Abstract
Variations in surface hydrophobicity of six Lactobacillus strains with and without an S-layer upon changes in ionic strength are derived from contact angle measurements with low- and high-ionic-strength aqueous solutions. Cell surface hydrophobicity changed in response to changes in ionic strength in three out of the six strains, offering these strains a versatile mechanism to adhere to different surfaces. The dynamic behavior of the cell surface hydrophobicity could be confirmed for two selected strains by measuring the interaction force between hydrophobic and hydrophilic tips with use of atomic force microscopy.
Several studies have shown that bacterial strains, such as lactobacilli, can protect the host against infection by invading pathogens in the upper gastrointestinal tract and the vagina. Lactobacilli are acid tolerant and produce high levels of lactic acid, thereby lowering the environmental pH and suppressing pathogens. The exact mechanism by which lactobacilli exert this protection is not fully understood, but adhesion is a commonly accepted prerequisite (6). Several lactobacillus species possess a surface layer protein (SLP) anchored to the cell envelope. This surface layer consists of a (glyco-)protein, the so-called S-protein, which assembles into characteristic two-dimensional crystalline layers at the cell surface (7). The function of the S-layer on these organisms is unknown, but S-layers of lactobacilli are important in their adhesion to surfaces, as SLP confers hydrophobicity on the lactobacillus cell surface (9). Yet, adhesion of lactobacilli to surfaces often does not proceed according to expectations based on their cell surface hydrophobicity and hydrophobic strains do not always adhere best to hydrophobic substrata (5), as outlined by surface thermodynamics (1). This suggests that cell surfaces of lactobacilli may adapt their cell surface hydrophobicity in response to environmental changes, such as in pH or ionic strength.
Macroscopic bacterial cell surface hydrophobicity is commonly inferred from water contact angle measurements on bacteria deposited on membrane filters (2). If water molecules have a greater tendency to surround each other than to contact a bacterial cell surface, the surface appears hydrophobic and water droplets do not spread. If water molecules favor a microbial cell surface rather than each other, the surface appears hydrophilic. Hydrophobic lactobacillus isolates with water contact angles above 100° (Lactobacillus acidophilus RC14) have been described, but so have extremely hydrophilic ones with water contact angles of 19° (Lactobacillus casei 36) (10). Although cell surface hydrophobicity arises from interactions at the molecular level, hydrophobicity has never been assessed at the level of molecular cell surface components.
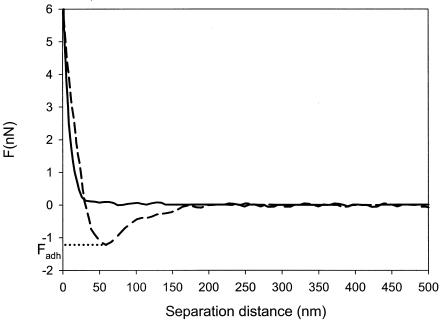
Atomic force microscopy (AFM) has emerged as a valuable tool for probing interaction forces at the molecular level with a high spatial resolution (4). A sharp tip located at the free end of a flexible cantilever is approached and retracted from the surface under study. Interaction forces between the tip and the sample surface cause the cantilever to deflect. The deflection signal during the approach and retraction process is acquired to provide so-called force-distance curves (Fig. 1 shows an example).
FIG. 1.
Force-distance curve for L. acidophilus ATCC 4356 interacting with a hydrophobic AFM tip at 10 mM KCl. The solid line represents the approach curve, while the dashed line indicates the retraction curve. The maximum adhesion force, Fadh, probed upon retraction is indicated on the graph.
In this paper, the macroscopic cell surface hydrophobicity of a collection of six lactobacillus strains with and without SLP was assessed by contact angle measurements with low- and high-ionic-strength solutions (10 mM and 100 mM KCl). Furthermore, the surfaces of two selected strains showing dynamic cell surface hydrophobicity were probed with regard to their interaction forces with chemically functionalized AFM tips, i.e., terminating in hydrophobic (CH3) and hydrophilic (OH) groups, also in low- and high-ionic-strength solutions.
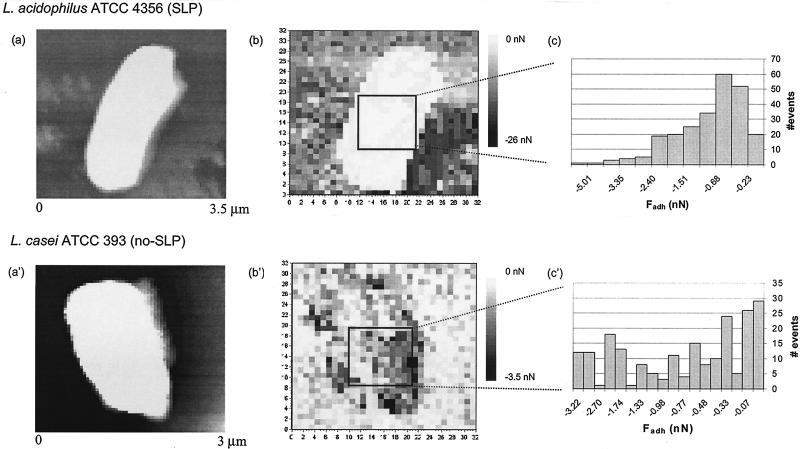
Bacterial strains were cultured in De Man-Rogosa-Sharpe medium (Merck, Darmstadt, Germany) at 37°C in an atmosphere containing 5% CO2. This culture was used to inoculate a second culture that was grown for 16 h prior to harvest. Bacteria were harvested by centrifugation (5 min at 10,000 × g), washed twice with demineralized water, and suspended in demineralized water-10 or 100 mM KCl solution. Contact angle measurements were performed on bacterial lawns prepared by depositing about 50 layers of bacteria suspended in demineralized water on a cellulose acetate membrane filter (pore diameter, 0.45 μm) (9). For AFM experiments, bacteria were attached to a positively charged poly-l-lysine-treated glass slide. V-shaped silicon nitride cantilevers with a spring constant of 0.06 N/m were functionalized by coating them with a thin layer of titanium and gold followed by their immersion in HS(CH2)11OH or HS(CH2)17CH3 solution (3). Functionalized probes were always used immediately after preparation. AFM measurements were made at room temperature under 10 and 100 mM KCl solution with an optical-level microscope (Nanoscope III Digital Instrument). An array of 32 by 32 force-distance curves were collected over the entire field of view, once a bacterium was imaged (Fig. 2a and a′ show examples). Adhesion maps were produced by taking the most negative force detected during the retraction curve (Fig. 1) and plotting that value against the x-y position of each force-distance curve (Fig. 2b and b′). From the adhesion maps, a selected area of ∼800 by 800 nm2 over the top of each bacterium was used to generate an adhesion distribution histogram (Fig. 2c and c′) from which an average adhesion force, Fadh, was calculated between functionalized AFM tips and the bacterial cell surfaces for each experimental condition studied. Three to five different organisms were studied in each particular case.
FIG. 2.
Array of 32 by 32 force-distance curves over the AFM field of view for L. acidophilus ATCC 4356 (SLP) (a) and L. casei ATCC 393 (no SLP) (a′) together with their corresponding adhesion maps (b and b′) obtained using a hydrophobic AFM tip at 10 mM KCl. Histograms (c and c′) show the distribution of adhesion forces over a selected area of about 800 by 800 nm2 on the bacterial cell surface.
The contact angles on the various lactobacillus strains measured with low- and high-ionic-strength solutions are shown in Table 1. The lactobacilli without SLP showed a lower contact angle when measured with a low-ionic-strength solution than when measured with a high-ionic-strength solution, albeit that this difference is not significant for Lactobacillus johnsonii LMG9436T. Lactobacillus crispatus JCM5810 and L. casei 393*/CA5A′, both with SLP, gave similar contact angles for the high- and low-ionic-strength solution. Only L. acidophilus ATCC4356, also having SLP, reduced its contact angle upon increase of the ionic strength.
TABLE 1.
Summary of contact angles (degrees) with aqueous, low- and high-ionic-strength solutions for lactobacillus strains with and without SLPa
| Lactobacillus strain | SLP | Contact angle (°)
|
|
|---|---|---|---|
| 10 mM KCl | 100 mM KCl | ||
| L. acidophilus ATCC 4356 | Yes | 76 ± 4 | 47 ± 3 |
| L. crispatus JCM 5810 | Yes | 66 ± 9 | 70 ± 13 |
| L. casei ATCC 393 | No | 32 ± 5 | 65 ± 5 |
| L. casei 393*/CA5A′ | Yes | 45 ± 7 | 50 ± 7 |
| L. gasseri LMG9203 | No | 45 ± 3 | 70 ± 9 |
| L. johnsonii LMG9436T | No | 42 ± 6 | 47 ± 4 |
Contact angles represent mean values of 10 measurements, equally distributed over two bacterial lawns, prepared out of different bacterial cultures.
For two selected strains showing dynamic cell surface hydrophobicity, L. acidophilus ATCC 4356 and L. casei ATCC 393, adhesion maps were made, indicating a heterogeneous surface distribution of interaction forces between the cell surfaces and functionalized tips for the two strains, regardless of ionic strength (examples are shown in Fig. 2b and b′). Histograms showing the distribution of these interaction forces over the top of each bacterium are presented in Fig. 2c and c′. The interaction forces detected by hydrophobic and hydrophilic AFM tips were averaged for each strain into an adhesion force, Fadh, and compared with contact angles measured with aqueous, low- and high-ionic-strength solutions (Tables 1 and 2). In general, high interaction forces with a hydrophilic tip were found to coincide with low contact angles, whereas a cell surface with a high contact angle showed the strongest interaction with a hydrophobic tip. In addition, both strains reversed their hydrophobic nature upon increase of the ionic strength from 10 to 100 mM KCl. The lactobacillus strain with SLP was found to be hydrophobic in 10 mM KCl and became more hydrophilic in 100 mM KCl, while the strain without SLP was hydrophilic in 10 mM KCl and became hydrophobic in 100 mM KCl.
TABLE 2.
Average adhesion force, Fadh, as probed with hydrophobic and hydrophilic AFM tips for two selected Lactobacillus strains with and without SLP and showing dynamic cell surface hydrophobicity upon changes in ionic strengtha
| Strain | Ionic strength (mM) | Contact angle (°) |
Fadh (nN) with tip:
|
|
|---|---|---|---|---|
| Hydrophobic | Hydrophilic | |||
| L. acidophilus ATCC 4356 with SLP | 10 | 76 ± 4 | −1.34 ± 0.19 | −0.11 ± 0.02 |
| 100 | 47 ± 3 | −0.88 ± 0.13 | −2.14 ± 0.17 | |
| L. casei ATCC 393 without SLP | 10 | 32 ± 5 | −0.91 ± 0.09 | −1.96 ± 0.33 |
| 100 | 65 ± 5 | −2.31 ± 0.45 | −0.47 ± 0.12 | |
AFM data are representative of results obtained from three to five cells, with use of different probes and independent preparations.
The structure of the S-layer on L. acidophilus ATCC 4356 is known to be composed of two subdomains: an external N-terminal region showing predominantly hydrophobic amino acid residues and a C-terminal region, serving to attach the S-layer to the cell wall, which is mainly composed of positively charged hydrophilic residues (8). The dynamic cell surface hydrophobicity observed may be explained by a shrinkage of the S-layer due to reduced intramolecular electrostatic repulsion at high ionic strength. Then, the inner hydrophilic region may become (partly) exposed at the aqueous periphery of the bacterial surface, rendering it more hydrophilic.
L. casei ATCC 393 on the other hand, does not possess an S-layer. Yet, its cell surface shows dynamic hydrophobicity as well. X-ray photoelectron spectroscopy indicated that the surface of L. casei ATCC 393 is rich in polysaccharides (11). At low ionic strength, this layer presents itself as a hydrophilic polyelectrolyte coating. At high ionic strength the polysaccharide layer is known to collapse, and this evidently results in exposure of a more hydrophobic surface.
It is interesting that the dynamic behavior of the cell surface hydrophobicity of the lactobacilli was measurable not only macroscopically by contact angles on bacterial lawns but also by AFM at a more microscopic level. Stronger interaction forces between the cell surfaces and hydrophobically or hydrophilically modified tips coincide with higher or lower contact angles with aqueous solutions. This is fully in line with surface thermodynamics, stating that hydrophobic surfaces favor interaction with hydrophobic surfaces. Analogously, hydrophilic surfaces show a greater affinity for hydrophilic surfaces.
In conclusion, this paper is the first to report the dynamic behavior of cell surfaces of lactobacilli with regard to their hydrophobicity in response to changes in environmental ionic strength. Dynamic cell surface hydrophobicity was demonstrated in three out of six strains by contact angles measured with low- and high-ionic-strength solutions, while confirmed for two strains at a more microscopic level by AFM. This dynamic behavior of bacterial cell surfaces upon changes in ionic strength offers certain lactobacillus strains a versatile mechanism to adhere to hydrophobic and hydrophilic surfaces in low- and high-ionic-strength solutions, respectively.
REFERENCES
- 1.Absolom, D. R., P. V. Lamberti, Z. Policova, W. Zingg, C. J. van Oss, and A. W. Neumann. 1983. Surface thermodynamics of bacterial adhesion. Appl. Environ. Microbiol. 46:90-97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Busscher, H. J., A. H. Weerkamp, H. C. van der Mei, A. W. J. van Pelt, H. P. de Jong, and J. Arends. 1984. Measurement of the surface free energy of bacterial cell surfaces and its relevance for adhesion. Appl. Environ. Microbiol. 48:980-983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dufrêne, Y. F. 2000. Direct characterization of the physicochemical properties of fungal spores using functionalized AFM probes. Biophys. J. 78:3286-3291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dufrêne, Y. F. 2002. Atomic force microscopy, a powerful tool in microbiology. J. Bacteriol. 184:5205-5213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Millsap, K. W., G. Reid, H. C. van der Mei, and H. J. Busscher. 1996. Adhesion of Lactobacillus species in urine and phosphate buffer to silicone rubber and glass under flow. Biomaterials 18:87-91. [DOI] [PubMed] [Google Scholar]
- 6.Sanders, M. E. 1993. Effect of consumption of lactic cultures on human health. Adv. Food Nutr. Res. 37:67-130. [DOI] [PubMed] [Google Scholar]
- 7.Sára, M., and U. B. Sleytr. 2000. S-layer proteins. J. Bacteriol. 182:859-868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Smit, E., D. Jager, B. Martinez, F. J. Tielen, and P. H. Pouwels. 2002. Structural and functional analysis of the S-layer protein crystallisation domain of Lactobacillus acidophilus ATCC 4356: evidence for protein-protein interaction of two subdomains. J. Mol. Biol. 324:953-964. [DOI] [PubMed] [Google Scholar]
- 9.Van der Mei, H. C., B. van de Belt-Gritter, P. H. Pouwels, B. Martinez, and H. J. Busscher. 2003. Cell surface hydrophobicity is conveyed by S-layer proteins—a study in recombinant lactobacilli. Colloids Surf. B 28:127-134. [Google Scholar]
- 10.Van der Mei, H. C., R. Bos, and H. J. Busscher. 1998. A reference guide to microbial cell surface hydrophobicity based on contact angles. Colloids Surf. B Biointerfaces 11:213-221. [Google Scholar]
- 11.Van der Mei, H. C., J. de Vries, and H. J. Busscher. 2000. X-ray photoelectron spectroscopy for the study of microbial cell surfaces. Surf. Sci. Rep. 39:1-24. [Google Scholar]