (miR-193b, miR-30b, miR-106b, miR-30c, miR-373, miR-24, miR-590-5p, miR-25, and miR-27b).
The results were further validated with single assays specific for miR-30c and for miR-203. Twin A mean Ct ± standard deviation (SD) was 34.9 ± 1.9 and 31.3 ± 1.1 for miR-30c and miR-203, respectively. Twin B mean Ct ± SD was 35.3 ± 2.2 and 30.8 ± 1.2 for miR-30c and miR-203, respectively. The ΔΔCt analyses showed no difference in the levels of miR-30c normalized on miR-203 in twins A versus B (FC: 1.9; P value: 0.7). When performing the same analysis in twins (A and B together) versus the historical control of SBMs from euploid implanted blastocysts, a statistically significant 93.2 × lower level was detected (P < 0.001) in the formers. No miRNA expression was found in negative controls.
Significantly lower expression levels of miR-30c in twins were detected with either of two normalization strategies
A statistically significant lower level of miRNA-30c detected in SBM from twin embryo culture than in SBM of control embryos was identified using either of two normalization strategies. It captured our interest, because this miRNA has been identified as a putative biomarker of blastocyst implantation potential when secreted at high levels in the SBM during IVF cycles [2]. Next, using a computational analysis, we investigated possible roles of miRNA-30c during early development. According to the miRNA-gene interaction data provided by the DIANA TarBase v7 (http://diana.cslab.ece.ntua.gr/tarbase), miRNA-30c has 1,643 putative gene targets. To identify which of them are expressed in TE and/or ICM of human blastocyst, we filtered these genes against our blastocyst transcriptome database, which contains a list of the genes detected by the RNA-seq method in isolated TE and ICM samples [12]. We found that 1,061 of these genes are expressed in ICM, 1,166 in TE, and 1,006 in both ICM and TE (Fig. 7A). Based on the pathway information provided by the Kyoto Encyclopedia of Genes and Genomes (KEGG; www.genome.jp/kegg), we found that the main pathways retrieved were ubiquitin mediated proteolysis and spliceosome (Fig. 7B).
FIG. 7.
Putative roles of miRNA-30c in human blastocysts. (A) From 1,643 miRNA-30c target genes, 1,061 were expressed in ICM, 1,166 in TE, and 1,006 in both ICM and TE of the human blastocysts. (B) The pathways involving miR-30c targeted genes in ICM and TE according to the KEGG database. KEGG, Kyoto Encyclopedia of Genes and Genome.
Discussion
miRNAs act mostly as posttranscriptional repressors of their target genes. The importance of miRNAs in early embryo development has been identified in a range of species, although relatively little is known about the miRNA regulatory network in mammalian preimplantation embryos [1,13–17]. Aberrant miRNA profiles in transferable blastocysts have been linked with male factor infertility and polycystic ovaries [5]. Since miRNAs are known to be secreted [18], attempts have been made to link specific miRNAs detected in the SBM with embryo ploidy status and reproductive competence [2–5].
miRNAs detected in the spent medium from early embryos might be the output of paracrine circuits, reflecting normal communication among cells of healthy embryos; alternatively, they might be a component of the communication pathway between the embryo and the uterine epithelium, influencing epithelial readiness for embryo apposition and implantation. However, it remains possible that the miRNAs detected in the SBM are products of degrading cells.
Significantly lower levels of miR-30c in twins perhaps indicate poor developmental prognosis for the twin embryos. miR-30c is known to interfere with epithelial-to-mesenchymal transition (EMT) in breast cancer through regulation of TWF1 and IL11 [19] and has been identified as a key indicator of reproductive competence [2]. EMT is an essential part of the first lineage fate decision when during asymmetric cell division one of the daughter cells is pushed inwards, losing polarity and forming ICM [20,21]. Whether miR-30c is directly involved in the ICM formation or is an essential part of the embryo/maternal dialogue at the time of implantation remains to be investigated.
Most of the miRNAs that were detected at significantly lower level in SBM of twin embryos using mean normalization were involved in various developmental processes and differentiation. For example, miR-30b is involved in development of embryonic ectoderm [22], miR-373 promotes mesendoderm differentiation [23], and miR-24 is required for hematopoietic differentiation [24]. However, only miR-24 [25,26] and miR-25 [3] were detected in preimplantation embryo culture media.
Evidence is emerging that miRNA plays a role in embryo-endometrium cross talk during implantation [25,27]. Microarray profiling revealed that six miRNAs were differentially expressed in the human endometrial epithelium during the implantation window and secreted by the endometrial glands into endometrial fluid. miR-30d, the most differentially secreted miRNA in this study, has been reported to be internalized as an exosome-associated molecule by embryo TE, leading to an increase in the expression of genes involved in adhesion [28]. miRNAs detected in follicular fluid, such as miR-320, are thought to influence embryo quality [29]. However, embryo development is almost certainly affected not only by miRNAs from surrounding tissues but also by miRNAs in the early embryos themselves. For instance, miR-29b negatively regulates DNMT3A/3B expression, altering DNA methylation levels in transition from morula to blastocyst stage [30], and miRNA Let-7a posttranscriptionally regulates the expression of ribonuclease type III Dicer1 altering microRNA profile and the implantation competency of the activated blastocysts [31].
Therefore, the most interesting finding was that 22.9% (11 out of 48) miRNAs secreted by twin embryos were never detected in normal, reproductively competent blastocysts (Fig. 6A). Since the TE cells of twin embryos express dual markers of both ICM and TE [9], such discrepancy in miRNA profile may not come as a surprise. miR-155, specifically detected in SBM of twin embryos, inhibits proliferation and migration of trophoblast-derived cell lines [32], whereas nothing is known about potential roles of the other ten “twin-specific” miRNAs in human preimplantation embryo development. Although some information concerning their potential roles in cancer diagnosis, prognosis, and therapy has been reported, extrapolating these data to draw conclusions on their function in embryo development would currently be too speculative.
Moreover, these findings support miRNA analysis from SBM as an effective approach to capture biological variability between embryos of different quality. In this study, the miRNA analysis from SBM was able to clearly distinguish between low quality manipulated and nonmanipulated high quality embryos. Accordingly, miRNA analysis from SBM holds the potential to have enough resolution to capture biological variation from reproductively competent and noncompetent blastocysts and to be used as new noninvasive biomarker of embryo selection.
In conclusion, we are only just starting to get a glimpse of the role of miRNA in development of human preimplantation embryos and therefore, although the overall significance of the data presented in this study is not yet clear, it may add important information to the very limited understanding of these mechanisms.
Supplementary Material
Acknowledgments
This project was supported by the Saudi Arabian Government studentship to L.N., the Merck Serono Grant for Fertility Innovation (2013) to A.C., and Y.K.'s and D.I.'s incentive funds. The authors also thank the staff at the Assisted Conception Unit of Guy's and St Thomas' NHS Foundation Trust for supporting the research program. The authors are especially indebted to patients who donated embryos.
Author Disclosure Statement
No competing financial interests exist.
References
- 1.Rosenbluth EM, Shelton DN, Sparks AE, Devor E, Christenson L. and Van Voorhis BJ. (2013). MicroRNA expression in the human blastocyst. Fertil Steril 99:855–861 [DOI] [PubMed] [Google Scholar]
- 2.Capalbo A, Ubaldi FM, Cimadomo D, Noli L, Khalaf Y, Farcomeni A, Ilic D. and Rienzi L. (2016). MicroRNAs in spent blastocyst culture medium are derived from trophectoderm cells and can be explored for human embryo reproductive competence assessment. Fertil Steril 105:225–235 [DOI] [PubMed] [Google Scholar]
- 3.Kropp J, Salih S. and Khatib H. (2014). Expression of microRNAs in bovine and human pre-implantation embryo culture media. Front Genet 5:91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rosenbluth EM, Shelton DN, Wells LM, Sparks AE. and Van Voorhis BJ. (2014). Human embryos secrete microRNAs into culture media-a potential biomarker for implantation. Fertil Steril 101:1493–1500 [DOI] [PubMed] [Google Scholar]
- 5.McCalli B, Schoolcraft WB. and Katz-Jaffe MG. (2010). Aberration of blastocyst microRNA expression is associated with human infertility. Fertil Steril 93:2374–2382 [DOI] [PubMed] [Google Scholar]
- 6.Thouas GA, Dominguez F, Green MP, Vilella F, Simon C. and Gardner DK. (2015). Soluble ligands and their receptors in human embryo development and implantation. Endocr Rev 36:92–130 [DOI] [PubMed] [Google Scholar]
- 7.Van de Velde H, Cauffman G, Tournaye H, Devroey P. and Liebaers I. (2008). The four blastomeres of a 4-cell stage human embryo are able to develop individually into blastocysts with inner cell mass and trophectoderm. Hum Reprod 23:1742–1747 [DOI] [PubMed] [Google Scholar]
- 8.Illmensee K, Levanduski M, Vidali A, Husami N. and Goudas VT. (2010). Human embryo twinning with applications in reproductive medicine. Fertil Steril 93:423–427 [DOI] [PubMed] [Google Scholar]
- 9.Noli L, Dajani Y, Capalbo A, Bvumbe J, Rienzi L, Ubaldi FM, Oglilve C, Khalaf Y. and Ilic D. (2015). Temporal control of blastulation compromises developmental competence of human twin blastocysts created by embryo splitting. Hum Reprod 30:2774–2784 [DOI] [PubMed] [Google Scholar]
- 10.Xiang M, Zeng Y, Yang R, Xu H, Chen Z, Zhong J, Xie H, Xu Y. and Zeng X. (2014). U6 is not a suitable endogenous control for the quantification of circulating microRNAs. Biochem Biophys Res Commun 454:210–214 [DOI] [PubMed] [Google Scholar]
- 11.Mestdagh P, Van Vlierberghe P, De Weer A, Muth D, Westermann F, Speleman F. and Vandesompele J. (2010). A novel and universal method for microRNA RT-qPCR data normalization. Genome Biol 10:R64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Noli L, Capalbo A, Ogilvie C, Khalaf Y. and Ilic D. (2015). Discordant growth of monozygotic twins starts at the blastocyst stage: a case study. Stem Cell Reports 5:946–953 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Goossens K, Mestdagh P, Lefever S, Van Poucke M, Van Zeveren A, Van Soom A, Vandesompele J. and Peelman L. (2013). Regulatory microRNA network identification in bovine blastocyst development. Stem Cells Dev 22:1907–1920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Maraghechi P, Hiripi L, Tóth G, Bontovics B, Bősze Z. and Gócza E. (2013). Discovery of pluripotency-associated microRNAs in rabbit preimplantation embryos and embryonic stem-like cells. Reproduction 145:421–437 [DOI] [PubMed] [Google Scholar]
- 15.Suh N, Baehner L, Moltzahn F, Melton C, Shenoy A, Chen J. and Blelloch R. (2010). MicroRNA function is globally suppressed in mouse oocytes and early embryos. Curr Biol 20:271–277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yang Y, Bai W, Zhang L, Yin G, Wang X, Wang J, Zhao H, Han Y. and Yao YQ. (2008). Determination of microRNAs in mouse preimplantation embryos by microarray. Dev Dyn 237:2315–2327 [DOI] [PubMed] [Google Scholar]
- 17.Mineno J, Okamoto S, Ando T, Sato M, Chono H, Izu H, Takayama M, Asada K, Mirochnitchenko O, Inouye M. and Kato I. (2006). The expression profile of miRNAs in mouse embryos. Nucleic Acids Res 34:1765–1771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Valadi H, Ekström K, Bossios A, Sjöstrand M, Lee JJ. and Lötvall JO. (2007). Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol 9:654–659 [DOI] [PubMed] [Google Scholar]
- 19.Bockhorn J, Dalton R, Nwachukwu C, Huang S, Prat A, Yee K, Chang YF, Huo D, Wen Y, et al. (2013). MicroRNA-30c inhibits human breast tumour chemotherapy resistance by regulating TWF1 and IL-11. Nat Commun 4:1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bruce AW. and Zernicka-Goetz M. (2010). Developmental control of the early mammalian embryo: competition among heterogeneous cells that biases cell fate. Curr Opin Genet Dev 20:485–491 [DOI] [PubMed] [Google Scholar]
- 21.Johnson MH. and Ziomek CA. (1981). The foundation of two distinct cell lineages within the mouse morula. Cell 24:71–80 [DOI] [PubMed] [Google Scholar]
- 22.Song PP, Hu Y, Liu CM, Yan MJ, Song G, Cui Y, Xia HF. and Ma X. (2011). Embryonic ectoderm development protein is regulated by microRNAs in human neural tube defects. Am J Obstet Gynecol 204:544.e9–e17 [DOI] [PubMed] [Google Scholar]
- 23.Rosa A, Papaioannou MD, Krzyspiak JE. and Brivanlou AH. (2014). miR-373 is regulated by TGFβ signaling and promotes mesendoderm differentiation in human embryonic stem cells. Dev Biol 391:1–88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Roy L, Bikorimana E, Lapid D, Choi H, Nguyen T. and Dahl R. (2015). miR-24 is required for hematopoietic differentiation of mouse embryonic stem cells. PLoS Genet 11:e1004959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dior UP, Kogan L, Chill HH, Eizenberg N, Simon A. and Revel A. (2014). Emerging roles of microRNA in the embryo-endometrium cross talk. Semin Reprod Med 32:402–409 [DOI] [PubMed] [Google Scholar]
- 26.Kropp J. and Khatib H. (2015). Characterization of microRNA in bovine in vitro culture media associated with embryo quality and development. J Dairy Sci 98:6552–6563 [DOI] [PubMed] [Google Scholar]
- 27.Galliano D. and Pellicer A. (2014). MicroRNA and implantation. Fertil Steril 101:1531–1544 [DOI] [PubMed] [Google Scholar]
- 28.Vilella F, Moreno-Moya JM, Balaguer N, Grasso A, Herrero M, Martínez S, Marcilla A. and Simón C. (2015). hsa-miR-30d, secreted by the human endometrium, is taken up by the pre-implantation embryo and might modify its transcriptome. Development 142:3210–3221 [DOI] [PubMed] [Google Scholar]
- 29.Feng R, Sang Q, Zhu Y, Fu W, Liu M, Xu Y, Shi H, Xu Y, Qu R, et al. (2015). miRNA-320 in the human follicular fluid is associated with embryo quality in vivo and affects mouse embryonic development in vitro. Sci Rep 5:8689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang J, Wang Y, Liu X, Jiang S, Zhao C, Shen R, Guo X, Ling X. and Liu C. (2015). Expression and potential role of microRNA-29b in mouse early embryo development. Cell Physiol Biochem 35:1178–1187 [DOI] [PubMed] [Google Scholar]
- 31.Cheong AW, Pang RT, Liu WM, Kottawatta KS, Lee KF. and Yeung WS. (2014). MicroRNA Let-7a and dicer are important in the activation and implantation of delayed implanting mouse embryos. Hum Reprod 29:750–762 [DOI] [PubMed] [Google Scholar]
- 32.Dai Y, Qiu Z, Diao Z, Shen L, Xue P, Sun H. and Hu Y. (2012). MicroRNA-155 inhibits proliferation and migration of human extravillous trophoblast derived HTR-8/SVneo cells via down-regulating cyclin D1. Placenta 33:824–829 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.