Abstract
In the developing mammalian brain, a large fraction of excitatory synapses initially contain only N-methyl-d-aspartate receptor and thus are “silent” at the resting membrane potential. As development progresses, synapses acquire α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid receptors (AMPA-Rs). Although this maturation of excitatory synapses has been well characterized, the molecular basis for this developmental change is not known. Here, we report that dendrite arborization and synapse maturation 1 (Dasm1), an Ig superfamily member, controls excitatory synapse maturation. Dasm1 is localized at the excitatory synapses. Suppression of Dasm1 expression by using RNA interference or expression of dominant negative deletion mutants of Dasm1 in hippocampal neurons at late developmental stage specifically impairs AMPA-R-mediated, but not N-methyl-d-aspartate receptor-mediated, synaptic transmission. The ability of Dasm1 to regulate synaptic AMPA-Rs requires its intracellular C-terminal PDZ domain-binding motif, which interacts with two synaptic PDZ domain-containing proteins involved in spine/synapse maturation, Shank and S-SCAM. Moreover, expression of dominant negative deletion mutants of Dasm1 leads to more immature silent synapses. These results suggest that Dasm1, as a transmembrane molecule, likely provides a link to bridge extracellular signals and intracellular signaling complexes in controlling excitatory synapse maturation.
Fast excitatory synaptic potentials in mammalian CNS are largely mediated by binding of presynaptic released glutamate onto postsynaptic ionotropic glutamate receptors (1), of which α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid (AMPA) type receptor (AMPA-R) and N-methyl-d-aspartate (NMDA) type receptor (NMDA-R) are the two major types of receptors that reside in the postsynaptic density (PSD) of excitatory synapses and mediate synaptic transmission. These receptors have different electrophysiological and pharmacological properties and play distinct roles in synaptic function. Although AMPA-Rs mediate rapid synaptic transmission, activation of NMDA-Rs triggers various forms of synaptic plasticity. An important aspect of excitatory synapse development concerns proper expression of these two types of receptors at postsynaptic sites. Recently, both electrophysiological and anatomical studies indicated that early in postnatal development, a significant fraction of excitatory synapses possesses only NMDA-Rs, and no AMPA-Rs (2-8). These immature synapses are “silent” at the resting membrane potentials, because they do not transmit signals because of the voltage-dependent Mg2+ blockage of NMDA-Rs. As development proceeds, these immature synapses acquire functional AMPA-Rs with little change in NMDA-R numbers, and, thus, the percentage of immature silent synapses decreases. Since its discovery in the hippocampus, silent synapses have been observed in many regions of the mammalian CNS, including the cortex (9, 10), spinal cord (11), and cerebellum (12). Indeed, conversion of silent synapses into functional ones is a common feature of postnatal development of brain circuits.
Synapse formation and maturation is the last and essential step of dendrite development for neurons in the mammalian CNS. The vast majority of excitatory synapses (>90%) are formed at dendritic spines (13), small protrusions along dendritic shafts that contain neurotransmitter receptors and other proteins of the PSD necessary for synaptic transmission (14). The intimate relationship between synapses and dendrites is manifested by the critical role of synaptic activity in shaping dendritic arbor (15, 16). We have shown that dendrite arborization and synapse maturation 1 (Dasm1) plays a critical role in dendritic arborization [see the companion article by Shi et al. (17) in this issue of PNAS]. Whether dendrite outgrowth and synapse development rely on common molecular mechanisms is an intriguing open question. In this study, we examined the role of Dasm1 in controlling excitatory synapse maturation in the hippocampus.
Materials and Methods
Rat Brain Subcellular Fractionation, Yeast Two-Hybrid Screen, and Coimmunoprecipitation Assay. Rat brain subcellular fractionation was performed according to the protocol in ref. 18. Yeast two-hybrid screen was performed as described in ref. 19. The bait plasmid expressing the last 60 aa of Dasm1 fused with Gal4 DNA-binding domain in pPC97 vector was used to screen a rat brain cDNA library fused to Gal4 transcriptional activation domain in pPC86 vector. For coimmunoprecipitation assay, mammalian COS-7 cells were transfected with either no DNA, enhanced GFP (EGFP)-tagged Dasm1 cytoplasmic tails, Myc-tagged Shank SH3/PDZ domains or S-SCAM PDZ domains subcloned into pRK5Myc vector, or both EGFP-tagged Dasm1 cytoplasmic tails and Myc-tagged Shank SH3/PDZ domains or S-SCAM PDZ domains. Two days later, the cells were collected and lysed. The solution was cleared by centrifugation at 10,000 × g for 5 min at 4°C. To the supernatant, protein G-Sepharose gel was added to preadsorb nonspecific gel binding, and the solution was again centrifuged at 5,000 × g for 1 min at 4°C. After exposure to antibody against EGFP (monoclonal anti-EGFP, 12 μg per sample; Boehringer Mannheim) at 4°C for 2 h, the immunocomplex was adsorbed onto protein G-Sepharose gel (50 μl) at 4°C for 2 h. Finally, the gel was washed twice with lysis buffer (20 mM Tris·HCl, pH 8.0/1 mM EDTA/150 mM NaCl/1.0% Nonidet P-40/1× protease inhibitor cocktail) and once with washing buffer (50 mM Tris·HCl/0.25 M NaCl/0.1% Nonidet P-40/0.05% deoxylate), subjected to SDS/PAGE, and immunoblotted with polyclonal anti-Myc (Cell Signaling Technology, Beverly, MA) and monoclonal anti-EGFP antibodies. In general, 10-20% of the immunoprecipitation sample was loaded as the input.
Hippocampal Organotypic Slice Culture and Electrophysiology. Hippocampal organotypic culture slices were prepared as described in ref. 20. Neurons in slices cultured in vitro for 3-6 days were either transfected with a gene gun (Bio-Rad) or infected with Sindbis virus carrying genes of interest, and recordings were performed ≈1-3 days later. The recording chamber was perfused with physiological solution (22-25°C), containing 119 mM NaCl, 2.5 mM KCl, 4 mM CaCl2, 4 mM MgCl2, 26 mM NaHCO3, 1 mM NaH2PO4, 11 mM glucose, 0.1 mM picrotoxin, 0.01 mM bicuculline, and 0.004 mM 2-chloroadenosine (pH 7.4) and gassed with 5% CO2/95% O2. The 2-chloroadenosine was included to prevent bursting. For recording miniature synaptic responses, 1 μM tetrodotoxin was included. Patch recording pipettes (3-6 MΩ) were filled with internal solutions containing 115 mM cesium methanesulfonate, 20 mM CsCl, 10 mM Hepes, 2.5 mM MgCl2, 4 mM Na2ATP, 0.4 mM Na3GTP, 10 mM sodium phosphocreatine, and 0.6 mM EGTA (pH 7.25). Simultaneous whole-cell recordings were obtained from pairs of transfected/infected and nearby untransfected/uninfected CA1 pyramidal neurons with Axopatch-1D amplifiers (Axon Instruments, Union City, CA) under visual guidance with fluorescence and transmitted light illumination. Synaptic transmission was evoked by using two bipolar electrodes with a single voltage pulse (≈400 μs, up to 10 V). Signals were filtered at 1 or 2 kHz and sampled by using programs written in igor (WaveMetrics, Lake Oswego, OR). Data were analyzed by using macros written in igor and excel (Microsoft). The amplitude of AMPA-R-mediated excitatory postsynaptic currents (EPSCs) recorded at -60 mV was measured around the peak of the response. The amplitude of NMDA-R-mediated EPSCs recorded at +40 mV was measured with a 3- to 5-msec window at ≈150 or 200 msec after synaptic stimulation. Failure rate of synaptic transmission was calculated from ≈100 trials at each potential (+55 mV and -60 mV), by using a method similar to that described in ref. 2. The fraction of silent synapses was calculated by the equation 1 - (ln Fd)/(ln Fh), where Fd and Fh are the failure rates at +55 and -60 mV, respectively. All results were reported as mean ± SEM, and statistical differences of the means were determined by using Wilcoxon and paired t test for paired measurements and Mann-Whitney and nonpaired t test for nonpaired measurements unless otherwise stated in the text. In all experiments where multiple tests were used, the level of significance matched all tests.
Results
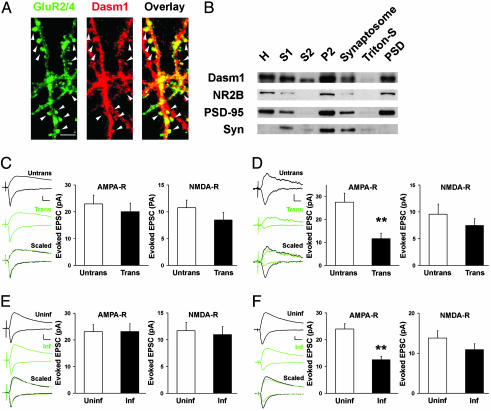
Dasm1 Is Localized at Synapses. The persistent expression of Dasm1 in the adult brain raises the question of whether it serves any function later in dendrite development. To explore this possibility, we first examined Dasm1 subcellular localization in mature neurons, which receive many synaptic inputs. Double labeling of hippocampal neurons cultured for ≈4 weeks with antibodies against Dasm1 and glutamate receptor 2/4 (GluR2/4) revealed a partial overlap between Dasm1 and GluR2/4 in a punctate pattern along the dendrites, especially at spine-like structures (Fig. 1A, arrowheads), suggesting that Dasm1 is localized at the excitatory synapses in mature neurons. Furthermore, subcellular fractionation of adult rat brains showed that Dasm1 was abundant in the PSD, in which postsynaptic proteins such as PSD-95 and NR2B were enriched (Fig. 1B).
Fig. 1.
Dasm1 is localized at the synapse and has a synaptic function. (A) Mature hippocampal neurons (cultured for ≈30 days in vitro) were double-labeled with antibodies against Dasm1 (red, Center) and glutamate receptor GluR2/4 (green, Left). As shown in the overlay (Right), Dasm1 partially colocalized with GluR2/4 along the dendrites, especially at spine-like structures (arrowheads). (Scale bar = 10 μm.) (B) Western blot analysis of the localization of Dasm1 in subcellular fractions of rat brain. Ten micrograms of protein from each fraction except the homogenate (H, ≈30 μg) were separated by electrophoresis on an SDS/polyacrylamide gel and immunoblotted with NR2B, PSD-95, and synaptophysin (Syn) antibodies. S, supernatant; P, pellet; Triton-S, supernatant after extraction with Triton X-100; PSD, postsynaptic density, pellet after extraction with Triton. (C) Neurons transfected with the control RNAi and EGFP showed no change in either AMPA-R-mediated (Center) or NMDA-R-mediated (Right) excitatory synaptic transmission. Nearby transfected neurons (Trans) expressing control RNAi/EGFP and untransfected controls (Untrans) were recorded simultaneously in pairs at -60 and +40 mV to measure AMPA-R- and NMDA-R-mediated evoked EPSCs, respectively. The sample traces of one such pair (black, untrans; green, trans) are shown in Left. Scaled, synaptic transmissions from transfected cell are scaled so that the NMDA-R-mediated synaptic transmission at +40 mV matches that of the untransfected cell. The same symbols and trace display conventions are used in subsequent figures. (Scale bars = 10 pA and 20 msec.) (D) Neurons transfected with the Dasm1 RNAi and EGFP showed an ≈50% decrease in AMPA-R-mediated synaptic transmission (**, P < 0.0001) (Center) with no significant change in NMDA-R-mediated synaptic transmission (Right). (Scale bars = 10 pA and 20 msec.) (E) Expression of EGFP alone had no effect on either AMPA-R-mediated (Center) or NMDA-R-mediated (Right) synaptic transmission. (Scale bars = 10 pA and 20 msec.) (F) Expression of Dasm1(delC)-EGFP at postsynaptic sites resulted in an ≈50% depression in AMPA-R-mediated (**, P < 0.0001) (Center), but no change in NMDA-R-mediated (Right), synaptic transmission. (Scale bars = 10 pA and 20 msec.)
Dasm1 Has a Synaptic Function. To determine whether Dasm1 plays a functional role at the synapse, we transfected Dasm1 RNA interference (RNAi) plasmid together with EGFP into CA1 pyramidal neurons in organotypic hippocampal slice culture by using a gene gun. The hippocampal slices were prepared from 6- to 8-day-old rats and cultured in vitro for 3-6 days before the transfection. Neurons at this stage in the preparation have rather mature dendritic arbors without rapid outgrowth of dendritic branches; instead, spine-like structures and synapses on the dendrites undergo dynamic developmental changes (21). To examine the effect of Dasm1 RNAi on synaptic transmission, we used a well established method (20, 22). We recorded evoked EPSCs simultaneously from transfected neurons expressing the RNAi constructs, and nearby untransfected neurons served as control. Although neurons transfected with control RNAi plasmid showed no change in either AMPA-R- or NMDA-R-mediated excitatory synaptic transmission [AMPA-R, untransfected neighbor (untrans), 23.0 ± 3.3 pA; transfected (trans), 20.0 ± 3.2 pA; n = 28; P = 0.4; NMDA-R, untrans, 10.8 ± 1.4 pA; trans, 8.5 ± 1.4 pA; n = 22; P = 0.1] (Fig. 1C), neurons transfected with Dasm1 RNAi plasmid showed an ≈50% decrease in AMPA-R-mediated, but no significant change in NMDA-R-mediated, synaptic transmission (AMPA-R, untrans, 27.5 ± 4.0 pA; trans, 11.7 ± 2.4 pA; n = 22; P < 0.0001; NMDA-R, untrans, 9.6 ± 1.9 pA; trans, 7.5 ± 1.2 pA; n = 18; P = 0.1) (Fig. 1D). As a result, the ratio of AMPA-R- to NMDA-R-mediated synaptic transmission was significantly smaller in neurons transfected with Dasm1 RNAi than in either nearby untransfected control or those transfected with control RNAi (control RNAi, untrans, 2.5 ± 0.3, n = 24; trans, 2.5 ± 0.6, n = 22; P = 0.9; Dasm1 RNAi, untrans, 2.8 ± 0.5, n = 14; trans, 1.5 ± 0.2, n = 17; P < 0.05). These results suggest that suppression of Dasm1 expression specifically impairs AMPA-R-mediated, but not NMDA-R-mediated, synaptic transmission in neurons undergoing dynamic synaptic maturation.
To further test the role of Dasm1 in synaptic maturation, we transiently expressed the putative dominant negative mutant protein Dasm1(delC)-EGFP in CA1 pyramidal neurons of organotypic hippocampal slice culture by using the Sindbis virus (23). Neurons expressing Dasm1(delC)-EGFP showed ≈50% depression in synaptic transmission mediated by AMPA-Rs, but no significant change in that mediated by NMDA-Rs [AMPA-R, uninfected neighbor (uninf), 24.0 ± 2.0 pA; infected (inf), 12.6 ± 1.2 pA; n = 101; P < 0.0001; NMDA-R, uninf, 11.4 ± 1.1 pA; inf, 9.8 ± 1.0 pA; n = 71; P = 0.1] (Fig. 1F), indicating that Dasm1(delC)-EGFP expression specifically reduces AMPA-R-mediated synaptic transmission. Consequently, the ratio of AMPA-R- vs. NMDA-R-mediated synaptic transmission was smaller in neurons expressing Dasm1(delC)-EGFP than in uninfected control (uninf, 2.5 ± 0.3, n = 66; inf, 1.5 ± 0.1, n = 65; P < 0.01). This effect was not an artifact of viral infection, because neurons expressing EGFP alone showed no change in either AMPA-R- or NMDA-R-mediated synaptic transmission (AMPA-R, uninf, 23.1 ± 2.6 pA, inf, 23.1 ± 3.0 pA; n = 36; P = 0.9; NMDA-R, uninf, 11.7 ± 1.6 pA; inf, 11.0 ± 1.5 pA; n = 33; P = 0.3) (Fig. 1E) or their ratio (uninf, 2.5 ± 0.3, n = 24; inf, 2.5 ± 0.4, n = 25; P = 0.4).
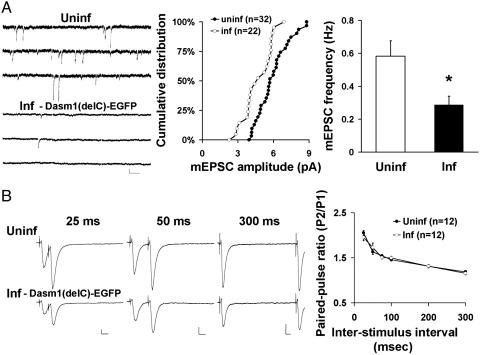
The comparable effects of Dasm1(delC)-EGFP and Dasm1 RNAi on synaptic function further suggest that Dasm1(delC) functions as a dominant negative. We next examined the effect of expressing this deletion mutant on miniature EPSCs (mEPSCs), another commonly used assay for synaptic function. Consistent with previous results, both the mean amplitude and frequency of mEPSC significantly decreased in neurons expressing Dasm1(delC)-EGFP when compared with the nearby uninfected control (amplitude, P < 0.05; frequency, uninf, 0.6 ± 0.1 Hz, n = 32; inf, 0.3 ± 0.1 Hz, n = 22; P < 0.01) (Fig. 2A). To assess whether perturbation of Dasm1 function also has an effect on presynaptic function, we examined paired-pulse facilitation, which is inversely correlated with the probability of neurotransmitter release (24). Expression of Dasm1(delC)-EGFP had no effect on this form of short-term synaptic plasticity (Fig. 2B). These results indicate that disrupting Dasm1 function at postsynaptic sites for 24-48 h in neurons at later developmental stages specifically reduces synaptic transmission mediated by AMPA-R, but not NMDA-R, without affecting the presynaptic function or the overall dendrite morphology (data not shown).
Fig. 2.
Effect of Dasm1(delC)-EGFP on mEPSCs and presynaptic function. (A) Decreases in both the mean amplitude (Center)(P < 0.05) and frequency (Right)(*, P < 0.05) of mEPSCs in neurons expressing Dasm1(delC)-EGFP compared with nearby uninfected control neurons. (Left) Sample traces of mEPSC recorded from neurons expressing Dasm1(delC)-EGFP (Inf) and uninfected control neuron (Uninf) are shown. (Scale bars = 10 pA and 200 msec.) (B) (Right) Expression of Dasm1(delC)-EGFP had no effect on paired-pulse facilitation at various interstimulus intervals. Pairs of sample traces from infected neurons expressing Dasm1(delC)-EGFP (Lower Left) and nearby uninfected neurons (Upper Left) at interstimulus intervals of 25, 50, and 300 msec are shown. (Scale bars = 10 pA and 25 msec.)
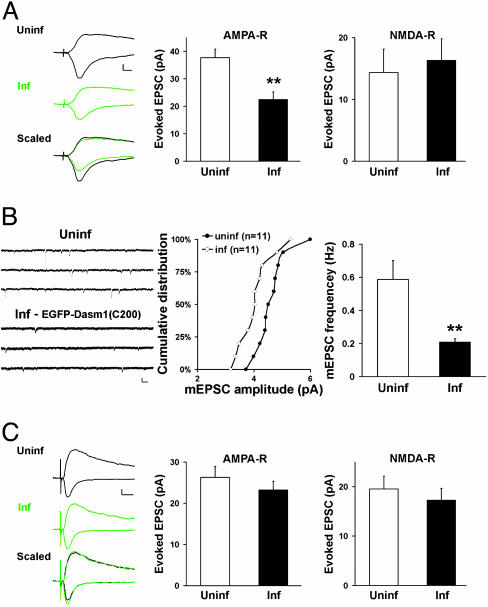
Having found similar synaptic defects induced by Dasm1 RNAi and the putative dominant negative mutant Dasm1(delC)-EGFP, we tested the effect of another potential dominant negative mutant, the last 200 aa (979-1178) of Dasm1 cytoplasmic tail with EGFP fused to its N terminus, EGFP-Dasm1(C200), to preserve the type I PDZ domain-binding motif at the C terminus. If the cytoplasmic tail of Dasm1 interacts with other proteins and mediates the signaling transduction, one would predict that overexpression of the isolated cytoplasmic tail in neurons also will perturb the endogenous Dasm1 function. Consistent with this prediction, neurons expressing EGFP-Dasm1(C200) exhibited depressed AMPA-R-mediated synaptic transmission as compared with nearby uninfected control neurons, whereas NMDA-R-mediated synaptic transmission was not affected (AMPA-R, uninf, 37.7 ± 3.1 pA; inf, 22.4 ± 2.8 pA; n = 35; P < 0.001; NMDA-R, uninf, 14.4 ± 3.8 pA; inf, 16.3 ± 3.5 pA; n = 20; P = 0.4) (Fig. 3A). As a result, the ratio of AMPA-R- vs. NMDA-R-mediated synaptic transmission was significantly reduced in EGFP-Dasm1(C200)-expressing neurons (uninf, 2.8 ± 0.4, n = 35; inf, 1.4 ± 0.3; n = 20; P < 0.05). We also examined its effect on mEPSCs and found that both the mean amplitude and frequency of mEPSCs were significantly decreased in neurons expressing EGFP-Dasm1(C200) (amplitude, P < 0.05; frequency, uninf, 0.6 ± 0.1 Hz, n = 11; inf, 0.2 ± 0.0 Hz, n = 11; P < 0.005) (Fig. 3B). These results reinforce the notion that Dasm1 regulates synaptic AMPA-R function and further suggest that the cytoplasmic tail of Dasm1 mediates its signaling at the synapse.
Fig. 3.
The PDZ domain-binding motif of Dasm1 is required for its synaptic function. (A) Expression of the last 200 aa of Dasm1 cytoplasmic tail [EGFP-Dasm1(C200,TLL)] caused a depression in AMPA-R-mediated (Center), but not NMDA-R-mediated (Right), synaptic transmission (**, P < 0.001). (Scale bars = 10 pA and 10 msec.) (B) Decreases in both the mean amplitude (Center) (P < 0.05) and frequency (Right) (**, P < 0.005) of mEPSCs in neurons expressing Dasm1(delC)-EGFP compared with nearby uninfected control neurons. (Scale bars = 10 pA and 100 msec.) (C) Expression of the last 200 aa of Dasm1 cytoplasmic tail with a single mutation at the -2 position in PDZ domain-binding motif [EGFP-Dasm1(C200,ALL)] had no effect on either AMPA-R-mediated (Center) or NMDA-R-mediated (Right) synaptic transmission. (Scale bars = 20 pA and 20 msec.)
PDZ Domain-Binding Consensus Sequence Is Required for the Synaptic Function of Dasm1. The cytoplasmic tail of Dasm1 contains a type I PDZ domain-binding motif (-TLL) at the end (25). To see whether the PDZ-binding motif of Dasm1 is important for its synaptic function, we expressed in neurons the cytoplasmic tail of Dasm1 with a single mutation at the -2 position, EGFP-Dasm1(C200,-ALL). Interestingly, neither AMPA-R-nor NMDA-R-mediated synaptic transmission, nor their ratio was changed in neurons expressing EGFP-Dasm1(C200,ALL), compared with the nearby uninfected control neurons (AMPA-R, uninf, 26.3 ± 2.7 pA; inf, 23.2 ± 2.1 pA; n = 42; P = 0.2; NMDA-R, uninf, 19.6 ± 2.6 pA; inf, 17.2 ± 2.4 pA; n = 35; P = 0.2; AMPA-R/NMDA-R, uninf, 2.5 ± 0.4, n = 37; inf, 2.5 ± 0.4, n = 35; P = 0.9) (Fig. 3C). These results indicate that interactions between Dasm1 and PDZ domain-containing proteins are required for its synaptic function.
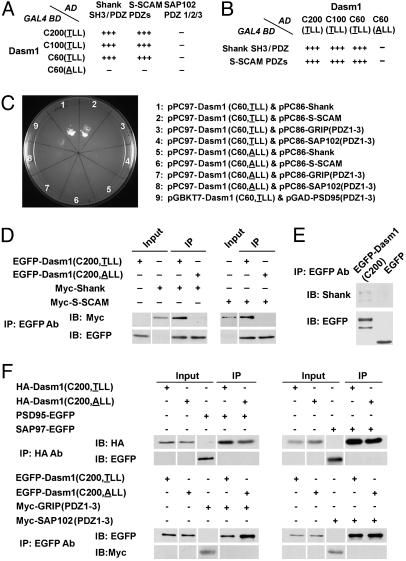
Interactions of Dasm1 with Synaptic PDZ Domain-Containing Proteins. To search for proteins that interact with the cytoplasmic tail of Dasm1, we carried out yeast two-hybrid screen by using as the bait the last 60 aa of Dasm1, which include the PDZ domain-binding motif (-TLL), and isolated two synaptic PDZ domain-containing proteins, Src homology 3 domain and ankyrin repeat-containing protein (Shank) (26, 27) and synaptic scaffolding molecule (S-SCAM) (28). We then confirmed the specificity of the interactions and characterized the domains responsible for these interactions in both yeast and mammalian systems. In yeast, the wild-type cytoplasmic tails of Dasm1 (C200, C100, and C60) interacted strongly and specifically with the Shank SH3/PDZ domains and the S-SCAM PDZ domains, but not PDZ domains of other prominent synaptic type I PDZ domain-containing proteins such as SAP102, PSD-95, and GRIP (Fig. 4 A-C). Moreover, the single mutation in the PDZ domain-binding motif (-ALL) of Dasm1 abolished these interactions. Similarly, in mammalian COS-7 cells, the wild-type cytoplasmic tail of Dasm1 coimmunoprecipitated with Shank and S-SCAM, whereas the mutant form lacking the PDZ domain-binding motif did not (Fig. 4D). Furthermore, the wild-type cytoplasmic tail of Dasm1 did not coimmunoprecipitate with PSD-95, GRIP, SAP102, or SAP97 (Fig. 4F). These results indicate that Dasm1 specifically interacts with the PDZ domains of two synaptic scaffolding proteins, Shank and S-SCAM, through its C-terminal PDZ domain-binding motif. The availability of antibodies against Shank permitted the demonstration that the cytoplasmic tail of Dasm1 could pull down endogenous Shank in hippocampal organotypic slice cultures (Fig. 4E).
Fig. 4.
The cytoplasmic tail of Dasm1 interacts with synaptic PDZ domain-containing proteins Shank and S-SCAM. (A and B) In the yeast two-hybrid assay, the cytoplasmic tails of Dasm1 (C200, C100, and C60) specifically interacted with the PDZ domains of Shank and S-SCAM, but not SAP102. A single mutation in the PDZ domain-binding motif (TLL to ALL) eliminated these interactions. The scores reflect the number of colonies grown in the triple selection plates (Leu-, Trp-, and His-). (C) The cytoplasmic tail of Dasm1 did not interact with type I PDZ domains of PSD-95, GRIP, and SAP102 in yeast two-hybrid assay. (D) Interactions between Dasm1 cytoplasmic tail and the PDZ domains of Shank and S-SCAM in COS-7 cells. IP, immunoprecipitation; IB, immunoblot. (E) Interaction between Dasm1 cytoplamic tail and endogenous Shank in hippocampal organotypic slice culture. The cytoplasmic tail of Dasm1 fused with EGFP, EGFP-Dasm1(C200), coprecipitated with endogenous Shank, whereas EGFP alone did not. The Shank antibody used recognizes different isoforms. The smaller band in the EGFP blot in the left lane might be the proteolytic product of EGFP-Dasm1(C200). (F) The cytoplasmic tail of Dasm1 did not interact with type I PDZ domains of PSD-95, GRIP, SAP102, and SAP97 in COS-7 cells.
Both Shank and S-SCAM are enriched in the PSD and have been implicated in synaptic function. Overexpression of Shank in neurons promotes the enlargement and maturation of dendritic spines, sites for most of the excitatory synapses, and increases mEPSC frequency (29). S-SCAM interacts with β-catenin, which plays a critical role in spine and synapse maturation (30, 31). The specific interactions between Dasm1 and these two synaptic proteins are consistent with the functional role of Dasm1 at the synapse and also suggest that Dasm1 likely is involved in spine and synapse maturation.
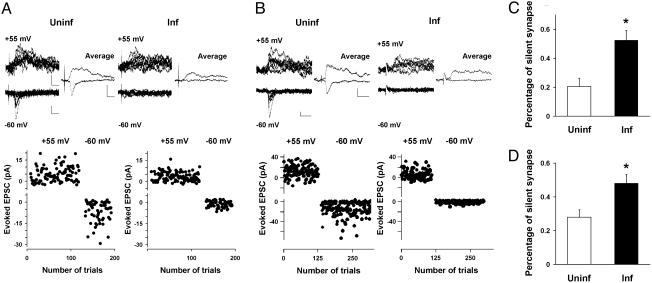
Dasm1 Controls Synapse Maturation. One emerging model of excitatory synapse maturation is the “silent synapse” hypothesis (2-5). In neurons in which Dasm1 function is perturbed by expressing Dasm1 RNAi, the cytoplasmic tail deletion mutant, or the isolated cytoplasmic tail, we found that AMPA-R-mediated, but not NMDA-R-mediated, synaptic transmission is specifically reduced, raising the possibility that Dasm1 is involved in synapse maturation, specifically the activation of silent synapses. This possibility was pursued by examining the fraction of synaptic transmission failures (failure rate) evoked with minimal stimulation at both hyperpolarized (-60 mV) and depolarized (+55 mV) potentials in neurons expressing the dominant negative proteins [either Dasm1(delC)EGFP or EGFP-Dasm1(C200)] and nearby uninfected control neurons (Fig. 5). Compared with nearby uninfected control neurons, the evoked synaptic transmission onto neurons expressing Dasm1(delC)-EGFP (Fig. 5A) showed a greater failure rate when neurons were clamped at the -60 mV, but not when clamped at +55 mV. Thus, there are more silent synapses in neurons expressing Dasm1(delC)-EGFP (uninf, 21 ± 6%, n = 39; inf, 53 ± 7%, n = 37; P < 0.001) (Fig. 5C). Similar results were obtained for EGFP-Dasm1(C200) (uninf, 28 ± 4%, n = 20; inf, 48 ± 5%, n = 27; P < 0.01) (Fig. 5 B and D). These results suggest that Dasm1 is involved in “awakening” the silent synapses and thus promoting synaptic maturation.
Fig. 5.
Increase in the percentage of silent synapse in neurons expressing either Dasm1(delC)-EGFP or EGFP-Dasm1(C200). (A and B)(Lower) Plot of evoked EPSCs amplitude vs. trial number at indicated membrane potentials for uninfected neurons (Left) and neurons expressing either Dasm1(delC)-EGFP (A) or EGFP-Dasm1(C200) (B)(Right). (Upper) A series of 10 consecutive synaptic responses recorded at the holding potentials of +55 and -60 mV (Left) evoked with minimal stimulation, and their averaged responses (Right) from uninfected control neuron and neuron expressing either Dasm1(delC)-EGFP (A) or EGFP-Dasm1(C200) (B) are shown. (Scale bars = 10 pA and 20 msec.) (C and D) Percentage of silent synapses (see Materials and Methods) for uninfected neurons and neurons expressing either Dasm1(delC)-EGFP (C) or EGFP-Dasm1(C200) (D) (*, P < 0.05).
Discussion
The studies reported in this article and the companion article (17) revealed that a previously uncharacterized member of the Ig superfamily, Dasm1, which controls dendrite outgrowth and synapse maturation, two essential processes of dendrite development (Fig. 6). Whereas dendritic outgrowth was specifically disrupted by expressing either Dasm1 RNAi or the truncated mutant Dasm1 for >7-8 days in early developing hippocampal neurons shortly after they were in culture, excitatory synapse maturation, but not dendritic outgrowth, was affected by transient expression of Dasm1 RNAi or the same truncation mutant Dasm1 protein in neurons at later developmental stages in the slice culture. Both electrophysiology and electron microscopy studies have shown that in early developmental stages, many synapses contain only NMDA-Rs, known as silent synapses because they are incapable of generating synaptic currents at the resting membrane potential. As development proceeds, more synapses contain both NMDA-Rs and AMPA-Rs. Such selective acquisition of AMPA-Rs has been proposed to be an essential mechanism for synapse maturation. Disrupting either extracellular or intracellular signaling of Dasm1 in the postsynaptic neuron through expression of either Dasm1 lacking the intracellular tail (extracellular and transmembrane domain only) or the intracellular tail alone caused a similar depression in AMPA-R-mediated, but not NMDA-R-mediated, synaptic transmission and an increase in the abundance of silent synapses. These results indicate that Dasm1 is directly involved in excitatory synapse maturation, the last and essential step of dendrite development. Recent studies show that Sidekicks, SynCAM, and SYG-1, which are members of the Ig superfamily, play critical roles in initiating synapse formation (32-34). It is conceivable that different types of Ig superfamily molecules are involved in different stages of synapse development.
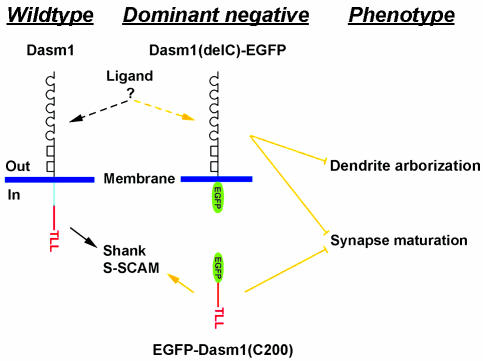
Fig. 6.
Dasm1, a previously unrecognized member of the Ig superfamily, controls dendrite outgrowth and synapse maturation. Expression of Dasm1 RNAi or a deletion mutant of Dasm1 lacking the cytoplasmic tail, Dasm1(delC), impairs dendrite outgrowth in early-stage neurons and prevents synapse maturation in later-stage neurons. The effect of Dasm1(delC) is likely because of the competition with endogenous Dasm1 for an as-yet-unknown ligand at the surface. The synaptic function of Dasm1 requires its C-terminal type I PDZ domain binding motif (-TLL), which interacts with two synaptic PDZ domain-containing proteins, Shank and S-SCAM.
How is Dasm1 involved in synapse maturation? Dasm1 in mature neurons is located at the excitatory synapse and contains an essential type I PDZ domain-binding motif (-TLL) at the C terminus (25). PDZ domain-containing proteins are critical components at the PSD and play instructive roles in many aspects of the synapse (35). A single mutation of the PDZ domain-binding consensus sequence (from TLL to ALL) rendered the last 200 aa of Dasm1 totally ineffective in altering synaptic transmission, suggesting that Dasm1 interacts with PDZ domain-containing protein(s) for its function at the synapse. Indeed, in our yeast two-hybrid screen, we identified two synaptic PDZ domain-containing proteins, Shank and S-SCAM, that interact with the Dasm1 cytoplasmic tail. These interactions seem to be rather specific, because Dasm1 does not interact with four other prominent synaptic type I PDZ domain-containing proteins tested, PSD-95, GRIP, SAP97, and SAP102. Interestingly, both Shank and S-SCAM have been shown to be involved in spine and synapse maturation. How the interactions between Dasm1 and Shank or S-SCAM may regulate synaptic AMPA-Rs and synapse maturation remains to be investigated, but the existence of these interactions provides a potential paradigm for extracellular signals to be transmitted to the signaling complexes in the postsynaptic neuron, possibly leading to changes in spine morphology and synaptic function.
Acknowledgments
We thank Drs. R. A. Nicoll, Y. Hayashi, Z. Z. Ma, and W. Grueber for helpful comments on an earlier version of the manuscript; Drs. P. Worley and A. Lanahan (The Johns Hopkins University, Baltimore) for the yeast two-hybrid screen library; Drs. B. Ye and Y. Hong for help with two-hybrid screen; and Drs. M. Sheng and A. Y. Hung (Massachusetts Institute of Technology, Cambridge) for the Shank antibody. This work was supported by a Helen Hay Whitney Fellowship (to S.-H.S.) and National Institutes of Health grants (to Y.-N.J. and L.Y.J.). S.-H.S. was a Research Associate and Y.-N.J. and L.Y.J. are Investigators of the Howard Hughes Medical Institute.
Abbreviations: Dasm1, dendrite arborization and synapse maturation 1; NMDA, N-methyl-d-aspartate; NMDA-R, NMDA receptor; AMPA, α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid; AMPA-R, AMPA receptor; EGFP, enhanced GFP; EPSC, excitatory postsynaptic current; mEPSC, miniature EPSC; RNAi, RNA interference; trans, transfected; untrans, untransfected; inf, infected; uninf, uninfected.
References
- 1.Hollmann, M. & Heinemann, S. (1994) Annu. Rev. Neurosci. 17, 31-108. [DOI] [PubMed] [Google Scholar]
- 2.Liao, D., Hessler, N. A. & Malinow, R. (1995) Nature 375, 400-404. [DOI] [PubMed] [Google Scholar]
- 3.Isaac, J. T., Nicoll, R. A. & Malenka, R. C. (1995) Neuron 15, 427-434. [DOI] [PubMed] [Google Scholar]
- 4.Durand, G. M., Kovalchuk, Y. & Konnerth, A. (1996) Nature 381, 71-75. [DOI] [PubMed] [Google Scholar]
- 5.Wu, G., Malinow, R. & Cline, H. T. (1996) Science 274, 972-976. [DOI] [PubMed] [Google Scholar]
- 6.Petralia, R. S., Esteban, J. A., Wang, Y. X., Partridge, J. G., Zhao, H. M., Wenthold, R. J. & Malinow, R. (1999) Nat. Neurosci. 2, 31-36. [DOI] [PubMed] [Google Scholar]
- 7.Nusser, Z., Lujan, R., Laube, G., Roberts, J. D., Molnar, E. & Somogyi, P. (1998) Neuron 21, 545-559. [DOI] [PubMed] [Google Scholar]
- 8.Takumi, Y., Ramirez-Leon, V., Laake, P., Rinvik, E. & Ottersen, O. P. (1999) Nat. Neurosci. 2, 618-624. [DOI] [PubMed] [Google Scholar]
- 9.Rumpel, S., Hatt, H. & Gottmann, K. (1998) J. Neurosci. 18, 8863-8874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Isaac, J. T., Crair, M. C., Nicoll, R. A. & Malenka, R. C. (1997) Neuron 18, 269-280. [DOI] [PubMed] [Google Scholar]
- 11.Li, P. & Zhuo, M. (1998) Nature 393, 695-698. [DOI] [PubMed] [Google Scholar]
- 12.Losi, G., Prybylowski, K., Fu, Z., Luo, J. H. & Vicini, S. (2002) J. Neurophysiol. 87, 1263-1270. [DOI] [PubMed] [Google Scholar]
- 13.Harris, K. M. & Stevens, J. K. (1989) J. Neurosci. 9, 2982-2997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kennedy, M. B. (2000) Science 290, 750-754. [DOI] [PubMed] [Google Scholar]
- 15.Cline, H. T. (2001) Curr. Opin. Neurobiol. 11, 118-126. [DOI] [PubMed] [Google Scholar]
- 16.Wong, R. O. & Ghosh, A. (2002) Nat. Rev. Neurosci. 3, 803-812. [DOI] [PubMed] [Google Scholar]
- 17.Shi, S.-H., Cox, D. N., Wang, D., Jan, L. Y. & Jan, Y.-N. (2004) Proc. Natl. Acad. Sci. USA 101, 13341-13345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Carlin, R. K., Grab, D. J., Cohen, R. S. & Siekevitz, P. (1980) J. Cell. Biol. 86, 831-845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dong, H., O'Brien, R. J., Fung, E. T., Lanahan, A. A., Worley, P. F. & Huganir, R. L. (1997) Nature 386, 279-284. [DOI] [PubMed] [Google Scholar]
- 20.Shi, S., Hayashi, Y., Esteban, J. A. & Malinow, R. (2001) Cell 105, 331-343. [DOI] [PubMed] [Google Scholar]
- 21.Maletic-Savatic, M., Malinow, R. & Svoboda, K. (1999) Science 283, 1923-1927. [DOI] [PubMed] [Google Scholar]
- 22.Hayashi, Y., Shi, S. H., Esteban, J. A., Piccini, A., Poncer, J. C. & Malinow, R. (2000) Science 287, 2262-2267. [DOI] [PubMed] [Google Scholar]
- 23.Malinow, R., Hayashi, Y., Maletic-Savatic, M., Zaman, S., Poncer, J.-C., Shi, S.-H. & Esteban, J. (2000) in Imaging Neurons: A Laboratory Manual, eds. Juste, R., Lanni, F. & Konnerth, A. (Cold Spring Harbor Lab. Press, Plainview, New York), pp. 58.1-58.8.
- 24.Zucker, R. S. (1989) Annu. Rev. Neurosci. 12, 13-31. [DOI] [PubMed] [Google Scholar]
- 25.Songyang, Z., Fanning, A. S., Fu, C., Xu, J., Marfatia, S. M., Chishti, A. H., Crompton, A., Chan, A. C., Anderson, J. M. & Cantley, L. C. (1997) Science 275, 73-77. [DOI] [PubMed] [Google Scholar]
- 26.Naisbitt, S., Kim, E., Tu, J. C., Xiao, B., Sala, C., Valtschanoff, J., Weinberg, R. J., Worley, P. F. & Sheng, M. (1999) Neuron 23, 569-582. [DOI] [PubMed] [Google Scholar]
- 27.Boeckers, T. M., Kreutz, M. R., Winter, C., Zuschratter, W., Smalla, K. H., Sanmarti-Vila, L., Wex, H., Langnaese, K., Bockmann, J., Garner, C. C. & Gundelfinger, E. D. (1999) J. Neurosci. 19, 6506-6518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hirao, K., Hata, Y., Ide, N., Takeuchi, M., Irie, M., Yao, I., Deguchi, M., Toyoda, A., Sudhof, T. C. & Takai, Y. (1998) J. Biol. Chem. 273, 21105-21110. [DOI] [PubMed] [Google Scholar]
- 29.Sala, C., Piech, V., Wilson, N. R., Passafaro, M., Liu, G. & Sheng, M. (2001) Neuron 31, 115-130. [DOI] [PubMed] [Google Scholar]
- 30.Murase, S., Mosser, E. & Schuman, E. M. (2002) Neuron 35, 91-105. [DOI] [PubMed] [Google Scholar]
- 31.Togashi, H., Abe, K., Mizoguchi, A., Takaoka, K., Chisaka, O. & Takeichi, M. (2002) Neuron 35, 77-89. [DOI] [PubMed] [Google Scholar]
- 32.Shen, K. & Bargmann, C. I. (2003) Cell 112, 619-630. [DOI] [PubMed] [Google Scholar]
- 33.Biederer, T., Sara, Y., Mozhayeva, M., Atasoy, D., Liu, X., Kavalali, E. T. & Südhof, T. C. (2002) Science 297, 1525-1531. [DOI] [PubMed] [Google Scholar]
- 34.Yamagata, M., Weiner, J. A. & Sanes, J. R. (2002) Cell 110, 649-660. [DOI] [PubMed] [Google Scholar]
- 35.Garner, C. C., Nash, J. & Huganir, R. L. (2000) Trends Cell Biol. 10, 274-280. [DOI] [PubMed] [Google Scholar]