Abstract
Development of both dendrites and axons is important for the formation of neuronal circuits, because dendrites receive information and the axon is responsible for sending signals. In the past decade, extensive studies have revealed many molecules underlying axonal outgrowth and pathfinding. In contrast, much less is known about the molecular mechanisms that control dendrite development. Here we report the identification of an evolutionarily conserved Ig superfamily member, dendrite arborization and synapse maturation 1 (Dasm1), which plays a critical role in dendrite development. Dasm1 contains five Ig domains and two fibronectin III domains in the extracellular N terminus, a single transmembrane domain, and an intracellular C-terminal tail with a type I PDZ domain binding motif at the end. It is highly expressed in the brain and localized at the dendrites. Suppression of Dasm1 expression in hippocampal neurons via RNA interference or expression of Dasm1 without its cytoplasmic tail specifically impairs dendrite, but not axon, outgrowth. Together with its orthologues in other species, Dasm1 defines a family of molecules likely involved specifically in dendrite arborization.
Communication between neurons involves synapses formed between axons of presynaptic neurons and dendrites of postsynaptic neurons. During development, the nervous system progresses from a large number of disconnected neurons to a network of neuronal circuitries that convey and process information and generate functional outputs. To form these circuits, axons of presynaptic neurons need to grow and navigate through often long distances to the correct region to meet their targets, usually the dendrites of postsynaptic neurons. Equally important, the dendrites of postsynaptic neurons need to grow and elaborate into the right shape, because the dendritic morphology is essential for receiving and computing neuronal signals. Indeed, different types of neurons in the nervous system develop distinct forms of dendritic arbors to fulfill their particular physiological functions, as first observed by Ramón y Cajal more than a century ago. Defects in dendritic morphology often impair neuronal function and thus lead to various neurological diseases. Whereas extensive studies over the past decade have identified many molecules underlying axonal outgrowth and navigation (1), including some transmembrane receptors and their ligands, e.g., Roundabout (Robo)/Dutt and Slit, and DCC/Frazzled/UNC-40 and Netrin, molecular mechanisms that control dendrite outgrowth and elaboration are less well understood.
Both intracellular and extracellular signals play a role in ensuring proper dendrite arborization (2-6). For example, regulators of cytoskeleton dynamics, components of signal transduction pathways, and transcriptional factors have been shown to regulate dendritic morphology. Besides these intracellular signals, extracellular signals such as the neurotrophin family of growth factors and ephrins also influence dendritic morphology. In addition, neuronal activity is critical in shaping dendritic morphology (7, 8). Recently, many signals originally discovered for their involvement in controlling axonal outgrowth and navigation, such as Slit/Robo and Semaphorins, have been shown to affect dendrite outgrowth (9-12), raising the question whether dendrite development is orchestrated by the same sets of growth and guidance signals as axon development. Given that dendrites differ from axons in many important aspects both morphologically and functionally (13), it seems likely that there are mechanisms specific for dendrite development.
An essential step toward understanding how dendrites develop is to identify key molecules specifically involved in dendrite morphogenesis. In this study, we identified and characterized an evolutionarily conserved member of the Ig superfamily (IgSF), dendrite arborization and synapse maturation 1 (Dasm1), and showed that it plays an important role in dendrite outgrowth in mammalian neurons. Dasm1 is highly expressed in the brain, including hippocampus, cerebral cortex, and cerebellum, and is localized in dendrites. Perturbation of Dasm1 function in neurons at early stages impaired the outgrowth of dendrites but not axons. Thus, our studies suggest that Dasm1, together with its orthologues in human, Drosophila, and other species, represents a previously uncharacterized family of the IgSF molecules that specifically control dendrite arborization.
Materials and Methods
Molecular Biology. Human KIAA1355 carboxyl sequence (≈1.5 kb) was used as a probe to screen a mouse embryonic cDNA library (Stratagene). A full-length cDNA of Dasm1 was identified after three rounds of screening. For in situ hybridization, embryonic day 18 mouse embryo sections were probed with digoxigenin-labeled probes against Dasm1 C-terminal sequence. The cDNA encoding the last 100 aa of Dasm1 was subcloned into vector pGEX(4T-2), and the fusion protein GST-Dasm1(C100) was purified and used to immunize the rabbits (Animal Pharm Services, Healdsburg, CA). A synthetic peptide corresponding to the last 10 aa of Dasm1 was separately used to immunize the rabbits. The polyclonal rabbit antisera were used for Western and immunohistochemistry analyses. The cytoplasmic tail deletion mutant of Dasm1 was generated as follows. A HindIII site was introduced 8 nt upstream of the start codon ATG and a SalI site was introduced to the position at amino acid 795 (after the transmembrane domain). The HindIII/SalI fragment was cloned into pEGFP-N2 (Clontech). Cytoplasmic tails (C200, C100, and C60) of Dasm1 were amplified by PCR and cloned into pEGFPC3 (Clontech). Various enhanced GFP (EGFP)-tagged Dasm1 sequences were then subcloned into pSinrep5 (Invitrogen) to generate Sindbis viruses. Point mutation of Dasm1 was generated by using the QuikChange mutagenesis kit (Stratagene). Dasm1 RNA interference (RNAi) plasmid was constructed as follows. Two oligos that form a hairpin structure corresponding to Dasm1 nucleotides 69-87 (5′-GATCCC gcctgaggtggtctctgtg TTCAAGAGA cacagaagaccacctcaggc TTTTTTGGAAA-3′ and 5′-AGCTTTTCCAAAAAA gcctgaggtggtctctgtg TCTCTTGAA cacagagaccacctcaggc GG-3′) were annealed together, and this fragment was then inserted into pSilencer 2.0 (Ambion, Austin, TX). We screened three Dasm1 RNAi plasmids against different regions of Dasm1 for a robust and specific effect on suppressing Dasm1 protein expression in Cos-7 cells. The other two Dasm1 RNAi sequences were against nucleotides 868-886 (5′-GATCCC gccactcagcctgatgacg TTCAAGAGA cgtcatcaggctgagtggc TTTTTTGGAAA-3′ and 5′-AGCTTTTCCAAAAAA gccactcagcctgatgacg TCTCTTGAA cgtcatcaggctgagtggc GG-3′) and nucleotides 3341-3359 (5′-GATCCC cttgagccagagttaggtg TTCAAGAGA cacctaactctggctcaag TTTTTTGGAAA-3′ and 5′-AGCTTTTCCAAAAAA cttgagccagagttaggtg TCTCTTGAA cacctaactctggctcaag GG-3′). We mainly used the first Dasm1 RNAi sequence because it showed the highest reduction in Dasm1 protein expression in both Cos-7 cells and neurons. The control RNAi sequence was a randomly scrambled sequence not found in the mouse, human, or rat genome databases (pSilencer handbook). All of the constructs were confirmed by sequencing.
Neuronal Culture, Immunohistochemistry, and Morphological Assay. Preparation of the hippocampal neuronal cultures was according to published protocols with some modifications (14). Around 1-3 days [Dasm1 RNAi, 1-2 days; Dasm1(delC)-EGFP, 2-3 days] after plating, neurons were transfected with various DNA constructs by using Lipofectamine 2000 (Invitrogen). Around either 6-8 or 12-14 days later, neurons were fixed with 4% paraformaldehyde/4% sucrose in PBS at 4°C for 20 min and processed for immunohistochemistry and morphological assay.
Freshly fixed neurons were first washed with PBS and blocked with 10% horse serum in PBS containing 0.1% Triton X-100 to reduce nonspecific antibody binding. Neurons were then incubated with the primary antibody [monoclonal antibody against MAP2 (Chemicon) or monoclonal antibody against GluR2/4 (Chemicon)] or polyclonal antibody against Dasm1 at 4°C overnight. After washing with PBS, a Cy5- or Cy3-conjugated secondary antibody (The Jackson Laboratory) was applied at room temperature for 1-2 h. Neuronal morphology was analyzed with a Nikon inverted microscope (Eclipse E800) equipped with epifluorescence and differential interference contrast optics. Images of neurons were taken with a Spot charge-coupled device camera real-time (RT) slider (Diagnostic Instruments, Sterling Heights, MI) and analyzed with imagej and photoshop (Adobe Systems, San Jose, CA). Each construct was tested at least three times by scoring neurons on multiple coverslips, and in each case all of the healthy transfected neurons with smooth processes in each transfection were included in analysis. Data are given as mean ± SE, and the statistical significance was assessed by using Student's t test.
Results
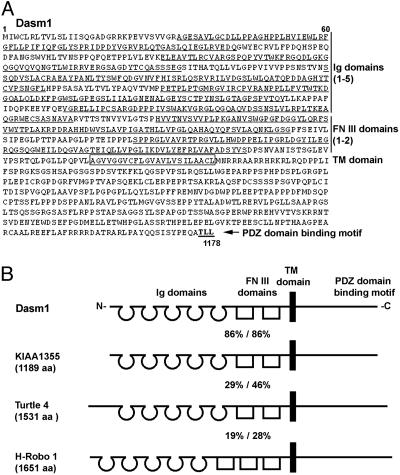
Cloning and Molecular Domain Structure of Dasm1. Genetic studies on dendrite development of multiple dendrite (md) neurons in the Drosophila peripheral nervous system (PNS) indicated that Turtle, an IgSF molecule (15), was involved in md neuron dendrite arborization (D.N.C., L.Y.J., and Y.-N.J., unpublished data). To test whether similar molecules also play a role in mammalian dendrite development, we searched protein databases for vertebrate proteins with sequence and domain structure similar to Drosophila Turtle and found human KIAA1355, which shows the greatest similarity to Drosophila Turtle (identity/similarity = 29%:46%) and is highly expressed in fetal brain tissue (16). We then screened a mouse brain cDNA library and isolated a full-length cDNA that encodes a 1,178-aa protein (86% identity to KIAA1355; Fig. 1). The same sequence has also been recently reported as a gene in the vicinity of the mouse loop tail gene (17). Based on its function on dendrite arborization (see below) and synapse maturation (see ref. 18), we named it dendrite arborization and synapse maturation 1 (Dasm1). The Dasm1 protein is a member of the IgSF and contains an extracellular N terminus with five Ig domains followed by two fibronectin type III domains, a single transmembrane segment, and a large C-terminal cytoplasmic tail with a type I PDZ domain binding motif (-TLL) at the end (Fig. 1 A), identical in domain structure to human KIAA1355 (16) (Fig. 1B). Proteins closely related to Dasm1 are also present in fugu, zebrafish, and chicken, as indicted in the database. Proteins more distantly related to Dasm1 include human Roundabout 1 (H-Robo 1, also known as DUTT1) (identity/similarity = 19%:28%), which contains five Ig and three fibronectin domains (1) and thus differs in domain structure from Dasm1. These results suggest that Dasm1, Turtle, and KIAA1355 represent a previously uncharacterized subfamily of the IgSF evolutionarily conserved from fruit flies to mammals.
Fig. 1.
Amino acid sequence and molecular domain structure of Dasm1. (A) Dasm1 encodes a 1,178-aa protein with an extracellular domain containing five Ig repeats and two fibronectin III repeats, a single transmembrane (TM) domain, and a large cytoplasmic domain with a type I PDZ domain binding motif at the end. Single-letter abbreviations for amino acid residues are as follows: A, Ala; C, Cys; D, Asp; E, Glu; F, Phe; G, Gly; H, His; I, Ile; K, Lys; L, Leu; M, Met; N, Asn; P, Pro; Q, Gln; R, Arg; S, Ser; T, Thr; V, Val; W, Trp; Y, Tyr. (B) An illustration of molecular domain structure of Dasm1 and its similarity to human KIAA1355, Drosophila Turtle, and human Roundabout 1 (H-Robo 1). The positions of the Ig, fibronectin III, and TM domains and the percentages of amino acid identity/similarity between Dasm1 and KIAA1355, Turtle, and H-Robo 1 are shown.
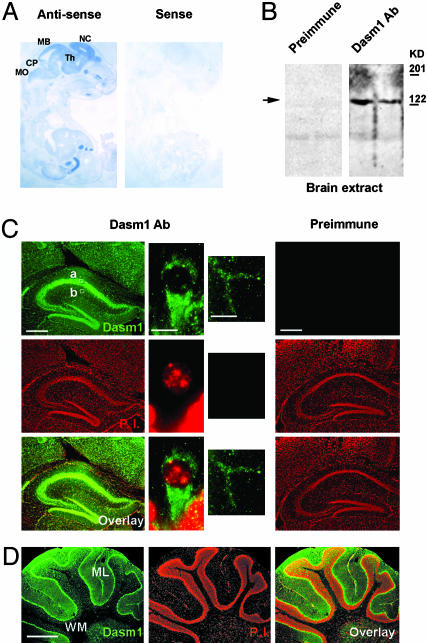
Expression of Dasm1 in the Mouse Brain. Northern analysis with a mixture of probes against both the N and C termini of Dasm1 revealed three mRNA transcripts of ≈4.2, 2.7, and 1.8 kb, with the 4.2-kb transcript specifically expressed in the brain (data not shown). These transcripts might be different splice variants of Dasm1, because there are 22 exons spanning the entire Dasm1 coding sequence but no other obvious homologues in the current mouse genome database. In situ hybridization with a digoxigenin-labeled probe against the Dasm1 C terminus revealed that Dasm1 was highly expressed in the brain of embryonic day 18 mouse embryo, especially in the forebrain (Fig. 2A). A polyclonal antibody generated against the C terminus of Dasm1 recognized a protein of ≈130 kDa, the predicted size of Dasm1, in Western analysis of rat-brain membrane extracts (Fig. 2B). Immunohistochemistry analysis in adult mouse brain sections revealed punctate staining of Dasm1 in both cell bodies and dendrites of cortical and hippocampal neurons (Fig. 2C). In addition, immunohistochemistry analysis showed that Dasm1 was also highly expressed in the cell bodies and dendrites (molecular layer), but not axons (white matter), of adult cerebellar Purkinje cells (Fig. 2D).
Fig. 2.
Expression of Dasm1 in the mouse brain. (A) In situ hybridization analysis of Dasm1 expression. Embryonic day 18 mouse embryo sections were probed with digoxigenin-labeled antisense (Left) and sense (Right) sequences of Dasm1. Note the high expression of Dasm1 in the brain, especially in the forebrain (neopallial cortex, NC), intermediate zone, and ventricular zone. CP, cerebellar primordium; MB, midbrain; MO, medulla oblongata; Th, thalamus. (B) Western blot analysis of Dasm1 expression in the brain. Crude membrane protein extracts from adult rat brains were immunoblotted with the polyclonal antiserum against the C terminus of Dasm1 (Right) and the preimmune serum (Left). The anti-Dasm1 serum strongly recognizes a band ≈130 kDa but not the preimmune serum (arrow). Molecular mass markers are listed on the left. (C) Adult mouse brain sections (≈7 μm) were stained with the polyclonal antiserum against Dasm1 (green, Top) and, propidium iodide (PI), a dye, labels the DNA (red, Middle). The overlay is shown in Bottom. Antibody against Dasm1 (green) showed punctate staining in both the cell bodies and dendrites (Left) of hippocampal and cortical neurons, whereas the preimmune serum gave no detectable staining (Right). High-magnification images of the staining in cell body (a) and apical dendrites (b) of hippocampal CA1 pyramidal neurons (squares in Left) are shown. [Scale bars (from left to right) = 250, 20, 20, and 250 μm.] (D) Immunohistochemistry analysis of Dasm1 in the cerebellum. ML, molecular layer; WM, white matter. (Scale bar = 500 μm.)
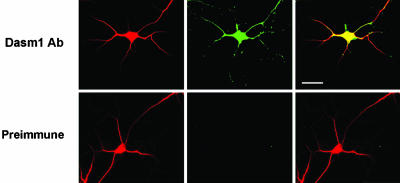
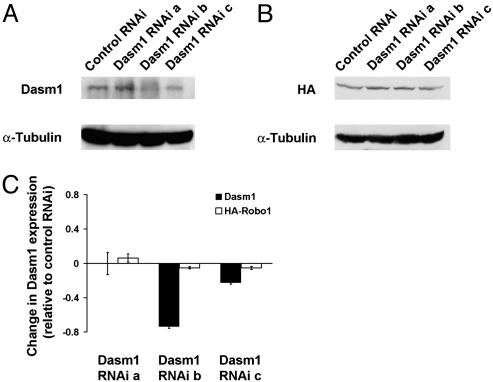
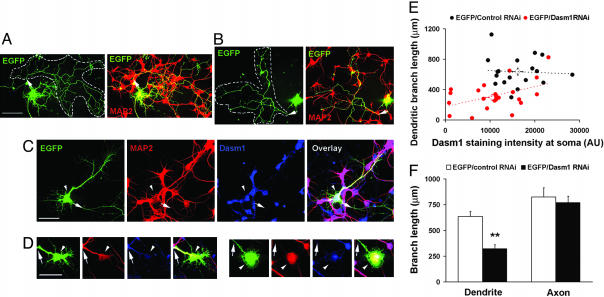
Dasm1 RNAi Impairs Dendrite Outgrowth. As a first step toward establishing the function of Dasm1, we used RNAi to knock down the expression of the Dasm1 protein (19) in rat dissociated hippocampal neuron culture, a well characterized system for studying mammalian dendrite morphogenesis (14, 20). Double labeling of hippocampal neurons cultured for 4-5 days in vitro (DIV) with antibodies against Dasm1 and the dendritic marker MAP2 showed that Dasm1 was expressed in the developing dendrites (Fig. 3), which undergo dynamic outgrowth and elaboration (20). Having screened three Dasm1 RNAi sequences against different regions of Dasm1 for their efficacy and specificity in suppressing Dasm1 expression in Cos-7 cells (Fig. 4), we chose the one that specifically reduced Dasm1 expression by ≈75%. To test whether Dasm1 plays a critical role in dendrite outgrowth, we cotransfected neurons with EGFP and the Dasm1 RNAi plasmid. Neurons were transfected ≈1-2 DIV, and their morphology was assessed ≈7-8 days later. To reveal the effect of RNAi on Dasm1 expression and dendrite morphology, neurons were double-labeled with antibodies against Dasm1 and the dendritic marker MAP2 (Fig. 5 A-D). Compared with control neurons transfected with control RNAi plasmid against a randomly scrambled sequence with no sequence correspondence in the database, neurons transfected with Dasm1 RNAi plasmid averaged an ≈36% reduction in Dasm1 expression (P < 0.05; Fig. 5 C-E). Furthermore, the dendritic arbor was drastically reduced in neurons transfected with Dasm1 RNAi plasmid (control RNAi, 636 ± 46 μm; Dasm1 RNAi, 325 ± 36 μm; P < 0.001; Fig. 5 C, D, and F). In fact, there was a correlation between the level of Dasm1 expression and the complexity of dendrite arbor (control RNAi, correlation index R2 = 0.004; Dasm1 RNAi, correlation index R2 = 0.5; Fig. 5E). Interestingly, there was no detectable effect of Dasm1 RNAi on the MAP2 negative axonal arbor revealed by EGFP fluorescence (control RNAi, 825 ± 89 μm; Dasm1 RNAi, 773 ± 60 μm; P = 0.6; Fig. 5 A, B, and F). Moreover, transfection of neurons with Dasm1 RNAi did not lead to increased apoptosis (data not shown). These results strongly suggest that Dasm1 is expressed in developing dendrites and specifically controls dendrite, but not axon, outgrowth.
Fig. 3.
Expression of Dasm1 in developing dendrites. Expression of Dasm1 in the dendrites of developing neurons is shown. Hippocampal dissociated neurons cultured for 4-5 days in vitro (DIV) were double-labeled with antibodies against Dasm1 (green) and MAP2 (red), a protein marker for the dendrites. Note that Dasm1 was expressed in the dendrites of these neurons, which undergo dynamic outgrowth at this stage. (Scale bar = 25 μm.)
Fig. 4.
Efficacy and specificity of Dasm1 RNAi. (A and B) Western analysis of Dasm1 RNAi plasmids on Dasm1 and Robo1 expression in Cos-7 cells. Compared with the control RNAi against a randomly scrambled sequence, Dasm1 RNAi b and c showed a suppression in Dasm1 expression, whereas Dasm1 RNAi a did not show an obvious effect on Dasm1 expression (A). In contrast, none of Dasm1 RNAi plasmids showed obvious effect on hemagglutinin-Robo1 expression compared with control RNAi (B). An equal amount of protein (≈25 μg) was loaded for each lane, and the same sample was blotted against α-tubulin antibody. (C) Quantification of Dasm1 RNAi plasmids on Dasm1 and Robo1 protein expression in Cos-7 cells.
Fig. 5.
Suppression of Dasm1 expression impairs dendrite outgrowth. (A and B) Dasm1 RNAi had no obvious effect on axonal arborization. Neurons transfected with either Dasm1 RNAi plasmid (B) or control RNAi plasmid (A) together with EGFP were stained with the dendritic marker MAP2 antibody. Extensive EGFP fluorescence-labeled axonal arbors lacking MAP2 staining (indicated with arrows and inside dotted lines) were seen in both groups. (Scale bar = 50 μm.) (C and D) Neurons transfected with Dasm1 RNAi/EGFP (D) showed defects in dendrite development, compared with those transfected with control RNAi/EGFP (C). Neurons were double-labeled with antibodies against MAP2 to visualize dendritic arbor (red) and Dasm1 to monitor the expression level of endogenous Dasm1 (blue) after RNAi treatments. The EGFP channel is shown in Left and the overlay is shown in Right. Arrows indicate the axon and arrowheads indicate the positions of cell body of transfected neurons. (Scale bar = 50 μm.) (E) Correlation between the expression level of Dasm1 and the complexity of dendritic arbor. The dendritic branch lengths of neurons transfected with EGFP/Dasm1 RNAi (red circle) or EGFP/control RNAi (black circle) were plotted against the Dasm1 staining intensity at the cell bodies. Note that there was a significant reduction in average Dasm1 staining intensity at the cell bodies with Dasm1 RNAi when compared with the control RNAi (P < 0.05; open circles). AU, arbitrary unit. (F) The average total dendritic, but not axonal, branch length was significantly reduced in neurons transfected with Dasm1 RNAi compared with that of the control neurons (**, P < 0.001).
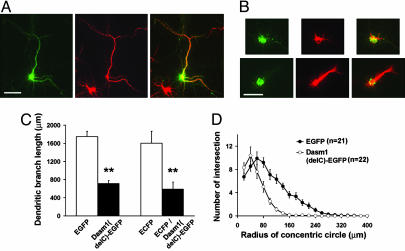
Dasm1(delC) Acts as a Dominant Negative and Impairs Dendrite Outgrowth. To further test the role of Dasm1 in dendrite outgrowth, we transfected neurons with a truncated form of Dasm1, Dasm1(delC)-EGFP, in which nearly the entire cytoplasmic tail (from amino acids 796 to 1178) was replaced with EGFP. We examined the effect of expressing this presumed dominant negative mutant protein on dendrite development. Neurons were transfected ≈2-3 DIV, and their morphology was assessed ≈12-14 days later. As a control, neurons transfected with EGFP alone showed normal development of the dendritic arbor, as visualized with MAP2 antibody staining (Fig. 6A) or enhanced cyan fluorescent protein (ECFP) fluorescence (data not shown). In contrast, the outgrowth of the dendritic arbor of neurons transfected with Dasm1(delC)-EGFP was severely impaired (Fig. 6B). The dendrites in those neurons were not as fully extended and elaborate as those in either EGFP transfected (Fig. 6 A and B) or untransfected (data not shown) control neurons. This effect was quantified as a decrease in total dendritic branch length [EGFP, 1,748 ± 118 μm, n = 21; Dasm1(delC)-EGFP, 719 ± 60 μm, n = 22; P < 0.001; ECFP, 1,603 ± 261 μm, n = 11; ECFP/Dasm1(delC)-EGFP, 595 ± 150 μm, n = 9; Fig. 6C] and by Sholl analysis, which measures the number of dendrites crossing a sphere centered on the cell body as a function of the radius [EGFP, n = 21; Dasm1(delC)-GFP, n = 22; Fig. 6D]. Similar to Dasm1 RNAi, the effect of Dasm1(delC)-EGFP was specific for dendrites without affecting the MAP2 negative axonal arbor (data not shown). These results suggest that Dasm1(delC)-EGFP acts as a dominant negative and further support that Dasm1 specifically controls dendrite outgrowth and elaboration.
Fig. 6.
Dasm1 lacking the intracellular tail acts as a dominant negative and impairs dendrite outgrowth. (A and B) Neurons expressing Dasm1(delC)-EGFP (B) showed defects in dendrite development when compared with control neurons expressing EGFP alone (A). The dendritic arbor was visualized with MAP2 antibody staining (Center). The EGFP channel is shown in Left, and the overlay is shown in Right. (Scale bar = 50 μm.) (C) The average total dendritic branch length was decreased in neurons expressing Dasm1(delC)-EGFP or enhanced cyan fluorescent protein (ECFP)/Dasm1(delC)-EGFP compared with control neurons expressing EGFP or ECFP alone (**, P < 0.001). (D) Sholl analysis showed that the dendritic arbor was severely reduced in neurons expressing Dasm1(delC)-EGFP (n = 22) compared with control neurons expressing EGFP alone (n = 21). Concentric circles with a 20-μm spacing were drawn around the cell body, and the number of intersections of all dendritic branches with the circles was counted.
Discussion
As the primary site for receiving and integrating synaptic inputs, dendrites play critical roles in neuronal function and circuit properties in the brain. Whereas dendrite arborization is known to depend on extrinsic and intrinsic factors (2-6) as well as neuronal activity (7, 8), little is known about molecular mechanisms specific for dendrite development. In this study we have identified a member of the IgSF, Dasm1, which controls dendrite arborization. The molecular domain structure of Dasm1 suggests that it may be a membrane receptor, similar to the cell-surface receptors Robo/Dutt1 and DCC/Frazzled/UNC-40 (1). Consistent with this notion, expression of the truncation mutant of Dasm1 lacking the cytoplasmic domain resulted in defective dendrite outgrowth similar to the RNAi phenotype. The effect of Dasm1(delC) is likely due to the competition with endogenous Dasm1 for its as yet unknown ligand and thus blocking the signal transducing into the neurons. Such perturbations of Dasm1 function have no obvious effect on axon outgrowth. Similarly, no obvious defect in axon development was observed in Drosophila Turtle mutants (15). This functional specificity in dendrite, but not axon, development, along with its sequence and domain structure divergence from other IgSF members, suggests that Dasm1, Drosophila Turtle, and human KIAA1355 represent a previously uncharacterized subfamily of the IgSF that controls dendrite arborization specifically.
Actin and microtubule are the essential structural components of dendrites, and the growth of dendrites entails the dynamic regulation of actin and microtubule. In fact, nearly all of the signals that regulate dendrite morphology in the end act through affecting actin and/or microtubule dynamics. Interestingly, there are several stretches of proline-rich sequences in the cytoplasmic tail of Dasm1. Many PRM (proline-rich motif)-containing proteins and their interacting partners have been implicated in delivering actin monomers to specific cellular localizations where actin-rich membrane protrusions form (21). Thus, they play important roles in axon outgrowth and navigation (22, 23). It should be interesting to explore the possibility that Dasm1 interacts with protein(s) involved in actin cytoskeleton dynamics.
Acknowledgments
We thank Jan laboratory members for insightful discussions, Dr. M. Schonemann for help with the cDNA library screen, and Drs. R. A. Nicoll, Y. Hayashi, Z. Z. Ma, and W. Grueber for helpful comments on an earlier version of this manuscript. This work was supported by a Helen Hay Whitney Fellowship (to S.-H.S.), the Jane Coffin Childs Memorial Fund for Medical Research (D.N.C.), and National Institutes of Health grants (to Y.-N.J. and L.Y.J.). S.-H.S. was a research associate and Y.-N.J. and L.Y.J. are investigators of the Howard Hughes Medical Institute.
Abbreviations: Dasm1, dendrite arborization and synapse maturation 1; IgSF, Ig super-family; EGFP, enhanced GFP; RNAi, RNA interference.
References
- 1.Grunwald, I. C. & Klein, R. (2002) Curr. Opin. Neurobiol. 12, 250-259. [DOI] [PubMed] [Google Scholar]
- 2.Whitford, K. L., Dijkhuizen, P., Polleux, F. & Ghosh, A. (2002) Annu. Rev. Neurosci. 25, 127-149. [DOI] [PubMed] [Google Scholar]
- 3.Van Aelst, L. & Cline, H. T. (2004) Curr. Opin. Neurobiol. 14, 297-304. [DOI] [PubMed] [Google Scholar]
- 4.Scott, E. K. & Luo, L. (2001) Nat. Neurosci. 4, 359-365. [DOI] [PubMed] [Google Scholar]
- 5.McAllister, A. K. (2000) Cereb. Cortex 10, 963-973. [DOI] [PubMed] [Google Scholar]
- 6.Jan, Y.-N. & Jan, L. Y. (2003) Neuron 40, 229-242. [DOI] [PubMed] [Google Scholar]
- 7.Cline, H. T. (2001) Curr. Opin. Neurobiol. 11, 118-126. [DOI] [PubMed] [Google Scholar]
- 8.Wong, R. O. & Ghosh, A. (2002) Nat. Rev. Neurosci. 3, 803-812. [DOI] [PubMed] [Google Scholar]
- 9.Whitford, K. L., Marillat, V., Stein, E., Goodman, C. S., Tessier-Lavigne, M., Chedotal, A. & Ghosh, A. (2002) Neuron 33, 47-61. [DOI] [PubMed] [Google Scholar]
- 10.Furrer, M. P., Kim, S., Wolf, B. & Chiba, A. (2003) Nat. Neurosci. 6, 223-230. [DOI] [PubMed] [Google Scholar]
- 11.Godenschwege, T. A., Simpson, J. H., Shan, X., Bashaw, G. J., Goodman, C. S. & Murphey, R. K. (2002) J. Neurosci. 22, 3117-3129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Polleux, F., Morrow, T. & Ghosh, A. (2000) Nature 404, 567-573. [DOI] [PubMed] [Google Scholar]
- 13.Craig, A. M. & Banker, G. (1994) Annu. Rev. Neurosci. 17, 267-310. [DOI] [PubMed] [Google Scholar]
- 14.Brewer, G. J., Torricelli, J. R., Evege, E. K. & Price, P. J. (1993) J. Neurosci. Res. 35, 567-576. [DOI] [PubMed] [Google Scholar]
- 15.Bodily, K. D., Morrison, C. M., Renden, R. B. & Broadie, K. (2001) J. Neurosci. 21, 3113-3125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nagase, T., Ishikawa, K., Kikuno, R., Hirosawa, M., Nomura, N. & Ohara, O. (1999) DNA Res. 6, 337-345. [DOI] [PubMed] [Google Scholar]
- 17.Doudney, K., Murdoch, J. N., Braybrook, C., Paternotte, C., Bentley, L., Copp, A. J. & Stanier, P. (2002) Genomics 79, 663-670. [DOI] [PubMed] [Google Scholar]
- 18.Shi, S.-H., Cheng, T., Jan, L. Y. & Jan, Y.-N. (2004) Proc. Natl. Acad. Sci. USA 101, 13346-13351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sui, G., Soohoo, C., Affar el, B., Gay, F., Shi, Y. & Forrester, W. C. (2002) Proc. Natl. Acad. Sci. USA 99, 5515-5520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Banker, G. & Goslin, K. (1988) Nature 336, 185-186. [DOI] [PubMed] [Google Scholar]
- 21.Holt, M. R. & Koffer, A. (2001) Trends Cell Biol. 11, 38-46. [DOI] [PubMed] [Google Scholar]
- 22.Wills, Z., Bateman, J., Korey, C. A., Comer, A. & Van Vactor, D. (1999) Neuron 22, 301-312. [DOI] [PubMed] [Google Scholar]
- 23.Wills, Z., Marr, L., Zinn, K., Goodman, C. S. & Van Vactor, D. (1999) Neuron 22, 291-299. [DOI] [PubMed] [Google Scholar]