Abstract
The dual-specificity CDC25 phosphatases are critical positive regulators of cyclin-dependent kinases (CDKs). Even though an antagonistic Arabidopsis thaliana WEE1 kinase has been cloned and tyrosine phosphorylation of its CDKs has been demonstrated, no valid candidate for a CDC25 protein has been reported in higher plants. We identify a CDC25-related protein (Arath;CDC25) of A. thaliana, constituted by a sole catalytic domain. The protein has a tyrosine-phosphatase activity and stimulates the kinase activity of Arabidopsis CDKs. Its tertiary structure was obtained by NMR spectroscopy and confirms that Arath;CDC25 belongs structurally to the classical CDC25 superfamily with a central five-stranded β-sheet surrounded by helices. A particular feature of the protein, however, is the presence of an additional zinc-binding loop in the C-terminal part. NMR mapping studies revealed the interaction with phosphorylated peptidic models derived from the conserved CDK loop containing the phosphothreonine-14 and phosphotyrosine-15. We conclude that despite sequence divergence, Arath;CDC25 is structurally and functionally an isoform of the CDC25 superfamily, which is conserved in yeast and in plants, including Arabidopsis and rice.
The cell-cycle progression is controlled by the activity of evolutionarily conserved complexes constituted by a catalytic subunit, the cyclin-dependent serine/threonine kinases (CDK), bound to the regulatory subunit cyclin. To ensure a proper timing of mitosis, CDK/cyclin kinase activity is regulated positively by the activating CAK kinase, which phosphorylates on an activating site equivalent to Thr-160 of human CDK2, and is regulated negatively by the inhibiting WEE1 and MYT1 kinases, which phosphorylate the sites equivalent to Thr-14 and Tyr-15 of CDK2. The kinase activity of the WEE1 and MYT1 kinases is counteracted by the dual-specificity phosphatase CDC25, which dephosphorylates both the Thr-14 and Tyr-15 residues (1, 2). In yeast, one major CDK (CDC28) controls the transition between the different phases of the cell cycle, and a single CDC25 was identified to dephosphorylate Tyr-15. The complexity increases in human cells, where three CDC25 isoforms (CDC25A, CDC25B, and CDC25C) control the specific activation of distinct CDK/cyclin complexes at different time points during the cell cycle by dephosphorylation of both Thr-14 and Tyr-15. CDC25A regulates the G1-S transition by dephosphorylating the cyclin E-CDK2 complex. CDC25B and CDC25C dephosphorylate the cyclin A-CDK2 and the cyclin B-CDK1 complexes, respectively, both required for the G2-M transition (3).
Crystal structures of the catalytic domains of human CDC25A and CDC25B show an α/β-domain with a central five-stranded β-sheet surrounded by α-helices, called a rhodanese-like 3D fold (4-6). The CDC25 phosphatases form a distinct clade in a neighbor-joining tree representing the rhodanese superfamily, which also includes the yeast ACR2 arsenate reductase protein (7). The catalytic cysteine residue of these proteins is located in a short loop, including the tyrosine phosphatase signature motif His-Cys-(Xaa)5-Arg. Except for this motif, however, the CDC25s do not show any sequence or structure homology with other protein tyrosine phosphatase such as PTP1B, low molecular weight tyrosine phosphatase, or dual-specificity phosphatase such as VH1. Nevertheless, the CDC25 proteins share with other protein tyrosine phosphatases the catalytic mechanism by which they dephosphorylate their substrate, namely a two-step mechanism including a phosphocysteine intermediate that is subsequently hydrolyzed to give an inorganic phosphate and the regenerated enzyme (8).
Compared with animals, plants have adapted their cell division cycle to a sedentary lifestyle particularly exposed to environmental conditions. The overall strategy that regulates the progression through the cell cycle in plants is similar to that in other eukaryotes, but there are many differences in regulation mechanisms. There are several CDK-cyclin complexes described in plants (9, 10), the CDKA;1-CYCB1;1 complex being equivalent to the CDK/cyclin B found at the G2/M transition in other eukaryotes. The Thr-14, Tyr-15, and Thr-160 residues are conserved in CDKA;1. In accordance, plant CAK kinases that complement Saccharomyces cerevisiae CAK mutations and that phosphorylate CDKs have been described in ref. 11. There is equally evidence for a functional WEE1 in Arabidopsis thaliana, for which the overexpression in Schizosaccharomyces pombe causes a cell-cycle arrest (12). Besides the cloning of the antagonistic WEE1 kinase, there is some biochemical evidence pointing to the regulation of plant CDK activity by Tyr dephosphorylation. Inactive CDK complexes purified from tobacco cell suspension arrested in G2 phase by cytokinin starvation can be activated upon incubation with the yeast CDC25 protein (13). Moreover, overexpression of the CDC25 protein of yeast in tobacco plants leads to a greater frequency of lateral root primordium formation and to division of cells in the primordia at a reduced size (14, 15). A classical CDC25 protein, able to rescue the Sch. pombe cdc25-22 conditional mutant, was recently identified in the Ostreococcus tauri, a green unicellular alga (16). However, to date, it has not been possible to identify a CDC25 protein in the fully sequenced A. thaliana (17, 18) and Oryza sativa (19) genomes. We present functional and structural data indicating that a small tyrosine phosphatase, Arath;CDC25, acts as a CDC25 phosphatase in A. thaliana.
Methods
Cloning, Recombinant Protein Expression, and Purification. The full-length Arath;CDC25 coding region was isolated by RT-PCR with RNA prepared from Arabidopsis cell suspensions and the Superscript RT II kit (Invitrogen). Mutations of codons cysteine to serine for Cys-72, Cys-131/Cys-72, and Cys-120/Cys-122/Cys-127/Cys-131 were introduced in the coding sequence by PCR amplification with mutagenic oligonucleotides. All sequences were cloned in NdeI-XhoI pET15b (Novagen), in frame with a sequence encoding an N-terminal six-histidine tag (MGSSH6SSGLVPRGSH). The sequence corresponding to the 14 N-terminal amino acids containing the putative nuclear localization signal are not included in the DNA construction. Escherichia coli BL21(DE3) strain (Novagen) transformed by the recombinant pET15b plasmid was used to produce Arath;CDC25 protein. For uniform labeling of Arath;CDC25.S72, cells were grown in M9 medium supplemented with [13C]glucose (Cambridge Isotope Laboratories, Cambridge, MA) and/or with [15N]ammonium chloride (Cambridge Isotope Laboratories). The protein was purified by affinity on nickel-nitrilotriacetic acid resin (Amersham Pharmacia). For activity assay, the protein was further purified by gel filtration chromatography on Superose 12 prep grade (Amersham Pharmacia).
Activity Assay. Arath;CDC25 protein (5-20 μM) was usually incubated with 2-100 mM para-nitrophenyl phosphate (pNPP) (Sigma) in 1 ml of buffer [25 mM Tris (pH 7.6)/250 mM NaCl/5 mM 2-mercaptoethanol] for 30 min at 30°C. The incubation was terminated by addition of NaOH, and the absorbance was measured at 410 nm. The absorbance was converted to para-nitrophenol product concentration by using a molar absorptivity of 1.78 × 104 M-1·cm-1. The enzyme concentration was determined by the Bradford assay with BSA as standard. Arabidopsis CDKs were purified by affinity by incubation of 600 μg of protein extract, from Arabidopsis cell suspension, with 50 μl of Arabidopsis p10CKS1At beads (20, 21). Various amounts of recombinant Arath;CDC25 were then added to the p10CKS1At beads before incubation. An assay was performed after addition of a specific inhibitor of CDC25 phosphatase (NSC-95397, Biomol, Plymouth Meeting, PA) (22). In vitro histone H1 kinase activity was assayed as described in ref. 23. Samples were analyzed by SDS/12% PAGE to control the loading and autoradiographed by phosphorimager to detect histone H1 phosphorylation. To normalize the samples, anti-CDKA Western blots were prepared in parallel with a PSTAIRE monoclonal mouse antibody (Sigma).
NMR Spectroscopy. The NMR samples contained 0.25-0.5 mM Arath;CDC25.S72 protein in 25 mM sodium phosphate (pH 7.6)/250 mM NaCl/5 mM 2-mercaptoethanol, in 90% H2O/10% 2H2O or 100% 2H2O. NMR measurements were performed at 293 K on a Bruker (Karlsruhe, Germany) DMX600 spectrometer with a triple resonance cryo-probehead. The sequential backbone assignment is based on pulse sequences published in ref. 24. The 1H-1H distance restraints were derived from a 15N-edited NOESY on a uniformly 15N-labeled protein (100 ms) and a 13C-edited NOESY recorded at 800 MHz on a uniformly 13C-labeled protein sample in 2H2O. Three-dimensional-spectra processing and peak picking were performed with the snarf program (Frank Van Hoesel, Groningen, The Netherlands).
Structure Calculation. A combination of the chemical shift of the 1Hα, 13Cα, and 13CO nuclei, and observation of characteristic nuclear Overhauser effect connectivities was used to define the secondary structure (25). Additional distance constraints were introduced as hydrogen bonds between HN(i) and Hα(i + 4) in identified helical segments and between β-strands when a characteristic β-sheet was observed. The talos program was used to impose dihedral angle restraints (26). Distances derived from nuclear Overhauser effect intensities were divided in three classes: 1.8-2.9 Å, 1.8-4.3 Å, and 1.8-5.0 Å according to the peak volume. The zinc atom was positioned at 2.3 Å of the Sγ of Cys-120, Cys-122, Cys-127, and Nδ1 of His-39 and 3.4 Å of the Cβ of these residues. A distance constraint of 3.6 Å was imposed between the Sγ of the zinc-binding cysteines and Nδ1 of His-39. The structures were calculated by using the CNS program (27). A family of 20 structures was calculated by a distance geometry-simulated annealing protocol. Statistics are shown in Table 1, which is published as supporting information on the PNAS web site.
Structure Analysis and Representation. The 14 N-terminal amino acids of the Arath;CDC25 were not taken into account in the amino acid numbering in the sequence and structure descriptions because they were not included in the recombinant Arath;CDC25 produced in E. coli and subsequent structure calculation. The rasmol program (28) was used for structure visualization, molmol (29) was used for structure representation and comparison, and procheck-nmr (30) was used for structure validation. The multalin program (31, 32) was used to obtain the amino acid sequence alignment and representation shown in Fig. 1.
Fig. 1.
Sequence comparison of the Arath;CDC25 protein with CDC25, small CDC25-like tyrosine phosphatases, and the yeast arsenate reductase ACR2. Identical amino acids in all of the sequences are shown in white on a black background. Conserved amino acids are in bold. Secondary structure elements are indicated above the alignment for human CDC25A (PDB ID 1C25) and below for A. thaliana Arath;CDC25. The catalytic loop is indicated by the HC(X)5R motif. CDC25ACDC25A_human, human CDC25A (accession number NP_001780); CDC25B_human, human CDC25B (NP_068658); CDC25C_human, human CDC25C (NP_073720); CDC25_pombe, Sch. pombe CDC25 (NP_013750); MIH1_cerevisiae, S. cerevisiae CDC25 (NP_013750); YGR203_cerevisiae, S. cerevisiae protein of unknown function (NP_011719); ACR2_cerevisiae, S. cerevisiae arsenate reductase (NP_015526); IBP1_pombe, Sch. pombe small CDC25-like protein (AL096796); Orysa;CDC25;1, O. sativa protein of unknown function (NP_922597); Arath;CDC25_Arabidop, this work (NP_568119).
Peptide Synthesis. Phosphorylated CDK peptides of sequence EKVEKIGEGT14Y15GVVYKT, corresponding to residues of the glycine-rich loop of the Arabidopsis CDKA;1 and phosphorylated on Thr-14 and/or Tyr-15, were synthesized by solid-phase with Fmoc chemistry and purified by reversed-phase HPLC with a C18 column. Matrix-assisted laser desorption ionization-time-of-flight MS confirmed the integrity of the peptide. A 31P 1D spectrum, a 2D-NOESY, and 2D-TOCSY spectrum were acquired for each peptide.
NMR Titration Experiments. The chemical shift perturbation of 1HN and 15N of [15N]Arath;CDC25.S72 were observed with 1H-15N-heteronuclear single quantum correlation (HSQC) spectra after addition of 1 mM CdCl2/3 mM EDTA to 250 μM protein. The Cd and Zn ions are slowly exchanged between the chelating EDTA and protein, equilibrium being reach after ≈10 days (33). The peptides were added to 100 μM [15N]Arath;CDC25.S72 protein in a 20-fold excess in 12.5 mM deuterated Tris (pH 7.6)/200 mM NaCl/5 mM 2-mercapthoethanol buffer. The combined 15N and 1HN shift changes were calculated as Δppm = [(Δ1HN)2 + 0.2 × (Δ15N)2]1/2.
Results
Sequence Analysis. Systematic homology searches for a CDC25 candidate in the A. thaliana genome database, by using the described mammalian or yeast CDC25 sequences, were unsuccessful. However, a few potential candidates were fetched when searching only for the catalytic domain harboring the conserved HC(X)5R motif of the active site loop of the tyrosinephosphatase proteins. Among these latter, the Arath;CDC25 gene (locus At5g03455) encodes a 146-aa protein annotated as a rhodanese-like domain containing protein. A nuclear localization signal is predicted at its N terminus (MGRSIFSFFTKKKK). In the rice genome two CDC25-related genes were found, Orysa;CDC25;1 and Orysa;CDC25;2. The Orysa;CDC25;1 protein is 137 aa long and displays 60% identity over 131 amino acid residues with the Arath;CDC25 protein (Fig. 1). Orysa;CDC25;2 encodes a 173-aa protein with 63% identity over 126 amino acid residues. Arath;CDC25 is also homologous to small CDC25-like proteins found in yeast such as YGR203w from S. cerevisiae or IBP1 from Sch. pombe, where the latter was recently shown experimentally to display a tyrosine-phosphatase activity (34). Another yeast protein homologous to Arath;CDC25 is the arsenate reductase ACR2 (29% identity over 130 aa). Finally, the Arath;CDC25 protein shows some homology with the catalytic domain of the three human CDC25 isoforms, CDC25A (26% identity/100 aa), CDC25B (28% identity/104 aa), and CDC25C (32% identity/65 aa) and the yeast CDC25 (Mih1 from S. cerevisiae and CDC25 from Sch. pombe). However, these latter proteins have a long nonconserved regulatory N-terminal extension to the catalytic domain that is absent in the Arath;CDC25 protein.
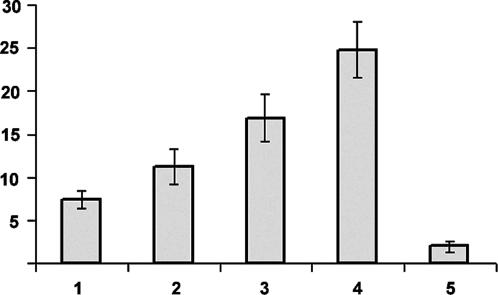
Arath;CDC25 Has a Tyrosine-Phosphatase Activity and Stimulates the Kinase Activity of Purified CDK Complexes of A. thaliana. Phosphatase activity of the Arath;CDC25 protein was first tested by using the phosphotyrosine analog para-nitrophenyl phosphate as substrate. We indeed observed hydrolysis of this substrate and calculated an approximate Km of 50 mM and a kcat of 0.15 sec-1 in the conditions of our enzymatic assay. To ensure that the phosphatase activity is dependent on the conserved HC72(X)5R78 motif, the Cysteine 72 was mutated to a serine. The mutated Arath;CDC25.S72 protein was unable to hydrolyze para-nitrophenyl phosphate (data not shown). We next investigated whether Arabidopsis CDK kinases could be activated by the Arath;CDC25 protein. Purified Arabidopsis CDK activity was measured by the incorporation of radioactive phosphate into added histone H1. A 1.5- to 1.8-fold increase of the histone H1 kinase activity in presence of 4 μg of the Arath;CDC25 protein was observed compared with the assay in the absence of the recombinant protein (Fig. 2, lanes 1 and 2). Kinase activity increased linearly with the amount of Arath;CDC25 protein added (Fig. 2, lanes 2-4). Addition of the NSC95397 specific inhibitor of CDC25 phosphatase (22) abolished the kinase activity associated with the CDK enzyme in the presence of the Arath;CDC25 protein (Fig. 2, lane 5). We conclude from these experiments that the Arath;CDC25 protein is able to activate CDK kinases of Arabidopsis, and that this activation is linked to the phosphatase activity of the Arath;CDC25 protein.
Fig. 2.
Activation of the A. thaliana CDK kinase activity by the Arath;CDC25 protein. The Arabidopsis CDK complexes were partially purified on p10CKS1At beads. The CDK-associated kinase activity was determined in the absence (lane 1) or presence of 4 (lane 2), 8 (lane 3), and 20 μg (lane 4) of Arath;CDC25 protein and 8 μg of Arath;CDC25 protein and 40 nM specific CDC25 inhibitor NSC95397 (lane 5). The graph illustrates the quantification of kinase activity by phosphorimager detection of the incorporation of radioactive phosphate by histone H1 normalized to the CDKA amount, control by Western blot with anti-PSTAIRE antibodies. Numbers on y axis are cpm × 10-3.
NMR Solution Structure of the Arath;CDC25.S72 Protein. Because expression of the WT Arath;CDC25 protein in minimal medium conditions required for isotope labeling gave a very poor yield, we concentrated on the Arath;CDC25.S72 mutant. This latter protein gave a substantially better yield, and the good chemical shift dispersion of the amide resonances in the HSQC spectrum indicated a stable fold. The 3D structure of Arath;CDC25.S72 was derived with a total of 775 unambiguous nuclear Overhauser effect-derived interproton distances, 47 talos-derived dihedral angle restraints, and 66 hydrogen-bond restraints (Table 1). Residues 1-6 have no regular secondary structure. The Arath;CDC25.S72 protein structure contains a five-stranded parallel β-sheet with a topology 15423 (Fig. 3). This β-sheet is very well defined by numerous long-range nuclear Overhauser effect distance restraints resulting in an rms deviation of 0.64 Å from the mean structure for the Cα, CO, and N atoms. Two α-helices, Ile-55-Asn-61 (α3) and Gly-79-Lys-93 (α4), are located on one side of the central β-sheet (Fig. 3). On the other side of the protein, there are two short helices, Leu-15-Leu-18 (α1) and Gln-32-Asn-35 (α2). The α5 helix (Asn-110-Ala-114) is pointing away from the core of the protein toward the C-terminal domain. The motif His71-Ser72(Xaa)5-Arg78 is in a helical loop between the β4 strand and the α4 helix (Fig. 3). Based on the multiple sequence alignment shown in Fig. 1, the backbone atoms (N, Cα, and CO) of residues from the five β-strands and the α4 helix of Arath;CDC25.S72 were superimposed on the corresponding residues of human CDC25A [Protein Data Bank (PDB) ID code 1C25] and CDC25B (PDB code 1QB0), resulting in an rms deviation of 2.5 Å. From the superposition of the backbone of these main secondary structures of Arath;CDC25.S72 and human CDC25A or CDC25B, it is clear that the core of the protein, consisting of the central β-sheet and the long α4 helix, is well conserved in Arath;CDC25.S72. Small differences between the Arath;CDC25.S72 protein and the CDC25A were observed at the level of the other helices with a longer α1 helix in the human CDC25A crystallographic structure and its α3 helix replaced by a helical loop, followed by a short α3 helix in the Arath;CDC25.S72 NMR structure. However, structures of the human CDC25A or CDC25B catalytic domain and the Arath;CDC25.S72 protein are completely different in what we define as a C-terminal domain, starting from the end of the α5 helix of the Arath;CDC25.S72 protein (Fig. 3). The presence of cysteines, which are conserved in the homologous Orysa;CDC25;1 (Fig. 1) and Orysa;CDC25;2 (sequence not shown) rice proteins, in the C-terminal stretch of the Arath;CDC25 protein hints at the presence of a metal-binding site. The metal was identified by atomic absorption spectroscopy as a zinc ion. A first indication of its precise location was obtained by the exchange of zinc for a different diamagnetic ion, cadmium, and subsequent monitoring of local chemical-shift perturbations. As expected, most of the chemical-shift perturbations due to the metal replacement were in the C-terminal domain of the Arath;CDC25.S72 protein, with major perturbations of the resonances of the amide group of the last 12 aa from Val-119 to Cys-131 and of the strand Gly-38-His-39-Ile-40-Ala-41-Gly-42. The side chain of His-39 equally shifted significantly. The involvement of His-39 in ion coordination suggested that only three of four Cys residues found in the C-terminal part of Arath;CDC25 participated in zinc binding. Recombinant Arath;CDC25.S72.S131 was well produced in E. coli and gave a 15N-HSQC spectrum with only a few perturbations of the amide resonances of the residues in the direct neighborhood of the Ser-131 mutation (namely Gly-129, Asp-130, Ala-132, Ala-41, and Val-119-Cys-120). The 3D 15N-NOESY spectrum of Arath;CDC25.S72 and Arath;CDC25.S72.S131 proteins were otherwise identical, demonstrating that the mutation of Cys-131 to Ser-131 did not cause any major structural modification, implying that the C-terminal Cys-131 is not implicated in metal binding. This finding unambiguously defined the quartet of residues coordinating the zinc as His-39, Cys-120, Cys-122, and Cys-127.
Fig. 3.
Ribbon representation in the same orientation of the backbone of a representative conformer of the Arath;CDC25 protein (Left) and the human CDC25A (Right) (PDB entry 1C25, ref. 4). The conserved five-stranded β-sheet and α4 helices of the structures have been superposed as described in Results. The catalytic loop is indicated by the HC(X)5R motif. The zinc ion is represented as a black sphere.
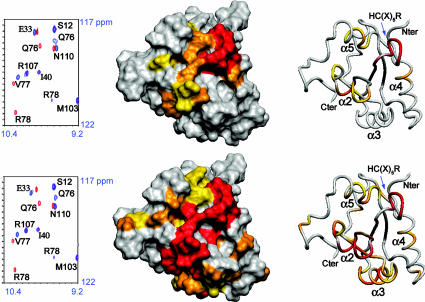
Interaction of the Arath;CDC25.S72 Protein with Phosphorylated CDKA;1 Model Substrate. The molecular interaction of 15N,13C-labeled Arath;CDC25.S72 with synthetic phosphopeptides of CDKA;1, corresponding to the CDK loop including the Thr-14 and Tyr-15 residues, was analyzed by NMR. The Arath;CDC25.S72 binds its substrates without phosphate hydrolysis as described for both the mutant human CDC25A and CDC25B proteins in ref. 35, allowing us to map the interaction sites of the phosphatase with its substrates. Upon addition of the phosphotyrosine (pTyr) peptide, the resonances of the amide groups of residues localized in the catalytic loop of Arath;CDC25.S72 showed large variations, with the amide group of Arg-78 being the most affected, similarly to the perturbations observed upon addition of para-nitrophenyl phosphate (data not shown). Chemical-shift perturbations were also observed for the amide resonances of the residues in the α5 helix, namely Phe-109 to Glu-113, and residues Asp-31 and Arg-34, which face the catalytic loop (Fig. 4). We next observed the interaction between the Arath;CDC25.S72 protein and an equivalent peptide containing phosphothreonine (pThr). No interaction was observed between the Arath;CDC25.S72 protein and the pThr14 peptide as was deduced from the absence of any chemical-shift modification. Upon addition of an excess of pThr14-pTyr15-containing peptide to the protein sample, however, we not only observed the same chemical-shift changes in the HSQC spectra as before with the pTyr15 peptide, but obtained equally perturbations in additional regions. For Gln-76, Val-77, and Arg-78 in the active site, the chemical shift perturbations are identical in presence of pTyr15 or pThr14-pTyr15 peptide (Fig. 4). However, for the doubly phosphorylated peptide, the perturbations of the auxiliary binding regions are more important (compare the perturbation of E33 chemical shift in Fig. 4 Upper and Lower), and there are additional residues involved in the binding site. Large chemical-shift modifications were indeed observed between bound and free state of the exposed Lys-54, His-57, and Gln-60 of the α3 helix (Fig. 4). In conclusion, the surface of interaction in the presence of the doubly phosphorylated peptide is more extended and almost completely covers one face of the protein (Fig. 4).
Fig. 4.
Map of the chemical-shift perturbation data on a surface and a ribbon representation in the same orientation of the Arath;CDC25 protein. Residues are colored according to the chemical-shift perturbation observed upon addition of an excess of pTyr15CDKA (Upper) or pThr14pTyr15CDKA (Lower) peptide. The color scale is as follows: yellow <0.04 ppm, orange between 0.04 and 0.1 ppm, and red >0.1 ppm combined 1H-15N chemical-shift difference between the free and the peptide-bound form of the Arath;CDC25 protein. The zinc ion is not represented. On the left of the structure representations, overlaid sections of 1HN-15N-HSQC spectra acquired on Arath;CDC25 before (in red contours) and after (in blue contours) addition of 20 molar equivalent of pTyr15CDKA and pThr14pTyr15CDKA peptides.
Discussion
Despite the availability of the complete genome sequence of both A. thaliana (17, 18) and O. sativa (19), no homologs of CDC25 have been reported for plants. Only recently, in the green monocellular algae Ostreococcus tauri, a protein was classified as a true CDC25 on the basis of its capacity to activate purified CDK-cyclin complexes and to suppress a cdc25-22 mutant in Sch. pombe (16).
Common to all of the CDC25 is the catalytic motif HC(X)5R. Because this motif is found in all tyrosine phosphatases and arsenate reductases, it is not sufficient to identify a CDC25. The tyrosine-phosphatase activity of the Arath;CDC25, which could potentially be associated with this catalytic motif, was demonstrated. The kcat/Km constant of the hydrolysis of the para-nitrophenyl phosphate substrate by the Arath;CDC25 protein of 3 M-1·s-1 is of the same order as the kinetic constant of 15-25 M--1·s-1 reported earlier for the CDC25A (36). Mutation of the Cys-72 residue abolished the tyrosine-phosphatase activity, confirming the C72(X)5R78 as the active site motif of the Arath;CDC25 protein. Arath;CDC25 was also found to stimulate the activity of purified Arabidopsis CDK complexes. The observed increase in CDK activity is linked to the phosphatase activity of the Arath;CDC25 protein because it can be abolished by the specific CDC25 inhibitor NSC95397 (22). These functional observations were underscored by the NMR observation of a direct physical interaction between the Arath;CDC25.S72 protein and peptide models mimicking the glycine-rich loop of the Arabidopsis CDKA;1. Phosphorylation on Tyr-15 is a necessary condition, but the equivalent addition of a phosphate to the Thr-14 side chain further strengthens this physical interaction. The interaction surface between the Arath;CDC25.S72 protein and the doubly phosphorylated peptide was larger than that determined for the singly phosphorylated Tyr-15 peptide, emphasizing the character of dual-specificity tyrosine phosphatase of the Arath;CDC25 protein.
The active site of the Arath;CDC25 protein was mapped by observing the modifications of the amide chemical shift of the residues involved in the interaction in the HSQC spectrum by adding the phospho-CDK peptides to the protein sample. As expected, the active site coincides with the conserved H71S72(X)5R78 loop located between β4 strand and α4 helix. Arg-78 is the most affected residue, implying direct association, probably with the phosphate moiety. The equivalent Arg-436 of CDC25A and Arg-493 of CDC25B are involved in the anion binding, as was revealed in the x-ray structure of the latter by the presence of a sulfate anion in the catalytic site (6). Two auxiliary regions are involved in the phospho-residue binding, the α2 and the α5 helices. The Phe-109, located in the α5 helix, is positioned close to the catalytic loop and could assist in tyrosine recognition, as was proposed earlier for the Phe-432 and Phe-489 located in the active site loop of CDC25A and CDC25B (4, 6). In the α2 helix, Asp-31 and Arg-34 seem to be involved in substrate binding, according to their chemical shift perturbation. In the presence of the doubly phosphorylated peptide, not only is the active site occupied by the phosphotyrosine residue but additional regions of interaction with the peptide were identified, including the α3 helix.
The 3D structure of Arath;CDC25 further confirms that the protein belongs to the CDC25 family. The Arath;CDC25.S72 protein has a fold similar to the catalytic domain of the CDC25A and CDC25B. The CDC25 phosphatase enzymes have a rhodanese-like topology, although they bear little sequence homology to this enzyme family (4-7). Other rhodanese domains have been predicted in Arabidopsis, although no experimental 3D structure of plant rhodanese domain is yet described. The C-terminal part of Arath;CDC25 contains a zinc-binding loop and is, as such, different from the human CDC25A and CDC25B (37). This zinc-binding loop could play a role of protein stabilization because the recombinant Arath;CDC25 protein containing only serines instead of the cysteines in its C-terminal part could not be produced in E. coli. However, the zinc-binding loop could equally be involved in substrate recognition because the zing fingers are well known interaction modules that bind to a variety of substrates, including protein or DNA (37). It would be functionally similar to the 17 final C-terminal residues of human CDC25B that were shown to confer substrate specificity and mediate protein substrate recognition (6, 38).
Although clear similarity between the Arath;CDC25 topology and substrate binding mode with the catalytic domain of the human CDC25 is observed, the Arath;CDC25 lacks the N-terminal regulatory region found in the human counterpart. The Arath;CDC25 catalytic domain is preceded only by a putative nuclear localization signal. In the classical CDC25, the nonconserved N-terminal domain undergoes multiple phosphorylations that regulate protein subcellular localization, stability, and catalytic activity. The different modular domain organization that we find here for Arath;CDC25 reminds us of our previous work on the PIN1At protein of A. thaliana with the plant enzyme lacking the N-terminal WW domain found in all of the other eukaryotic homologs (39). In both cases, the regulatory function could be controlled in trans by a distinct protein that awaits further identification.
Recently, a small CDC25-like phosphatase, IBP1, was reported for Sch. pombe (34). IBP1, like the Arath;CDC25 protein, is closely related to the catalytic domain of the CDC25 and has phosphatase activity in vitro (34). Overproduction of IBP1 suppresses a mutation in the DNA replication initiation kinase HSK1p, a Cdc7 family protein kinase essential for the initiation of DNA replication (34). IBP1 cannot suppress a Sch. pombe cdc25-22 mutant, and disruption of IBP1 in Sch. pombe has no obvious phenotype except disordered mitosis in <5% of the cells. Arath;CDC25 is also homologous to a similar small CDC25-like protein found in S. cerevisiae, named YGR203w. Deletion of YGR203w is viable but alters yeast fitness after 20 generations (40). In our hands, inducible overexpression of the Arath;CDC25 gene in Sch. pombe did not induce cells to divide faster, nor was the gene able to complement the temperature sensitive cdc25-22 mutant (unpublished results). Thus, Arath;CDC25 function may not be essential under normal growth conditions but might rather be specialized to some specific processes of the cell cycle such as genome duplication and stability. Similarly, the regulation of CDC28 kinase in S. cerevisiae by MIH1, the classical CDC25 protein, is not required under normal conditions of growth or even for the DNA replication checkpoint but is essential when the bud checkpoint is activated.
The functional and structural data presented here indicate that Arath;CDC25 protein of A. thaliana is a member of the CDC25 family and define an isoform of the CDC25 enzymes, conserved in yeast. Further experiments are now needed to elucidate the cellular function and the regulatory network of this protein.
Supplementary Material
Acknowledgments
We thank Caroline Smet and Julien Michel for peptide synthesis, Mathieu Sauty for the absorption spectroscopy, and Hervé Drobecq for mass spectrometry. The 600-MHz NMR facility used in this study was funded by the Fond Européen de Développement Régional (FEDER), the Région Nord-Pas de Calais, the Centre National de la Recherche Scientifique, and the Institut Pasteur de Lille. The spectrum at 800 MHz was acquired in Gif sur Yvette, France.
Abbreviations: CDK, cyclin-dependent kinase; HSQC, heteronuclear single quantum coherence; PDB, Protein Data Bank; pThr, phosphothreonine; pTyr, phosphotyrosine.
Data deposition: The coordinates of the final 20 structures of Arath;CDC25 have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 1T3K). A complete table of the 1H, 15N, and 13C chemical shifts has been deposited in the BioMagResBank, www.bmrb.wisc.edu (accession nos. 6195 and 6196).
References
- 1.Dunphy, W. G. & Kumagai, A. (1991) Cell 67, 189-196. [DOI] [PubMed] [Google Scholar]
- 2.Kumagai, A. & Dunphy, W. G. (1991) Cell 64, 903-914. [DOI] [PubMed] [Google Scholar]
- 3.Nilsson, I. & Hoffmann, I. (2000) Prog. Cell Cycle Res. 4, 107-114. [DOI] [PubMed] [Google Scholar]
- 4.Fauman, E. B., Cogswell, J. P., Lovejoy, B., Rocque, W. J., Holmes, W., Montana, V. G., Piwnica-Worms, H., Rink, M. J. & Saper, M. A. (1998) Cell 93, 617-625. [DOI] [PubMed] [Google Scholar]
- 5.Hofmann, K., Bucher, P. & Kajava, A. V. (1998) J. Mol. Biol. 282, 195-208. [DOI] [PubMed] [Google Scholar]
- 6.Reynolds, R. A., Yem, A. W., Wolfe, C. L., Deibel, M. R., Jr., Chidester, C. G. & Watenpaugh, K. D. (1999) J. Mol. Biol. 293, 559-568. [DOI] [PubMed] [Google Scholar]
- 7.Bordo, D. & Bork, P. (2002) EMBO Rep. 3, 741-746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jackson, M. D. & Denu, J.M. (2001) Chem. Rev. (Washington, D.C.) 101, 2313-2340. [DOI] [PubMed] [Google Scholar]
- 9.Vandepoele, K., Raes, J., De Veylder, L., Rouze, P., Rombauts, S. & Inze, D. (2002) Plant Cell 14, 903-916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.De Veylder, L., Joubes, J. & Inze, D. (2003) Curr. Opin. Plant Biol. 6, 536-543. [DOI] [PubMed] [Google Scholar]
- 11.Shimotohno, A., Matsubayashi, S., Yamaguchi, M., Uchimiya, H. & Umeda, M. (2003) FEBS Lett. 534, 69-74. [DOI] [PubMed] [Google Scholar]
- 12.Sorrell, D. A., Marchbank, A., McMahon, K., Dickinson, J. R., Rogers, H. J. & Francis, D. (2002) Planta 215, 518-522. [DOI] [PubMed] [Google Scholar]
- 13.Zhang, K., Letham, D. S. & John, P. C. (1996) Planta 200, 2-12. [DOI] [PubMed] [Google Scholar]
- 14.Bell, M. H., Halford, N. G., Ormrod, J. C. & Francis, D. (1993) Plant Mol. Biol. 23, 445-451. [DOI] [PubMed] [Google Scholar]
- 15.McKibbin, R. S., Halford, N. G. & Francis, D. (1998) Plant Mol. Biol. 36, 601-612. [DOI] [PubMed] [Google Scholar]
- 16.Khadaroo, B., Robbens, S., Ferraz, C., Derelle, E., Eychenie, S., Cooke, R., Peaucellier, G., Delseny, M., Demaille, J., Van De Peer, Y., et al. (2004) Cell Cycle 3, 513-518. [PubMed] [Google Scholar]
- 17.The Arabidopsis Genome Initiative (2000) Nature 408, 796-815. [DOI] [PubMed] [Google Scholar]
- 18.Wigge, P. A. & Weigel, D. (2001) Curr. Biol. 11, 112-114. [DOI] [PubMed] [Google Scholar]
- 19.Buell, C. R. (2002) Plant Physiol. 130, 1585-1586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Landrieu, I., Casteels, P., Odaert, B., De Veylder, L., Portetelle, D., Lippens, G., Van Montagu, M. & Inze, D. (1999) Protein Expression Purif. 16, 144-151. [DOI] [PubMed] [Google Scholar]
- 21.Brizuela, L., Draetta, G. & Beach, D. (1987) EMBO J. 6, 3507-3514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lazo, J. S., Nemoto, K., Pestell, K. E., Cooley, K., Southwick, E. C., Mitchell, D. A., Furey, W., Gussio, R., Zaharevitz, D. W., Joo, B. & Wipf, P. (2002) Mol. Pharmacol. 61, 720-728. [DOI] [PubMed] [Google Scholar]
- 23.Magyar, Z., Meszaros, T., Miskolczi, P., Deak, M., Feher, A., Brown, S., Kondorosi, E., Athanasiadis, A., Pongor, S., Bilgin, M., et al. (1997) Plant Cell 9, 223-235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Grzesiek, S., Bax, A., Hu, J. S., Kaufman, J., Palmer, I., Stahl, S. J., Tjandra, N. & Wingfield, P. T. (1997) Protein Sci. 6, 1248-1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wishart, D. S., Sykes, B. D. & Richards, F. M. (1992) Biochemistry 31, 1647-1651. [DOI] [PubMed] [Google Scholar]
- 26.Cornilescu, G., Delaglio, F. & Bax, A. (1999) J. Biomol. NMR 13, 289-302. [DOI] [PubMed] [Google Scholar]
- 27.Brunger, A. T., Adams, P. D., Clore, G. M., DeLano, W. L., Gros, P., Grosse-Kunstleve, R. W., Jiang, J. S., Kuszewski, J., Nilges, M., Pannu, N. S., et al. (1998) Acta Crystallogr. D 54, 905-921. [DOI] [PubMed] [Google Scholar]
- 28.Sayle, R. A. & Milner-White, E. J. (1995) Trends Biochem. Sci. 20, 374. [DOI] [PubMed] [Google Scholar]
- 29.Koradi, R., Billeter, M. & Wüthrich, K. (1996) J. Mol. Graphics 14, 51-59. [DOI] [PubMed] [Google Scholar]
- 30.Laskowski, R. A., Rullmannn, J. A., MacArthur, M. W., Kaptein, R. & Thornton, J. M. (1996) J. Biomol. NMR 8, 477-486. [DOI] [PubMed] [Google Scholar]
- 31.Corpet, F. (1988) Nucleic Acids Res. 16, 10881-10890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gouet, P., Courcelle, E., Stuart, D. I. & Metoz, F. (1999) Bioinformatics 15, 305-308. [DOI] [PubMed] [Google Scholar]
- 33.Hanzawa, H., de Ruwe, M. J., Albert, T. K., van der Vliet, P. C., Timmers, H. T. & Boelens, R. (2001) J. Biol. Chem. 276, 10185-10190. [DOI] [PubMed] [Google Scholar]
- 34.Snaith, H. A., Marlett, J. & Forsburg, S. L. (2003) Curr. Genet. 44, 38-48. [DOI] [PubMed] [Google Scholar]
- 35.Xu, X. & Burke, S. P. (1996) J. Biol. Chem. 271, 5118-5124. [DOI] [PubMed] [Google Scholar]
- 36.Rudolph, J., Epstein, D. M., Parker, L. & Eckstein, J. (2001) Anal. Biochem. 289, 43-51. [DOI] [PubMed] [Google Scholar]
- 37.Krishna, S. S., Majumdar, I. & Grishin, N. V. (2003) Nucleic Acids Res. 31, 532-550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wilborn, M., Free, S., Ban, A. & Rudolph, J. (2001) Biochemistry 40, 14200-14206. [DOI] [PubMed] [Google Scholar]
- 39.Landrieu, I., De Veylder, L., Fruchart, J. S., Odaert, B., Casteels, P., Portetelle, D., Van Montagu, M., Inzé, D. & Lippens, G. (2000) J. Biol. Chem. 275, 10577-10581. [DOI] [PubMed] [Google Scholar]
- 40.Giaever, G., Chu, A. M., Ni, L., Connelly, C., Riles, L., Vernneau, S., Dow, S., Lucau-Danila, A., Anderson, K., Andre, B., et al. (2002) Nature 418, 387-391. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.