Abstract
Fragile X syndrome is caused by the absence of the fragile X mental retardation protein (FMRP). This RNA-binding protein is widely expressed in human and mouse tissues, and it is particularly abundant in the brain because of its high expression in neurons, where it localizes in the cell body and in granules throughout dendrites. Although FMRP is thought to regulate trafficking of repressed mRNA complexes and to influence local protein synthesis in synapses, it is not known whether it has additional functions in the control of translation in the cell body. Here, we have used recently developed approaches to investigate whether FMRP is associated with the translation apparatus. We demonstrate that, in the brain, FMRP is present in actively translating polyribosomes, and we show that this association is acutely sensitive to the type of detergent required to release polyribosomes from membranous structures. In addition, proteomic analyses of purified brain polyribosomes reveal the presence of several RNA-binding proteins that, similarly to FMRP, have been previously localized in neuronal granules. Our findings highlight the complex roles of FMRP both in actively translating polyribosomes and in repressed trafficking ribonucleoparticle granules.
The RNA-binding proteins play pivotal roles in posttranscriptional regulation of gene expression. These proteins are involved in all subsequent steps in RNA function, from maturation and nucleocytoplasmic transport to subcellular localization and mRNA translation and stability (1-3). Proteins that coat RNA contain regions or domains essential for RNA recognition and binding. In neurons, in addition to being present in the cell body, a small fraction of mRNA is found in dendrites and axons at considerable distances from the nucleus (4-7). This differential distribution implies mechanisms of sorting, targeting, transport, and delivering of specific mRNA to these particular distal subcellular domains, where local protein synthesis is required (8-11).
The fragile X mental retardation protein (FMRP) is thought to be a key player in the control of mRNA transfer to distal locations such as dendrites (12). This protein is widely expressed in human and mouse tissues and is particularly abundant in neurons (13). The absence of FMRP causes fragile X syndrome, the most common monogenic form of mental retardation (14, 15). Studies on brain of fragile X patients and Fmr1 knockout mice strongly suggest that FMRP is involved in the proper development of neuronal spines (16-20). These abnormalities have been postulated to be at the basis of the mental retardation that results from defects in the process of neurite extension, guidance, and branching.
FMRP is an RNA-binding protein present in messenger ribonucleoparticle (mRNP) complexes associated with the translation machinery (21-24); however, the exact role of FMRP in translation remains unclear. High levels of FMRP act as a negative regulator of translation in vitro and in vivo (25-28). We have proposed that, in nonneural cells, FMRP is dispensable, whereas in neurons, a small fraction of FMRP acts as a repressor for the RNA to be transported and to be delivered at the budding dendrites (28). Indeed, although the great majority of FMRP has been observed in the neuron cell body (13, 29), a small fraction was detected either by immunofluorescent staining or immunoelectron microcopy at distal locations such as neurites, dendrites, and synaptosomes (22, 29-32). A series of neuronal mRNAs has been isolated either by immunoprecipitation approaches (33) or using antibody positioned RNA amplification (34). Although initial cell fractionation studies have shown that proteins from the FXR family are associated with polyribosomal mRNPs derived from rodent brain (29, 35), recent results have proposed that, in mouse brain, FMRP behavior is unique because it was not detected at the level of polyribosomes. Instead, FMRP was found exclusively associated with other classes of slow sedimenting ribonucleoparticles (RNPs) (36). These RNPs have been inferred to correspond to repressed mRNPs that are distinct from those present in the translation machinery.
Here, we report that, by using recently developed approaches, we reproducibly found FMRP to be associated with polyribosomal mRNPs in brain, as is the case in nonneuronal cells grown in culture. Also, we show that the association of FMRP as well as of several populations of RNA-binding proteins with these structures is acutely sensitive to the type of detergent used to release polyribosomes from membranous structures. These approaches enabled us to characterize a series of RNA-binding proteins that are associated with the translation apparatus.
Materials and Methods
Preparation of Homogenates. Adult (2-4 months old) and young (10-12 days old) CD1 mice were used throughout this study. Anesthetized animals were killed by cervical dislocation, and their brains and livers were quickly removed, immediately chilled in ice-cold PBS containing 50 μg/ml cycloheximide, and finely minced with scissors. The fragments from two brains were pooled and transferred to 7 ml of a buffer containing 20 mM Tris·HCl (pH 7.4), 100 mM KCl, 1.25 mM MgCl2, 1 mM DTT, 10 units/ml RNasine (Amersham Pharmacia), and protease inhibitors (Mini Complete, Roche Biochemicals). Tissues were slowly homogenized by hand (7-10 strokes) in a Kontes glass homogenizer (Vineland, NJ) fitted with the loose-B pestle. A postmitochondrial supernatant was prepared by centrifuging the homogenate at 9,000 × g for 15 min.
Polyribosomes Studies. To concentrate polyribosomes, 1% Nonidet P-40 was added to the postmitochondrial supernatant, and 7 ml of the solution was layered over a 3-ml pad made of 45% (wt/wt) sucrose in an 11-ml tube and centrifuged in a Sorval TH-641 rotor at 34,000 rpm (105,000 × g) for 3 h. The ribosomal pellets were then resuspended in a buffer (20 mM Tris·HCl, pH 7.4/100 mM KCl/1.25 mM MgCl2) containing the appropriate anionic or nonionic detergents as described in Results. Resuspended polyribosomes were analyzed by 15-45% (wt/wt) isokinetic sucrose gradients composed of 25 mM Tris·HCl, pH 7.4/100 mM KCl/5 mM MgCl2. After centrifugation in a Sorvall TH-641 rotor for 2 h at 34,000 rpm and 4°C, gradients were fractionated by upward displacement using an ISCO UA-5 flow-through spectrophotometer set at 254 nm and connected to a gradient collector. Each fraction was precipitated overnight at -20°C after addition of 2 volumes of ethanol. The precipitated material was collected by centrifugation at 12,000 rpm for 20 min and solubilized in SDS sample buffer before immunoblot analyses.
Protein Studies. Fractions of 150-500 S from three sucrose density gradients were pooled, and the polyribosomes were recovered after ultracentrifugation. Polyribosomes were resuspended in a buffer containing 20 mM Tris·HCl (pH 7.4), 100 mM KCl, 1.25 mM MgCl2, 1 mM DTT, 10 units/ml Rnasine, and protease inhibitors, treated either with 1% Nonidet P-40 or 1% deoxycholate (DOC) for 30 min at 4°C and recentrifuged at 105,000 × g for 2 h. The supernatants containing the Nonidet P-40 or DOC labile proteins as well as the recovered polyribosome pellets were denatured in SDS sample buffer and analyzed by immunoblotting or by SDS/PAGE, and the protein was revealed after Coomassie blue staining.
Immunoblot analyses were carried out as described (23). FMRP was detected with mAb1C3 (13) and mAb7B8 (obtained from A. Tartakoff, Case Western Reserve University, Cleveland), FXR1P was detected with mAb2FX specific for the 78- to 80-kDa isoforms (35), FXR2P was detected with mAbA42 and PABP1 was detected with mAb10E10 (both obtained from G. Dreyfuss, University of Pennsylvania, Philadelphia), NUFIP1 was detected with rabbit Ab1375 (37), p50 was detected with rabbit αYB-1 (from N. Sonenberg, McGill University, Montréal), 82-FIP was detected with rabbit Ab1666 (38), Stauffen1 and -2 were detected with rabbit αSt1 and αSt2, respectively (from L. DesGroseillers, University of Montréal), Sam68 was detected with rabbit SC-333 (Santa Cruz Biotechnology), and the ribosomal S6 and L7 proteins were detected with their respective antisera (from A. Ziemiecki, University of Bern, Switzerland).
Proteomics Analyses. Analyses were performed at the McGill University/Genome Québec Innovation Centre facility (Montréal). Samples were run on SDS/PAGE (10% acrylamide), and the gels were stained with Coomassie blue, scanned, and analyzed by using imagemaster software (Amersham Pharmacia). Gel slices were excised, proteins were digested with trypsin, and the resulting tryptic peptides were analyzed on a liquid chromatography quadrupole time-of-flight (Micromass, Manchester, U.K.) mass spectrometer. Peptides were electrosprayed as they exited the column, and double or triple charged ions were selected for passage into a collision cell. Fragmentation was induced by collision with argon gas, and data were collected in 1-s scans for up to 5 s. Peak lists of tandem MS data were prepared by using masslynx software (Micromass) and submitted to Mascot (Matrix Science, Boston) for identification by analysis against the National Center for Biotechnology Information nonredundant human and mouse databases.
Results
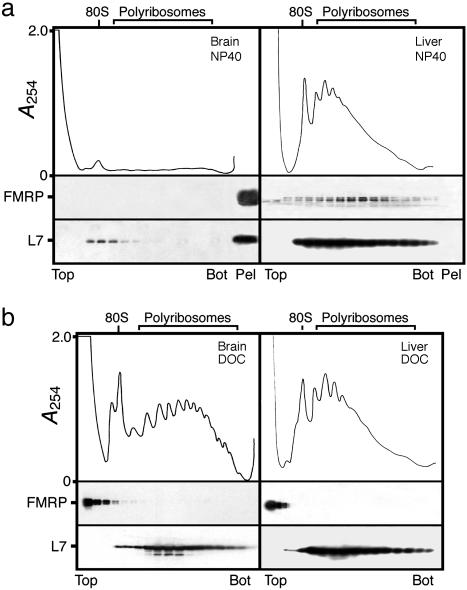
Isolation of Brain Polyribosomes from Adult Mouse. To prepare FMRP-containing mRNPs, we first isolated membranous structures by differential centrifugation of total brain lysate using buffers without detergents as reported (22, 29). We consistently observed that substantial amounts of FMRP could be recovered in a fraction corresponding to the rough endoplasmic reticulum (data not shown). In view of these results, we examined whether FMRP was associated with polyribosomal mRNPs or was part of different RNP complexes in brain. Because ≈40% of the polyribosomes are bound to membranes in rodent brain (39), the choice of the detergent was critical to release polyribosomes from these structures. Cytoplasmic extracts prepared in the presence of Nonidet P-40, a nonionic detergent, were analyzed by velocity sedimentation through linear sucrose density gradients. In repeated analyses, we were unable to obtain a UV absorption profile corresponding to polyribosomes because of contaminating light scattering, masking the distribution of subcellular components that may have penetrated the gradients. To eliminate or to reduce most of these unwanted contaminating UV-absorbing materials, the cytoplasmic fractions were first layered over a 45% (wt/wt) sucrose pad, and the polyribosomes were concentrated by ultracentrifugation as described for rat liver (40). The resulting opalescent pellets were resuspended in the extraction buffer containing MgCl2 and Nonidet P-40, and further analyzed by velocity sedimentation through sucrose density gradients. Unexpectedly, we constantly observed that only trace amounts of the 80S ribosomes could be detected as sedimenting materials. This observation was confirmed when the collected fractions were analyzed for the presence of the L7 ribosomal protein. In addition, we observed the presence of an opalescent pellet in the bottom of the centrifuge tubes where high levels of L7 and FMRP were detected (Fig. 1a). In contrast, liver polyribosomes that were used as control were clearly separated as shown in Fig. 1a, and FMRP was detected in fractions corresponding to polyribosomes containing L7 proteins. The difference of behavior between brain and liver extracts indicates that unknown materials that sediment together with brain polyribosomes have been compacted at the high g values used trapping these structures. We have not been able to resuspend intact polyribosomes in the presence of Nonidet P-40, Triton X-100, or Igepal CA-630, which are all related nonionic surfactants from the polyoxyethene p-t-octylphenol family. However, when the pellets were treated with buffers containing 1% DOC, an anionic detergent, the polyribosomes were released, and typical UV profiles with peaks corresponding to the 80S monomere and high levels of polyribosomes were obtained after centrifugation in sucrose density gradients (Fig. 1b). Under these experimental conditions, FMRP was restricted to the first fractions of the gradients, indicating that the protein was no longer associated with polyribosomes. The same distribution was observed also with liver polyribosomes treated in the same way, as FMRP remained at the top of the gradients (Fig. 1b).
Fig. 1.
Analyses of brain polyribosomes prepared from adult mice. (a) Postmitochondrial fraction was treated with 1% nonionic detergent Nonidet P-40, and total polyribosomes were first concentrated by ultracentrifugation, resuspended, and analyzed by sedimentation velocity throughout sucrose density gradients. Liver polyribosomes treated in the same way were analyzed in parallel. Note the presence of brain polyribosomes and FMRP at the bottom of the centrifuge tube, whereas the distribution of liver polyribosomes is different. (b) Treatment with 1% DOC allows brain and liver polyribosomes to distribute throughout the gradients according to their sedimentation values, whereas FMRP is detected at the top of the gradients. The integrity and distribution of polyribosomes were based on the UV profile as well as the presence of L7, a core protein of the large ribosomal subunit. Distribution of FMRP in different fractions was revealed by immunoblotting with mAb1C3.
We also tested a different procedure for the preparation of free and bound polyribosomes in the presence of a combination of nonionic and ionic detergents (1% Triton X-100 plus 1% DOC) that has previously been used for rat brain tissue (39). This approach allowed us to successfully isolate polyribosomes; however, under these conditions, FMRP could not be detected at the level of polyribosomes, but was instead present in the upper part of the sucrose gradients (data not shown).
Isolation of Brain Polyribosomes from Young Mouse. The results presented above clearly showed that under the conditions used, we were not able to prepare brain polyribosomes that carry FMRP from adult mice. Attempts to homogenize the brains more vigorously to fragment the endoplasmic reticulum resulted in breakage of the high molecular weight polyribosomes, which were shifted to the upper parts of the gradients. Because we have reported that brains from young animals contain 2- to 3-fold more FMRP as compared with adult (41) and that these tissues are easier to homogenize, we reasoned that using brains from younger animals would facilitate polyribosome extraction and reduce mechanical stress. A representative analysis of polyribosome preparations from young animals is presented in Fig. 2 and shows high levels of heavy sedimenting structures containing FMRP. These structures corresponded to polyribosomes because they were sensitive to EDTA, which causes the dissociation of the polyribosomes into the large and small ribosomal subunits and the release of the mRNPs. Polyribosomes were also destroyed after RNase treatment, and FMRP was found free floating in the top fractions of the gradient (data not shown). When polyribosome preparations were treated with 1% of DOC instead of Nonidet P-40, the displacement of FMRP was evident as it was detected in the upper fractions of the gradient, whereas the UV profile remained unaffected. Similar results were obtained for 3T3 and HeLa cells grown in culture (data not shown).
Fig. 2.
Effects of Nonidet P-40 (Left) and DOC (Right) on brain polyribosomes prepared from young animals. Although no major differences can be detected between the two polyribosomal profiles, the distribution of FMRP is clearly affected after treatment with DOC.
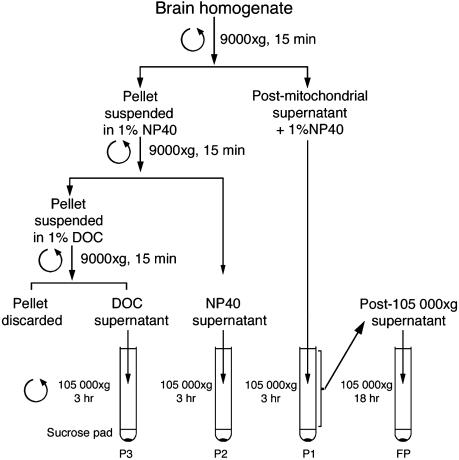
To estimate the yield of polyribosomes recovered, different subcellular fractions were prepared by using sequential extractions with nonionic followed by ionic detergents (schematically illustrated in Fig. 3) as adapted from the classic procedure of Blobel and Potter (42) for total liver polyribosomes preparation. Fig. 4 illustrates the results obtained after sedimentation analyses of the resuspended pellets and depicts the distribution of polyribosomes and FMRP. Densitometric and planometric analyses of the polyribosomal absorption profiles of three independent experiments indicated that ≈98% of FMRP was detected in the postmitochondrial supernatants treated with Nonidet P-40, whereas only trace amounts were detected after subsequent washing of the pellets with DOC. Finally, when the postribosomal fraction was centrifuged at 105,000 × g for 18 h, no enrichment of free RNPs was seen (Fig. 4). These results clearly show that the vast majority of FMRP was detected in association with polyribosomes, whereas only minute amounts of nonsedimentable FMRP were present in the upper fractions of the gradients loaded with materials that had been collected after a long ultracentrifugation for 18 h. The free FMRP likely correspond to molecules that had been released by the long and severe treatments imposed to the polyribosomes that are relatively fragile structures.
Fig. 3.
Schematic diagram of the steps used to prepare polyribosomes from the postmitochondrial supernatant and the residual fractions derived from homogenates of young mouse brain.
Fig. 4.
Quantitative distribution of brain polyribosomes and FMRP from the fractions prepared by differential sedimentations, as described in Fig. 3. Note that most of FMRP is associated with heavy sedimenting polyribosomes (P1), whereas only trace amounts are detected at the top of the gradient in panel FP, which stands for the final pellet obtained after centrifugation of the postribosomal pellet for 18 h at 105,000 × g.
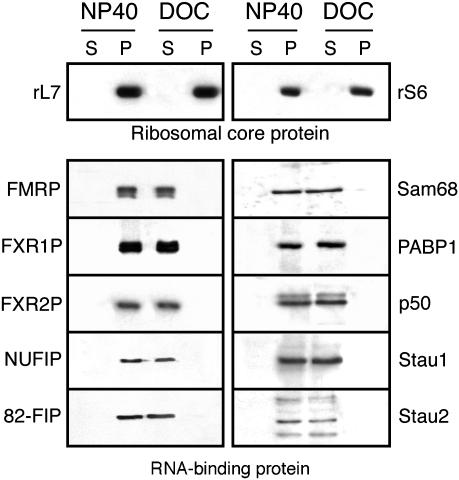
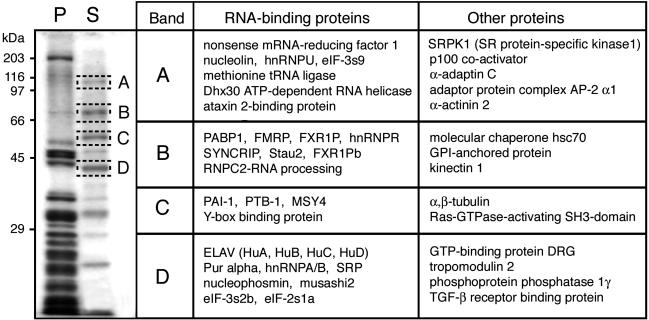
RNA-Binding Proteins Are Released from Polyribosomes After DOC Treatment. In view of the altered sedimentation properties of FMRP stripped off from the polyribosomes, we wondered whether this observation was unique to FMRP or was a generalized phenomenon that affected other RNA-binding proteins. Fractions corresponding to 150-500 S from sucrose density gradients were first pooled, and the polyribosomes were concentrated by ultracentrifugation in duplicate. One series of purified polyribosomes was resuspended in a buffer containing 1% Nonidet P-40, whereas the other was incubated in buffer containing 1% DOC. After a 30-min incubation period at 4°C, both samples were recentrifuged at 33,000 rpm to pellet the polyribosomes. The supernatants and pellets were analyzed by immunoblotting using a series of antibodies. Fig. 5 shows the results that can be summarized as follows. After Nonidet P-40 treatment, all RNA-binding proteins tested remained associated with the polyribosomes as monitored by using the ribosomal proteins S6 and L7. In contrast, although polyribosomes treated with DOC still contained the ribosomal proteins, all RNA-binding proteins tested were absent and, instead, were detected in the supernatants. These results clearly prove that the effects of DOC are not selective, because many other proteins associated with polyribosomes are stripped off similarly to FMRP. Furthermore, SDS/PAGE analyses of resistant and labile proteins detected after Coomassie brilliant blue staining showed that ≈15% of the total polyribosomal protein complement were extracted after DOC treatment. To identify the nature of the major polypeptide bands indicated by boxes in Fig. 6, these were cut, subjected to trypsin digestion, and analyzed by MS. The results of these analyses are listed in Fig. 6, and it can be seen that the majority of the released proteins that have been identified are RNA-binding proteins.
Fig. 5.
FMRP and several other RNA-binding proteins are released from polyribosomes after treatment with DOC, whereas Nonidet P-40 has no deleterious effects on the purification of polyribosomes that still carry these RNA-binding proteins. Immunoblot analyses were performed with the indicated specific antibodies. Note that neither Nonidet P-40 nor DOC has an effect on the core ribosomal proteins S6 and L7. P, polyribosomes recovered after ultracentrifugation; S, protein soluble either in Nonidet P-40 or DOC.
Fig. 6.
Identification of proteins extracted from polyribosomes after treatment with 1% DOC. The major Coomassie brilliant blue-stained bands in lane S were eluted and analyzed by MS. Identified proteins are classified as RNA-binding proteins and other proteins. P, polyribosomal proteins resistant to DOC treatment; S, soluble proteins after DOC treatment.
Discussion
Previous studies have shown that FMRP was absent from polyribosomes in mouse brain and instead was exclusively associated with light-sedimenting RNP complexes inferred to correspond to repressed mRNPs (36). These unexpected localizations led the authors to propose an original mechanism of action for FMRP in neurons. In the present study, we demonstrate that considerable amounts of FMRP cosediment with large polyribosomes isolated from young mouse brain, whereas no FMRP was found associated with light-sedimenting RNP complexes. These discrepancies between the association of FMRP with heavy sedimenting structures (this report) and the presence of FMRP in slow sedimenting RNPs (36) are likely due to the different procedures used to prepare polyribosomes from brain tissue. We found that the use of the anionic detergent DOC was necessary to release polyribosomes from adult mouse brain, presumably because of the chemical composition of the membranous structures at this age, whereas this harsh treatment was not required for other tissues such as liver. However, the complexes prepared in the presence of DOC seem to represent naked polyribosomes, as this detergent has been shown to selectively remove proteins associated with mRNA (43), including FMRP (22). Moreover, when added to the total homogenate, DOC releases slow sedimenting nuclear RNPs that contaminate the cytoplasmic fraction and that could be confounded with small cytoplasmic mRNPs (44, 45). However, we showed that native brain polyribosomes that carry RNA-binding complement can be prepared from young animals by using the nonionic detergents Nonidet P-40 or Triton X-100 of the polyoxyethene p-t-octylphenol family (43, 46). Under these experimental conditions, FMRP that has been assigned to neuronal granules (31, 32) and that has been reported to be exclusively present in light-sedimenting structures (36) was consistently present in polyribosomes. This was also the case for proteins that interact directly with FMRP, such as FXR1P, FXR2P, NUFIP, and 82-FIP (14). Furthermore, proteins interacting with the translation apparatus such as p50 (47), PABP1 (48), Sam68 (49), and Staufen 1/2 (50) were present.
Because DOC has the property to leave the polyribosome skeleton intact, we were able to selectively extract and characterize by proteomic analyses nonribosomal proteins. Two classes of proteins extracted with DOC were identified: the RNA-binding proteins and those that interact with the cytoskeleton framework, the local support of translation (51, 52). In addition to confirming the results obtained by immunoblotting, these analyses also revealed the presence of a series of RNA-binding proteins that directly interact with the translation machinery, such as the different eIFs, tRNA ligases, and RNA helicases (53, 54). Also, RNA-binding proteins known to be predominantly localized in the nucleus (3), such as the hnRNPs A/B/U/R and the ELAV, were also detected associated with polyribosomes.
Our findings that FMRP is associated with actively translating polyribosomes, in addition to its neuronal granule localization (32), strongly suggest a dual role for FMRP reminiscent to YB-1/p50, a nucleic acid chaperone (47, 55). Although the stoichiometry of FMRP to mRNA is not known, it is possible to propose that levels of FMRP above a certain threshold, yet to be determined, induce repression of translation by remodeling in concert with other interactor proteins that may influence FMRP function(s) (14), the conformational status of mRNAs in RNPs that have to be translocated to distal locations in neurons (56, 57). However, lower levels of FMRP might have complex roles in translation control because, when absent, the steady-state levels of many brain mRNAs are altered, some being increased and some being decreased (33, 34, 58). Finally, recent results suggest that FMRP may regulate neuronal translation via microRNAs (59), which in turn are found associated with polyribosomes (60).
It is tempting to speculate whether the recently observed FMRP-containing granules trafficking throughout dentrites (32) correspond to the class of repressed granules described earlier (61). Interestingly, these later repressed structures are many hundreds, if not thousands, of Svedbergs (S20.w) away from the small repressed mRNPs described by Zalfa et al. (36) because they sediment faster and ahead of polyribosomes. One of the key issues to unravel FMRP function(s) is to understand its RNA-binding selectivity and protein interaction properties. It seems that the nature of the approaches used to analyze the localization(s) and function(s) of FMRP has direct impacts on the interpretations of the results. On the basis of these considerations, we believe that our results will provide a useful basis for the future isolation and purification of brain FMRP-containing RNPs.
Acknowledgments
We thank Enzo Lalli, Urs-Peter Roos, Timothy Rose, Annette Schenk, Paul de Koninck, and Susan James for helpful discussions; Paul Naccache and Richard Kinkead for critically reading the manuscript; Luc DesGroseillers, Gideon Dreyfuss, Nahum Sonenberg, Alan Tartakoff, and Andrew Ziemiecki for providing antibodies; and anonymous referees for their constructive comments and suggestions. This work was supported by the Canadian Institutes of Health Research (E.W.K.), the National Institutes of Health Human Frontiers Science Program, and the Institut National de la Santé et de la Recherche Médicale (B.B.). L.D. holds a postdoctoral fellowship from the FRAXA Research Foundation, R.M. was the recipient of a postdoctoral fellowship from the Fragile X Research Foundation of Canada/Canadian Institutes of Health Research Partnership Challenge Fund program, and M.-E.H. holds a scholarship from the Canadian Institutes of Health Research.
Abbreviations: FMRP, fragile X mental retardation protein; mRNP, messenger ribonucleoparticle; DOC, deoxycholate; RNP, ribonucleoparticle.
References
- 1.Darnell, R. B. (2002) Cell 110, 545-550. [DOI] [PubMed] [Google Scholar]
- 2.Dever, T. E. (2002) Cell 108, 545-556. [DOI] [PubMed] [Google Scholar]
- 3.Dreyfuss, G., Kim, V. N. & Kataoka, N. (2002) Nat. Rev. Mol. Cell Biol. 3, 195-205. [DOI] [PubMed] [Google Scholar]
- 4.Kuhl, D. & Skehel, P. (1998) Curr. Opin. Neurobiol. 8, 600-606. [DOI] [PubMed] [Google Scholar]
- 5.Kiebler, M. A. & DesGroseillers, L. (2000) Neuron 25, 19-28. [DOI] [PubMed] [Google Scholar]
- 6.Jansen, R.-P. (2001) Nat. Rev. Mol. Cell Biol. 2, 247-256. [DOI] [PubMed] [Google Scholar]
- 7.Richter, J. D. & Lorenz, L. J. (2002) Curr. Opin. Neurobiol. 12, 300-304. [DOI] [PubMed] [Google Scholar]
- 8.Campenot, R. B. & Eng, H. (2000) J. Neurocytol. 29, 793-798. [DOI] [PubMed] [Google Scholar]
- 9.Martin, K. C., Barad, M. & Kandel, E. R. (2000) Curr. Opin. Neurobiol. 10, 587-592. [DOI] [PubMed] [Google Scholar]
- 10.Brittis, A. P., Lu, Q. & Flanagan, J. G. (2002) Cell 110, 223-235. [DOI] [PubMed] [Google Scholar]
- 11.Steward, O. & Schuman, E. M. (2003) Neuron 40, 347-359. [DOI] [PubMed] [Google Scholar]
- 12.Antar, L. N. & Bassell, G. J. (2003) Neuron 37, 555-558. [DOI] [PubMed] [Google Scholar]
- 13.Devys, D., Lutz, Y., Rouyer, N., Bellocq, J.-P. & Mandel, J.-L. (1993) Nat. Genet. 4, 335-340. [DOI] [PubMed] [Google Scholar]
- 14.Bardoni, B. & Mandel, J.-L. (2002) Curr. Opin. Genet. Dev. 12, 284-293. [DOI] [PubMed] [Google Scholar]
- 15.O'Donnell, W. T. & Warren, S. T. (2002) Annu. Rev. Neurosci. 25, 315-338. [DOI] [PubMed] [Google Scholar]
- 16.Hinton, V. J., Brown, W. T., Wisniewski, K. & Rudelli, R. D. (1991) Am. J. Med. Genet. 41, 289-294. [DOI] [PubMed] [Google Scholar]
- 17.Comery, T. A., Harris, J. B., Willems, P. J., Oostra, B. A., Irwin, S. A., Weiler, I. J. & Greenough, W. T. (1997) Proc. Natl. Acad. Sci. USA 94, 5401-5404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Greenough, W. T., Klintsova, A. Y., Irwin, S. A., Galvez, R., Bates, K. E. & Weiler, I. J. (2001) Proc. Natl. Acad. Sci. USA 98, 7101-7106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Irwin, S. A., Patel, B., Idupulapati, M., Harris, J. B., Crisostomo, R. A., Larsen, B. P., Kooy, F., Willems, P. J. Cras, P., Kozlowski, P. B., et al. (2001) Am. J. Med. Genet. 98, 161-167. [DOI] [PubMed] [Google Scholar]
- 20.Nimchinsky, E. A., Oberlander, A. M. & Svoboda, K. (2001) J. Neurosci. 21, 5139-5146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Eberhart, D. E., Malter, H. E., Feng, Y. & Warren, S. T. (1996) Hum. Mol. Genet. 5, 1083-1091. [DOI] [PubMed] [Google Scholar]
- 22.Khandjian, E. W., Corbin, F., Woerly, S. & Rousseau, F. (1996) Nat. Genet. 12, 91-93. [DOI] [PubMed] [Google Scholar]
- 23.Corbin, F., Bouillon, M., Fortin, A., Morin, S., Rousseau, F. & Khandjian, E. W. (1997) Hum. Mol. Genet. 6, 1465-1472. [DOI] [PubMed] [Google Scholar]
- 24.Feng, Y., Absher, D., Eberhart, D. E., Brown, V., Malter, H. E. & Warren, S. T. (1997) Mol. Cell 1, 109-118. [DOI] [PubMed] [Google Scholar]
- 25.Laggerbauer, B., Ostareck, D., Keidel, E. M., Ostareck-Lederer, A. & Fischer, U. (2001) Hum. Mol. Genet. 10, 329-338. [DOI] [PubMed] [Google Scholar]
- 26.Li, Z., Zhang, Y., Ku, L., Wilkinson, K. D., Warren, S. T. & Feng, Y. (2001) Nucleic Acids Res. 29, 2276-2283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schaeffer, C., Bardoni, B., Mandel, J.-L., Ehresmann, B., Ehresmann, C. & Moine, H. (2001) EMBO J. 20, 4803-4813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mazroui, R., Huot, M.-E., Tremblay, S., Filion, C., Labelle, Y. & Khandjian, E. W. (2002) Hum. Mol. Genet. 11, 3007-3017. [DOI] [PubMed] [Google Scholar]
- 29.Feng, Y., Gutekunst, C. A., Eberhart, D. E., Yi, H., Warren, S. T. & Hersch, S. M. (1997) J. Neurosci. 17, 1539-1547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Weiler, I. J., Irwin, S. A., Klintsova, A. Y., Spencer, C. M., Brazelton, A. D., Miyashiro, K., Comery, T. A., Patel, B., Eberwine, J. & Greenough, W. T. (1997) Proc. Natl. Acad. Sci. USA 94, 5395-5400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.De Diego Otero, Y., Severijnen, L.-A., van Cappellen, G., Schrier, M., Oostra, B. & Willemsen, R. (2002) Mol. Cell. Biol. 22, 8332-8341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Antar, L. N., Afroz, R., Dictenberg, J. B., Carroll, R. C. & Bassell, G. J. (2004) J. Neurosci. 24, 2648-2655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Brown, V., Jin, P., Ceman, S., Darnell, J. C., O'Donnell, W. T., Tenenbaum, S. A., Jin, X., Feng, Y., Wilkinson, K. D., Keene, J. D., et al. (2001) Cell 107, 477-487. [DOI] [PubMed] [Google Scholar]
- 34.Miyashiro, K. Y., Beckel-Mitchener, A., Purk, T. P., Becker, K. G., Barret, T., Liu, L., Carbonetto, S., Weiler, I. J., Greenough, W. T. & Eberwine, J. (2003) Neuron 37, 417-431. [DOI] [PubMed] [Google Scholar]
- 35.Khandjian, E. W., Bardoni, B., Corbin, F., Sittler, A., Giroux, S., Heitz, D., Tremblay, S., Pinset, C., Montarras, D., Rousseau, F. & Mandel, J.-L. (1998) Hum. Mol. Genet. 7, 2121-2128. [DOI] [PubMed] [Google Scholar]
- 36.Zalfa, F., Giorgi, M., Primerano, B., Moro, A., Di Penta, A., Reis, S., Oostra, B. & Bagni, C. (2003) Cell 112, 317-327. [DOI] [PubMed] [Google Scholar]
- 37.Bardoni, B., Willemsen, R., Weiler, I. J., Schenck, A., Severijnen, L.-A., Hindelang, C., Lalli, E. & Mandel, J.-L. (2003) Exp. Cell Res. 289, 95-107. [DOI] [PubMed] [Google Scholar]
- 38.Bardoni, B., Castets, M., Huot, M.-E., Schenck, A., Adinolfi, S., Corbin, F., Pastore, A., Khandjian, E. W. & Mandel, J.-L. (2003) Hum. Mol. Genet. 12, 1689-1698. [DOI] [PubMed] [Google Scholar]
- 39.Ramsey, J. C. & Steele, W.J. (1977) J. Neurochem. 28, 517-527. [DOI] [PubMed] [Google Scholar]
- 40.Wettstein, F. O., Staehelin, T. & Noll, H. (1963) Nature 197, 430-435. [DOI] [PubMed] [Google Scholar]
- 41.Khandjian, E. W., Fortin, A., Thibodeau, A., Tremblay, S., Côté, F., Devys, D., Mandel, J.-L. & Rousseau, F. (1995) Hum. Mol. Genet. 4, 783-789. [DOI] [PubMed] [Google Scholar]
- 42.Blobel, G. & Potter, V. R. (1967) J. Mol. Biol. 26, 279-292. [DOI] [PubMed] [Google Scholar]
- 43.Olsnes, S. (1970) Eur. J. Biochem. 15, 464-471. [DOI] [PubMed] [Google Scholar]
- 44.Moulé, Y. & Chauveau, J. (1968) J. Mol. Biol. 33, 465-481. [DOI] [PubMed] [Google Scholar]
- 45.Olsnes, S. (1970) Biochim. Biophys. Acta 213, 149-158. [DOI] [PubMed] [Google Scholar]
- 46.Borun, T. W., Scharff, M. D. & Robbins, E. (1967) Biochim. Biophys. Acta 149, 302-304. [DOI] [PubMed] [Google Scholar]
- 47.Davydova, E. K., Evdokimona, V. M., Ovchinnikov, L. P. & Hershey, J. W. B. (1997) Nucleic Acids Res. 25, 2911-2916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Adam, S. A., Nakagawa, T., Swanson, M. S., Woodruff, T. K. & Dreyfuss, G. (1986) Mol. Cell. Biol. 6, 2932-2943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Grange, J., Boyer, V., Fabian-Fine, R., Ben Fredj, N., Sadoul, R. & Goldberg, Y. (2004) J. Neurosci. Res. 75, 654-666. [DOI] [PubMed] [Google Scholar]
- 50.Duchaîne, T. F., Hemraj, I., Furic, L., Deitinghoff, A., Kiebler, M. A. & DesGroseillers, L. (2002) J. Cell Sci. 115, 3285-3295. [DOI] [PubMed] [Google Scholar]
- 51.Jansen, R.-P. (1999) FASEB J. 13, 455-466. [PubMed] [Google Scholar]
- 52.Bassell, G. & Singer, R. H. (1997) Curr. Biol. 9, 109-115. [DOI] [PubMed] [Google Scholar]
- 53.Gingras, A.-C., Rasught, B. & Sonenberg, N. (1999) Annu. Rev. Biochem. 68, 913-963. [DOI] [PubMed] [Google Scholar]
- 54.Valásek, L., Mathew, A. A., Shin, B.-S., Nielsen, K. H., Szamecz, B. & Hinnebusch, A. G. (2003) Genes Dev. 17, 786-799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kohno, K., Izumi, H., Uchiumi, T., Ashizuka, M. & Kuwano, M. (2003) BioEssays 25, 691-698. [DOI] [PubMed] [Google Scholar]
- 56.Gabus, C., Mazroui, R., Tremblay, S., Khandjian, E. W. & Darlix, J.-L. (2004) Nucleic Acids Res. 32, 2129-2137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mazroui, R., Huot, M.-E., Tremblay, S., Boilard, N., Labelle, Y. & Khandjian, E. W. (2003) Hum. Mol. Genet. 12, 3087-3096. [DOI] [PubMed] [Google Scholar]
- 58.D'Agata, V., Warren, S. T., Zhao, W., Torre, E. R., Alkon, D. L. & Cavallaro, S. (2002) Neurobiol. Dis. 10, 211-218. [DOI] [PubMed] [Google Scholar]
- 59.Jin, P., Zarnescu, D. C., Ceman, S., Nakamoto, M., Mowrey, J., Jongens, T. A., Nelson, D. L., Moses, K. & Warren, S. T. (2004) Nat. Neurosci. 7, 113-117. [DOI] [PubMed] [Google Scholar]
- 60.Kim, J., Krichevsky, A., Grad, Y., Hayes, G. D., Kosik, K. S., Church, G. M. & Ruvkun, G. (2004) Proc. Natl. Acad. Sci. USA 101, 360-365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Krichevsky, A. M. & Kosik, K. S. (2001) Neuron 32, 683-696. [DOI] [PubMed] [Google Scholar]