Abstract
Although the ability of Bifidobacterium spp. to grow on fructose as a unique carbon source has been demonstrated, the enzyme(s) needed to incorporate fructose into a catabolic pathway has hitherto not been defined. This work demonstrates that intracellular fructose is metabolized via the fructose-6-P phosphoketolase pathway and suggests that a fructokinase (Frk; EC 2.7.1.4) is the enzyme that is necessary and sufficient for the assimilation of fructose into this catabolic route in Bifidobacterium longum. The B. longum A10C fructokinase-encoding gene (frk) was expressed in Escherichia coli from a pET28 vector with an attached N-terminal histidine tag. The expressed enzyme was purified by affinity chromatography on a Co2+-based column, and the pH and temperature optima were determined. A biochemical analysis revealed that Frk displays the same affinity for fructose and ATP (Kmfructose = 0.739 ± 0.18 mM and KmATP = 0.756 ± 0.08 mM), is highly specific for d-fructose, and is inhibited by an excess of ATP (>12 mM). It was also found that frk is inducible by fructose and is subject to glucose-mediated repression. Consequently, this work presents the first characterization at the molecular and biochemical level of a fructokinase from a gram-positive bacterium that is highly specific for d-fructose.
The phosphorylation of monosaccharides is highly important for carbohydrate metabolism in the prokaryotic cell. This enzymatic reaction is required not only to allow sugars to be assimilated by a particular metabolic pathway, but also as a means of uptake by the cell. For example, prior to being catabolized by the bacterial cell, fructose needs to be phosphorylated either by specific fructose phosphoenolpyruvate (PEP)-phosphotransferase (PTS) enzymes or by a nucleotide-dependent fructokinase (EC 2.7.1.4). The phosphotransferase system involves two classes of proteins, sugar-specific (EII) and general (EI and HPr) enzymes, which supply the cell with fructose-1-P (16, 32, 35), whereas fructokinases (Frk or ScrK; ATP:β-d-fructose 6-phosphotransferases) catalyze the transfer of γ-phosphate from ATP to an intracellular molecule of fructose, leading to fructose-6-P (F6P) (3, 39, 40, 66). For the majority of bacteria, genes encoding fructokinases are either isolated on the chromosome (frk) (17, 64, 65, 66) or members of a sucrose utilization gene cluster (scrK, sacK, or cscK) (3, 5, 6, 28, 38, 53).
Although the ability of Bifidobacterium spp. to grow on fructose as a unique carbon source has been demonstrated (36), the enzyme(s) needed to incorporate fructose into a catabolic pathway has hitherto not been defined. Currently, bifidobacteria are reported to possess only one route for the metabolism of glucose, the F6P phosphoketolase (F6PPK) pathway (the so-called bifid shunt) (14, 59). The substrate for this pathway, F6P, links several metabolic routes to the bifid shunt, including N-acetyl hexose fermentation, galactose catabolism (the Leloir pathway), and peptidoglycan biosynthesis. A bioinformatic analysis of Bifidobacterium longum genome sequences revealed neither a fructose-specific PEP-PTS, which would deliver phosphorylated fructose to the cell, nor a complete sucrose utilization gene cluster. Thus, our hypothesis was that fructose is the sole hexose that is capable of incorporation into the bifid shunt by one single reaction leading to F6P (Fig. 1A). Consequently, the aim of this work was to identify the enzyme catalyzing the initial step of fructose metabolism and to demonstrate that this enzyme is necessary and sufficient to integrate fructose into the F6PPK pathway.
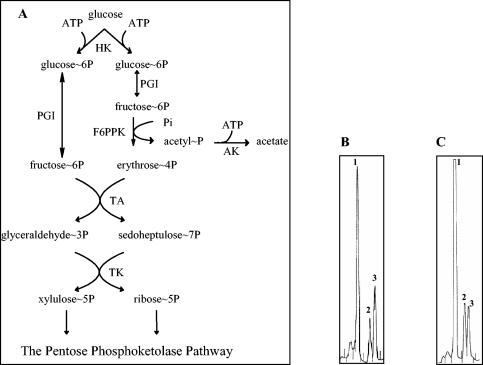
FIG. 1.
(A) Intermediates and enzymes of the F6PPK pathway for d-glucose catabolism in the genus Bifidobacterium. HK, hexokinase (EC 2.7.1.1); PGI, glucose-phosphate isomerase (EC 5.3.1.9); F6PPK, F6P phosphoketolase (EC 4.1.2. 9.); AK, acetokinase (EC 2.7.2.1); TA, transaldolase (EC 2.2.1.2); TK, transketolase (EC 2.2.1.1). HPAEC-PAD profiles of d-glucose (B) and d-fructose (C) fermentation by a crude extract of B. longum A10C are shown. Peaks of glucose-6-phosphate (1), F6P (2), and sedoheptulose-7-phosphate (3) are indicated.
MATERIALS AND METHODS
Bacterials strains, plasmids, and growth conditions.
The bacterial strains and plasmids used for this study are listed in Table 1. All B. longum strains were kindly provided by F. Gavini (INRA, Villeneuve d'Ascq, France). B. longum A10C and DSM20219 were used as sources of genomic DNA and for crude cell extract production. The Escherichia coli host strain BL21 (DE3) was obtained from Stratagene (La Jolla, Calif.).
TABLE 1.
Bacterial strains, plasmids, and oligonucleotides used for this study
| Strain, plasmid, or oligonucleotide | Relevant characteristics or nucleotide sequence (5′→3′) | Reference or source |
|---|---|---|
| Strains | ||
| B. longum DSM 20219 | Type strain | 46 |
| B. longum A 10C | Wild-type straina | This study |
| E. coli BL21 (DE3)/pLysS | F−ompT hsdSB(rB− mB−) gal dcm (DE3)/pLys (Cmr) | Stratagene; 50 |
| Plasmids | ||
| pET-28a(+) | Kanr LacI; expression vector; 5.4 kb | Novagen |
| pFrk | Kanr; 897-bp PCR-derived fragment from strain A10C in pET-28a(+); 6.3 kb | This study |
| Oligonucleotides | ||
| frkATG | GGAAAGGAAACCACATATGACTACCCCGbd | This study |
| frkTAA | GCTTGGGGTGGGATCCGATTACTTGCGcd | This study |
| glkATG | ATGAACAACAGGGGAGGACTGATG | This study |
| glkTAA | GCCCCGGCACTATGCCAGGCTACG | This study |
| Primer 1 | CGGGGTTGGTGATCAGCGAG | This study |
| Primer 2 | CGGCCCGGCCTATGATCGTG | This study |
| Primer 3 | CATCAGCATCGACCTGACGCAG | This study |
| Primer 4 | CGATGGCCTCGCCCTGTACG | This study |
| Primer 7 | CCGACGGTCTGGGACACGTCAATG | This study |
| Primer 8 | CCCGGCGCATGGGTGCCGTCCG | This study |
| Primer BILO1 | TCCAGTTGATCGCATGGTCde | 31 |
| Primer BILO2 | GGGAAGCCGTATCTCTACGAde | 31 |
This strain is able to ferment fructose as a unique carbon source (F. Gavini, personal communication).
The underlined sequence corresponds to an NdeI sites.
The underlined sequence corresponds to a BamHI restriction site.
Primer used for gene expression analysis (RT-PCR).
Primer specific to sequences within 16S rDNA of B. longum (31) and designed to produce a cDNA of 831 bp.
Unless otherwise indicated, B. longum strains were grown anaerobically at 37°C in Trypticase-Phytone-yeast medium as previously described (43). For RNA isolation and gene expression investigations, B. longum A10C cells were grown for 4 to 20 h in a modified Garches medium supplemented with hexoses (glucose or fructose, 2 g liter−1, or an equimolar mix of glucose and fructose containing 1 g of each liter−1). The modified Garches medium contained the following: 10 g of Casamino Acids (Difco) liter−1, 3.67 g of sodium acetate liter−1, 0.2 g of l-cysteine hydrochloride liter−1, 0.5 g of MgSO4 · 7H2O liter−1, 2.33 g of Na2HPO4 · 12H2O liter−1, 0.9 g of KH2PO4 liter−1, 10−3 g of p-aminobenzoic acid liter−1, 10−5 g of biotin liter−1, and 10−3 g of calcium pantothenate liter−1 (pH 6.1). As previously described (26), this is a semisynthetic medium that does not allow bifidobacterial growth without the addition of a carbon source. None of the B. longum strains used in this study were able to grow in control experiments performed on modified Garches medium without sugar. Stationary-phase cultures grown on the same medium were used as the inocula for this study. Bacterial growth was monitored spectrophotometrically at 600 nm.
E. coli was routinely grown at 37°C in Luria-Bertani medium (42). If required, the following supplements were added to the medium: kanamycin, 30 μg ml−1; 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal), 20 μg ml−1; and isopropyl-β-d-thiogalactopyranoside (IPTG), 240 μg ml−1 (all purchased from Sigma). All other chemicals were of analytical purity.
General DNA methods.
Total DNAs were obtained from B. longum strains as described by Tanaka and coworkers (51). DNA manipulations, including restriction enzyme analysis, ligation, and transformation, were done by standard methods (42) or according to the instructions supplied by the manufacturers of the reagents used. PCRs were routinely performed in a Master thermal cycler (Eppendorf, Hamburg, Germany). DNA sequencing was performed with Autoread Thermo Sequenase sequencing kits (Amersham Pharmacia Biotech, Roosendall, The Netherlands), Cy5 primers, and an ALF Express DNA sequencer (Amersham Pharmacia Biotech). The oligonucleotide primers used for this study are listed in Table 1. For Southern hybridization, B. longum DNA (4 μg per lane) was transferred from 0.7% (wt/vol) agarose gels to Hybond N+ membranes (Amersham Pharmacia Biotech). Probe labeling, hybridization, and washing were done with a digoxigenin DNA labeling and detection kit (Roche Diagnostics, Mannheim, Germany). The sizes of the hybridizing fragments of the chromosomal digestion products were determined by using digoxigenin-labeled ladder III (Roche Molecular Biochemicals).
The BLAST program was used for sequence homology searches of the National Center for Biotechnology Information GenBank database (http://www.ncbi.nih.nlm.gov) (1). The accession numbers of the two available B. longum genome sequences used for this study are NC_004307 (B. longum NCC2705 complete genome sequence) and NZ_AABM00000000 (B. longum DJO10A unfinished sequence from a whole-genome shotgun sequencing project).
Isolation of total RNA.
Total RNAs were extracted from B. longum cultures by use of an RNeasy mini kit (Qiagen), including treatment with a bacterial protecting reagent (Qiagen), as recommended by the manufacturer. Contaminating DNAs in the RNA preparations were digested with RNase-free DNase (Qiagen), and the RNA concentrations were subsequently determined spectrophotometrically at 260 nm.
Gene expression analysis by semiquantitative RT-PCR.
The relative levels of abundance of frk mRNAs under different conditions of cell growth were determined by semiquantitative reverse transcription-PCR (RT-PCR). Transcription was studied with a OneStep RT-PCR kit (Qiagen), with 5 ng of total RNA used as the template. The conditions were as follows: reverse transcription, 50°C for 30 min; initial PCR activation step, 95°C for 5 min; amplification, 25 to 40 cycles of 95°C for 1 min, 55°C for 1 min, and 72°C for 3 min; final extension step, 72°C for 3 min. Specific target RT-PCR products were normalized to an established endogenous internal control transcript (a primer pair designed to amplify a fragment of 831 bp from the B. longum 16S ribosomal DNA [rDNA] transcript), the expression of which is relatively constant in bacteria (12). The RT-PCR primers used for the detection of the 16S rDNA transcript were described by Matsuki and coworkers (31). The primers used for RT-PCR assays (Table 1) were designed to generate PCR products of comparable sizes. Negative controls were included and consisted of PCRs without each of the two sets of primers and Taq polymerase (Promega) to confirm the absence of contaminating DNAs in the RNA preparations. Aliquots (10 μl) from each reaction product were analyzed by electrophoresis in 2% agarose gels and were stained with ethidium bromide. The results were analyzed with Quantity One quantitation software (Bio-Rad) as previously described (9, 20).
Expression and purification of Frk protein.
The putative frk gene was amplified by PCR from B. longum A10C genomic DNA with primers frkATG and frkTAA, which contained an NdeI and a BamHI restriction site, respectively (Table 1). The amplicon was cleaved and inserted between the same two sites (NdeI-BamHI) in the expression vector pET28-a(+) (Novagen, Madison, Wis.), creating an in-frame fusion between the 5′ end of the putative frk gene and the six histidine codons. The resulting plasmid, pFrk, was transformed into E. coli BL21 (DE3). The presence of the correct insert and orientation was confirmed by restriction enzyme digestion and sequencing.
The recombinant His-tagged Frk protein was overproduced in cells growing exponentially at 37°C in Luria-Bertani medium containing kanamycin (50 μg ml−1). At an optical density at 600 nm of 1.0, IPTG was added to a final concentration of 1 mM. Incubation was continued for 2 h, after which the cells were harvested by centrifugation. The cells were resuspended in 50 mM sodium phosphate buffer (pH 8)-0.3 M NaCl and sonically disrupted, followed by removal of the cellular debris by centrifugation. The supernatant was applied to a 10-ml cobalt-based affinity column (BD Biosciences, Palo Alto, Calif.) equilibrated in sonication buffer, and the column was washed with 10 volumes of the same buffer. The Frk protein was eluted with 150 mM imidazole in sonication buffer and subsequently dialyzed against sonication buffer to remove the imidazole. The removal of the six-histidine residue tag was performed with a thrombin cleavage and capture kit (Novagen) according to the manufacturer's instructions. The resulting peptide was removed by ultrafiltration through a 10-kDa membrane (Amicon, Beverly, Mass.).
Preparation of B. longum cell extracts.
All procedures were performed at 4°C. Exponential-phase cultures were harvested by centrifugation (10,000 × g for 20 min). The cells were washed twice in 50 mM phosphate buffer (pH 7) and then resuspended at a concentration of 2 mg ml−1 (wet weight) in the same buffer. The cells were then disrupted on ice by a sonic treatment (15 min; duty cycle, 30%). Subsequently, the suspension was centrifuged at 10,000 × g for 10 min to remove nondisrupted cells, and the resulting supernatant was centrifuged at 20,000 × g for 20 min to pellet the cell debris. The supernatants (cell extracts) were filter sterilized and assayed for enzyme activity. Protein concentrations in the cell extracts were determined by the method of Bradford, with bovine serum albumin as a standard (8).
Fructokinase assays and activity unit definition.
Two procedures were used to measure fructokinase activity. Method I was the spectrometric method of Sebastian and Asensio (45), which couples the formation of F6P to the reduction of NADP+ via glucose phosphate isomerase (PGI; EC 5.3.1.9) and glucose-6-phosphate dehydrogenase. The standard assay mixture (200 μl at 50°C) contained 100 mM sodium phosphate, 2.7 mM fructose, 10 mM ATP, and 10 mM MgCl2 (pH 6.5). After preincubation at 50°C, the reaction was started with the addition of an aliquot of fructokinase (purified protein). This mixture was incubated for 5 min, and the reaction was then stopped by boiling at 80°C for 5 min. The F6P concentration was quantified by the addition of 3 mM NADP+, 0.2 U of PGI, and 1 U of glucose-6-phosphate dehydrogenase in 100 mM Tris-HCl (pH 7.8) to a final volume of 1 ml and a subsequent incubation at room temperature. The reduction of NADP+ at 340 nm was detected 10 min later. This assay was used throughout the entire study. Unless otherwise specified, enzyme activity is expressed in international units (micromoles of F6P formed per minute) and specific activity is expressed in international units per microgram of protein.
Method II involved a high-performance anion-exchange chromatography (HPAEC) technique that was used to identify the phosphorylated sugars produced during the incubation of fructose with a B. longum cell extract. The same method allowed us to investigate the total fructokinase activity in B. longum A10C cells grown in modified Garches medium in the presence of various carbohydrates and to measure the amount of nonfermented sugars in the supernatants of B. longum A10C cultures. This analytical system consisted of a Bio-LC GPM-II quaternary gradient module (Dionex Corporation, Sunnyvale, Calif.) associated with an eluent degas (He) module and a Dionex Carbopac PA-100 column (4 by 250 mm) in combination with a Carbopac PA-100 guard column (3 by 25 mm). The intermediary products of fructose catabolism were eluted in the order of increasing negative charges and were analyzed by use of the following gradient of sodium acetate in 0.1 M NaOH: 0 to 5 min, 0 M; 5 to 25 min, 0 to 0.5 M; and 25 to 35 min, 0.5 to 1 M. All gradients were terminated with 1 M sodium acetate for 5 min, followed by reequilibration with 0.1 mM NaOH for 15 min. The effluent (flow rate of 1 ml min−1) was monitored with a Dionex PED detector containing a gold electrode with an Ag-Ag-Cl reference electrode in the pulsed amperometric detection (PAD) mode. Routine assays were performed at 37°C with 1 g of substrate (glucose or fructose) liter−1 in 25 mM imidazole buffer (pH 6.5) containing 5 mM MgCl2 and 10 mM ATP. Boiling of the incubation mixture for 5 min stopped the reactions. The phosphorylated sugars were identified by using standards (d-fructose, F6P, fructose-1-phosphate, d-glucose, glucose-6-phosphate, glucose-1-phosphate, fructose-1,6-bisphosphate, sedoheptulose-7-phosphate, imidazole, and ATP). F6P formed during the incubation of fructose with a B. longum crude extract was quickly transformed by the bacterial PGI (Fig. 1) to glucose-6-phosphate, which accumulated in the reaction mixture. Thus, for experiments with the B. longum crude extract, the fructokinase specific activity was expressed in micromoles of hexose-6-phosphate (F6P plus glucose-6-phosphate) formed per minute per microgram of protein at 37°C.
Frk substrate specificity.
The Frk carbohydrate substrate specificity was investigated by thin-layer chromatography (TLC) and by HPAEC. TLC was performed on Silica Gel 60 plates (Merck, Darmstadt, Germany) with a solvent system composed of butanol, acetic acid, and water (2:2:1 [vol/vol/vol]). The sugars were visualized after being sprayed with an orcinol-sulfuric acid solution and heated at 120°C (60). Standards of d-glucose, d-fructose, d-sucrose, l-arabinose, d-mannose, d-xylose, F6P, and glucose-6-phosphate (Sigma) were used for comparative purposes. The nucleotide substrates used for specificity assays (ATP, GTP, ITP, CTP, and TTP) were each supplied at a concentration of 10 mM, and fructokinase activity was examined as described for method I.
pH and temperature optima of Frk.
The optimal pH was determined at 50°C in 100 mM citrate buffer over a pH range of 3 to 5.5 and in 100 mM sodium phosphate buffer over a pH range of 5 to 9. The effect of temperature on the activity was determined by measuring the enzyme activity at different temperatures ranging from 6 to 80°C. Thermal stability was determined by maintaining the enzyme solution in 20 mM Tris-HCl buffer (pH 7.8) for 15 min at various temperatures (from 6 to 60°C). The residual activity was estimated by standard procedures (method I).
Kinetic parameters of Frk.
Kinetic parameters were studied with a range of fructose concentrations (0.28 to 14 mM) plus 10 mM ATP and with a range of ATP concentrations (0.6 to 16 mM) plus 10 mM fructose. Activities were estimated by standard procedures (method I). The kinetic constants Km and Vmax were calculated from Hanes-Wolff plots.
Nonfermented carbohydrate analysis.
For measurements of the amounts of nonfermented hexoses in the supernatants of B. longum A10C cultures, samples were taken at regular time intervals from the bacterial suspension and were immediately cooled on ice to prevent further metabolic conversion. The samples were then centrifuged, and filtered supernatants were analyzed by HPAEC-PAD as described above.
RESULTS
Degradation of glucose and fructose by B. longum.
HPAEC was used to determine the profile of glucose and fructose transformation by a crude extract of B. longum A10C. A glucose degradation profile analysis (Fig. 1B) allowed the identification of three intermediates of the bifid shunt: glucose-6-phosphate, F6P, and sedoheptulose-7-phosphate. These results agree with the previously reported ability of bifidobacteria to ferment glucose via the hexose-6-phosphate phosphoketolase pathway (14, 59) (Fig. 1A). The same intermediate metabolites were identified (Fig. 1C) when the fructose degradation profile was analyzed, indicating that intracellular fructose and glucose are catabolized via the same degradation pathway. This observation points to F6P as one of the first metabolites produced during fructose fermentation and strongly suggests that a fructokinase is involved in fructose catabolism.
In both experiments, the main peak corresponded to glucose-6-phosphate, confirming that the reaction catalyzed by the glucose phosphate isomerase (EC 5.3.1.9) is reversible and that the equilibrium lies toward the left (glucose-6-phosphate) as long as the amount of F6P exceeds the amount of glucose-6-phosphate formed.
Identification and sequence analysis of frk and its genomic context.
In this study, we addressed the possibility that a fructokinase (namely, Frk) may be responsible for the incorporation of intracellular fructose into the F6PPK pathway and analyzed the two available B. longum genome sequences (DJO10A and NCC2705) for the presence of a fructokinase-encoding gene.
One putative fructokinase-encoding gene was revealed by bioinformatic analysis. This 894-bp open reading frame (ORF) is located at 80 min on the bacterial chromosome and corresponds to genes BL1339 (accession no. NP_696503.1) and 1774 (accession no. ZP_00120666.1) on the B. longum NCC2705 and DJO10A genomes, respectively. This DNA fragment encodes a predicted protein of 298 amino acids with a calculated molecular mass of 32.5 kDa. A database inquiry (BLASTP) identified it as a PfkB family carbohydrate kinase, sharing only 34% identity with a putative fructokinase from the archaebacteria Methanosarcina acetivorans.
Cloning of frk from B. longum A10C and protein purification.
The ORF encoding the putative fructokinase Frk was amplified by a PCR with B. longum A10C genomic DNA as a template. The amplicon was cloned between the NdeI and BamHI sites in the plasmid pET28a(+) and then was sequenced. A comparison between the B. longum A10C frk sequence and the sequences obtained from the two other B. longum strains, namely, NCC2705 and DJO10A, was carried out. Alignments indicated 97.5 and 97.2% DNA sequence identities with the NCC2705 and DJO10A frk sequences, respectively. Most of the base substitutions had no consequences on the protein's primary sequence. The A10C Frk protein shares 97.7% (seven amino acid substitutions) and 97.3% (eight amino acid substitutions) amino acid identity, respectively, with the NCC2705 and DJO10A predicted proteins.
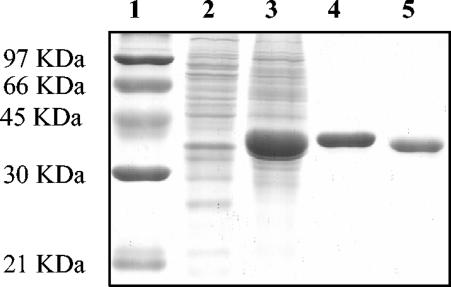
In order to further characterize this enzyme, we overexpressed the protein as an N-terminal histidine-tagged fusion and purified it by Co2+ affinity chromatography (Fig. 2). The overall yield from a 200-ml culture was 10 mg of pure Frk. The molecular mass estimated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis was 35 kDa (Fig. 2). This compares well with the predicted mass (32.4 kDa) calculated from the protein's primary sequence (Mwcalc [www.infobiogen.fr]).
FIG. 2.
Purification of the fructokinase Frk. Samples (1 μg of protein) were separated in a sodium dodecyl sulfate-10% polyacrylamide gel and stained with Coomassie brilliant blue R-250. Lane 1, molecular mass standard; lane 2, negative control [a crude extract of E. coli BL21 (DE3) containing pET28a(+)]; lane 3, crude extract of IPTG-induced cells of E. coli BL21 (DE3) containing pFrk; lane 4, eluate with sonication buffer containing 150 mM imidazole from cobalt-based affinity column; lane 5, purified Frk after elimination of the six-His tag by thrombin cleavage.
Biochemical characteristics of recombinant Frk activity. (i) pH and temperature optima and thermal stability.
The characteristics of the purified Frk protein were determined by using a coupled spectrophotometric assay as described in Materials and Methods. The specific activity was linear for protein concentrations in the range of 0.04 to 0.3 μg ml−1. The optimum pH was slightly acidic (6.0), with a marked decrease in fructokinase activity at pH values below 5.0 and above 8.0.
For the temperature range of 6 to 80°C, Frk exhibited the highest activity at 50°C. The thermostability of the purified enzyme was determined at 6 to 80°C in 0.1 M potassium phosphate buffer (pH 6.5) containing 10 mM ATP and 10 mM MgCl2. The Frk stability gradually decreased from 6 to 40°C, with a residual enzyme activity of about 70% after 15 min of incubation at 37 to 39°C, which corresponded to the bacterial growth temperature. At 50°C, the residual activity of the enzyme was 55% after 15 min of incubation, whereas the residual activity at 55°C was <20%. A complete lose of enzyme activity was observed within 15 min when it was incubated at 60°C or above.
(ii) Substrate specificity.
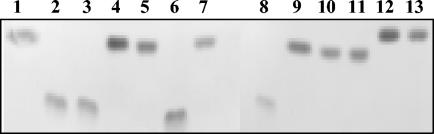
The carbohydrate (acceptor) and nucleotidic (donor) substrate specificities of the purified enzyme were determined. Several carbohydrate substrates that are likely to be phosphorylated by a hexokinase (d-fructose, d-glucose, d-mannose, d-xylose, l-arabinose, and sucrose) were investigated by TLC. No phosphotransferase activity was observed when assays were performed with carbohydrates other than d-fructose (Fig. 3). The unique specificity of Frk for d-fructose was also confirmed by HPAEC-PAD (data not shown).
FIG. 3.
TLC analysis of Frk carbohydrate substrate specificity. d-Fructose (lane 1), F6P (lane 2), d-mannose (lane 4), mannose-6-phosphate (lane 6), d-glucose (lane 7), glucose-6-phosphate (lane 8), and sucrose (lane 12) were used as standards (5 μg/lane). The phosphorylated carbohydrates formed during the incubation of Frk with d-fructose (lane 3), d-mannose (lane 5), d-glucose (lane 9), l-arabinose (lane 10), d-xylose (lane 11), and sucrose (lane 13) are shown.
The potential of five ribonucleoside triphosphates to act as nucleotidic substrates for B. longum Frk was investigated by using the enzyme-coupled assay. The γ-phosphoryl residue donors tested decreased in efficiency in the following order: ATP (100%) > ITP (78.24%) > GTP (23.01%) > CTP (6%) > TTP (0.18%).
(iii) Catalytic properties.
The purified enzyme exhibited typical Michaelis-Menten kinetics when assayed with increasing concentrations of the investigated substrate (d-fructose or ATP). An excess of ATP (>12 mM) had an inhibitory effect on the enzymatic activity. Therefore, we chose 10 mM as the optimum ATP concentration for the study of enzyme kinetics.
Kinetic analysis with the Hanes-Wolf equation indicated that the purified fructokinase had an apparent Km for fructose of 0.739 ± 0.18 mM and a Vmax of 0.84 ± 0.12 U mg of protein−1. The apparent Km for ATP was 0.756 ± 0.08 mM, and the Vmax for this substrate was 1.8 ± 0.11 U mg of protein−1. The Km values for d-fructose and ATP were quite similar, indicating that the enzyme displays the same affinity for both substrates.
Regulation of frk expression in response to the carbon source. (i) Semiquantitative measurement of frk transcription.
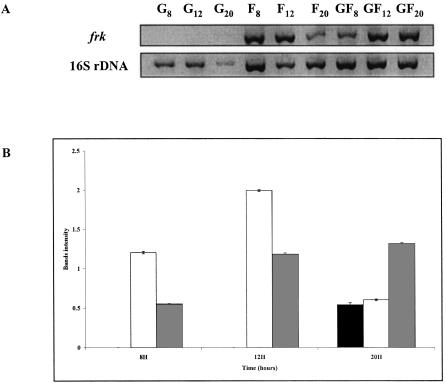
In order to investigate the involvement of the carbohydrate substrate on frk expression, we cultivated B. longum A10C in modified Garches medium containing either d-glucose (2 g liter−1), d-fructose (2 g liter−1), or a mix of fructose and glucose (1 g liter−1 of each) as the sole carbon source. The optical densities of the cultures were monitored spectrophotometrically, and carbohydrate utilization profiles were determined. In parallel, total RNAs were isolated from samples taken after 8, 12, and 20 h of growth and were used as templates for frk expression analyses by RT-PCR. Comparisons with an internal control (16S rDNA) provided a semiquantitative measurement of the level of frk expression. For each set of primers (frk and 16S rDNA), the number of PCR cycles was defined in order to ensure specific amplification under unsaturated conditions (data not shown). Consequently, 16S rDNA transcripts were analyzed after 25 cycles, while frk transcripts were analyzed after 40 cycles of amplification. The intensities of the bands, as measured with Quantity One software (Bio-Rad), were close approximations of their relative abundance.
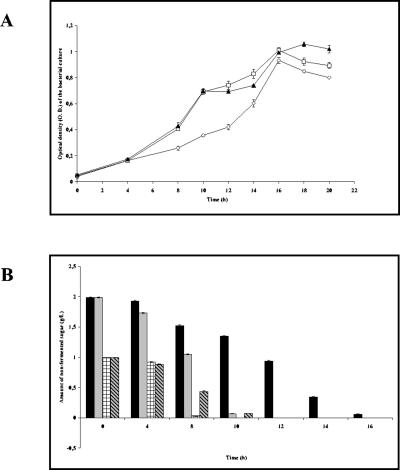
An analysis of the bacterial growth curves and the corresponding carbohydrate consumption profiles (Fig. 4) showed that in all cases, the substrate was totally fermented within 16 h of culture, indicating the end of the exponential growth phase.
FIG. 4.
Profile of carbohydrate utilization by B. longum A10C throughout the growth curve. (A) B. longum A10C growth curve. Cells were cultivated in modified Garches medium supplemented with glucose (diamonds), fructose (squares), or an equimolar mix of the two hexoses (triangles) for the indicated times. (B) Amounts of nonfermented sugars, as measured by HPAEC-PAD. Black bars, d-glucose; gray bars, d-fructose; checked bars, d-glucose in mixed GF medium; hatched bars, d-fructose in mixed GF medium.
RNA templates isolated after 8, 12, and 20 h of culture from cells grown in the presence of glucose provided specific bands derived from the 16S rDNA transcript (Fig. 5A), while no significant specific bands relating to the frk transcript could be detected by agarose gel electrophoresis (Fig. 5A). In contrast, both RT-PCR products were obtained when RNAs from cells grown in the presence of fructose were used as the template (Fig. 5A). For comparisons, the calculated ratios of frk to 16S rDNA (RT-PCR band intensities, in arbitrary units) were determined as a function of the carbohydrate substrate and time of growth. As depicted in Fig. 5B, the results of these calculations demonstrate a higher level of frk transcription in cells grown with fructose than in those grown with glucose, suggesting that frk expression is subject to glucose-mediated repression and/or fructose induction.
FIG. 5.
RT-PCR analysis of frk transcripts produced in B. longum A10C cells grown on different carbohydrate sources. (A) Primers specific for frk sequence and 16S rDNA were used to amplify fragments by RT-PCR. (B) Calculated ratios of frk to 16S rDNA (RT-PCR band intensities of specific products). Total RNAs were isolated from B. longum A10C cultivated for 8, 12, and 20 h on modified Garches medium containing 2 g of the following monosaccharides liter−1: glucose (G; black bars), fructose (F; white bars), or an equimolar mix of glucose and fructose (GF; gray bars).
To investigate this further, we performed the same experiment with B. longum A10C cells grown in modified Garches medium supplemented with an equimolar mix of glucose and fructose (denoted GF) (Fig. 5). In this case, in contrast to the case for glucose-grown cells, frk transcripts were detected after 8, 12, and 20 h of growth (Fig. 5B). Since after 8 h of growth both hexoses were present in the GF medium (0.03 g of glucose liter−1 and 0.43 g of fructose liter−1) (Fig. 4), the detection of an RT-PCR product specific for the frk transcript must mean that there is a significant increase in the quantity of frk mRNA and thus a fructose induction of frk expression (Fig. 5A). On the other hand, the calculated frk-to-16S rDNA ratio (Fig. 5B) for the 8-h GF culture was lower (0.55) than that for the corresponding cells grown in fructose (1.20). These data demonstrate the glucose-mediated catabolite repression of frk transcription.
(ii) Quantification of Frk activity.
Independent of the transcriptional approach, we estimated the level of Frk biosynthesis by using a biochemical method. For this purpose, B. longum A10C was cultivated in modified Garches medium as described above. The amount of residual sugar was measured after 16 h of bacterial growth. The cells were harvested during the late logarithmic phase (16 h) and lysed by sonication, and the three crude extracts were independently assayed for Frk activity by the HPAEC-PAD method (see Materials and Methods). Fructokinase activity, expressed as the sum of the two detected phosphorylated sugar peak areas (glucose-6-P and F6P), reflects the rate of Frk biosynthesis.
After 16 h of growth, sugar consumption was complete. Cells grown in glucose displayed the lowest Frk activity (0.26 ± 0.03 U mg of protein−1), while the presence of fructose as a unique carbon source led to a sevenfold increase in fructokinase activity (1.55 ± 0.10 U mg of protein−1). An intermediate Frk activity (4.5-fold induction) was obtained when the cells were grown with the mix of the two sugars (1.19 ± 0.17 U mg of protein−1). Taken together, these data confirm the fructose induction and glucose-mediated repression of Frk synthesis.
Analysis of a fructose-negative strain. (i) Mutant isolation.
An analysis of the fermentation pattern of 200 B. longum strains isolated from human feces revealed that 20% of them are unable to grow on fructose as a sole carbon source (i.e., they are fructose negative) (F. Gavini, personal communication). To confirm that the fructokinase Frk is the sole enzyme allowing the incorporation of fructose into the bifid shunt, we screened a set of 13 B. longum strains to isolate a fructose-negative mutant. For this purpose, these 13 strains were cultivated in modified Garches medium containing fructose (2 g liter−1) as the sole carbon source, and bacterial growth was monitored after 24 to 48 h (data not shown). In addition, the presence of frk was analyzed by PCRs performed with the genomic DNAs (data not shown). Only three strains displayed a fructose-negative phenotype, of which only one (the type strain DSM20219) did not yield a frk PCR product (data not shown), thereby suggesting a deletion event.
(ii) Mapping of the deletion.
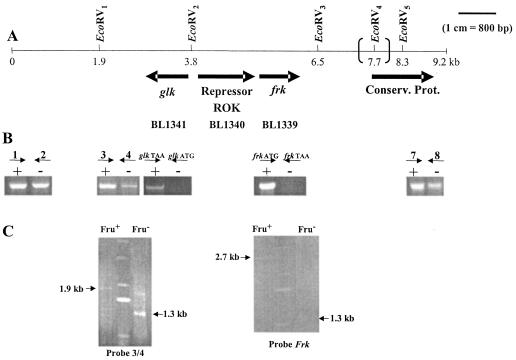
The mutant strain DSM20219 was thus chosen for further characterization. The genomic context surrounding the frk loci of the A10C and DSM20219 strains was analyzed by PCR and Southern blotting and compared to the two available genome sequences (DJO10A and NCC2705). The genetic organization of the locus surrounding frk differs slightly between the DJO10A and NCC2705 genome sequences. In NCC2705, the frk gene is flanked downstream by an ORF encoding an unidentified conserved protein and upstream by an ORF encoding a putative transcriptional regulator of the ROK (transcriptional regulators, ORF, kinases) family (Fig. 6A). A putative glucokinase-encoding gene (glk) reading in the opposite orientation to that of frk was also detected 1.5 kb upstream of the frk locus (Fig. 6A). The major difference between the two genome sequences consists of the presence of an additional 1,119-bp noncoding sequence separating frk from the downstream ORF in the DJO10A genome.
FIG. 6.
Genetic context of frk locus in two B. longum strains (NCC2705 and DJ010A) and genotype analysis of the fructose-negative strain DSM20219. (A) EcoRV restriction map of a 9.2-kb fragment of the chromosome of B. longum NCC2705 surrounding the frk locus. The additional 1,119-bp noncoding sequence separating frk from the downstream ORF on the DJO10A genome and the additional EcoRV4 site are indicated with large parentheses. (B) Analysis by agarose gel electrophoresis of amplicons generated by PCRs performed with fructose-positive A10C (+) and fructose-negative DSM20219 (−) genomic DNAs. The following sets of primers were designed to amplify DNA fragments located around the frk locus: 1-2 (861 bp), 3-4 (768 bp), glkATG-glkTAA (908 bp), frkATG-frkTAA (894 bp), and 7-8 (784 bp). (C) (Left) Southern blot of EcoRV-digested DNA from the A10C (Fru+) and DSM20219 (Fru−) strains hybridized with probes 3 and 4. (Right) Southern blot of EcoRV-digested DNA from the A10C (Fru+) and DSM20219 (Fru−) strains hybridized with the frk probe. Arrows indicate the specific bands detected with probes amplified from B. longum A10C genomic DNA. On both gels, the central lane corresponds to DNA ladder X (Roche).
PCRs and Southern blotting were conducted with five different sets of primers, namely 1-2, 3-4, glkATG-glkTAA, frkATG-frkTAA, and 7-8 (Fig. 6B and C). For these experiments, the genomic DNA of the wild-type strain A10C served as a positive control. Only two sets of primers (glkATG-glkTAA and frkATG-frkTAA) failed to yield an amplification product when PCRs were performed with the mutant strain DNAs (Fig. 6B). Taken together, these five PCR profiles allowed us to delineate a deleted region of 5.7 kb located between the 3′ end of primer 4 and the 5′ end of primer 7.
Southern blots performed with DSM20219 genomic DNA restricted by EcoRV revealed the presence of a common 1.3-kb fragment that was detected with both the frk and the 3-4 probes. In the case of the A10C strain, two separate restriction fragments, of 1.9 and 2.7 kb, were revealed by the 3-4 and frk probes, respectively. These data indicate that one EcoRV restriction site (named EcoRV2) (Fig. 6) is missing from the investigated DNA sequence of the mutant strain. Thus, the previously identified 1.3-kb fragment must correspond to the entire B. longum DSM20219 DNA sequence comprised of the region between the two other EcoRV sites (EcoRV1 and EcoRV3). Therefore, the estimated deleted region has a length of 3.3 kb and includes frk, glk, and an ORF encoding a putative ROK repressor.
(iii) Enzymatic complementation.
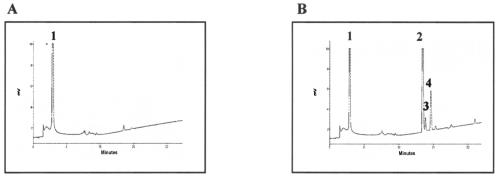
Concomitant to this genomic study, the ability of a B. longum DSM20219 crude extract to phosphorylate fructose was assayed in vitro by HPAEC-PAD. An analysis of the chromatographic profile (Fig. 7A) showed no phosphorylated products, indicating that fructose is not a substrate for any enzyme present in the lysate of the B. longum mutant strain.
FIG. 7.
(A) HPAEC-PAD analysis of fructose degradation by B. longum DSM20219 crude extract. (B) HPAEC-PAD analysis of fructose degradation by B. longum DSM20219 crude extract supplemented with 0.1 U of purified Frk. Peaks corresponding to d-fructose (1), glucose-6-phosphate (2), F6P (3), and sedoheptulose-7-phosphate (4) are indicated.
The same experiment was performed after the addition of 0.1 U of purified Frk to the DSM20219 crude extract. An examination of the HPAEC-PAD profile (Fig. 7B) indicated not only the expected peak related to F6P, but also two other bifid shunt phosphorylated intermediates (glucose-6-P and sedoheptulose-7-P). Complementation of a B. longum DSM20219 crude extract with Frk was sufficient to allow the complete degradation of fructose in vitro.
DISCUSSION
The ability of Bifidobacterium spp. to survive and persist in a competitive environment (i.e., the mucosal layer of the human gastrointestinal tract) is correlated with particular genetic features, such as numerous predicted exo- and endo-glycosyl hydrolases, high-affinity oligosaccharide transporters (degree of polymerization, <8), and the predominant use of negative transcriptional regulation as a flexible control mechanism in response to nutrient availability and diversity (43). Bioinformatic analyses complement biochemical data reporting the purification and characterization of Bifidobacterium sp. enzymes that are capable of hydrolyzing various glycosidic bonds from plant-derived carbohydrates and are often inducible by the released monomeric molecules (15, 22, 29, 33, 41, 48, 56, 58, 63). Moreover, several studies conducted on fructose-containing polymers as potential selective substrates for colonic bacteria have provided evidence that bifidobacteria are able to ferment these carbohydrates, in particular the short linear chains of β-2-1-linked fructosyl units (7, 24, 30, 36, 37). Indeed, three intracellular β-fructofuranosidases [(β-2-1) fructan hydrolases] that are capable of degrading inulin and oligofructose have been characterized in Bifidobacterium spp. (15, 22, 63). Recently, bfrA, a gene encoding an intracellular β-fructofuranosidase from Bifidobacterium lactis, was cloned and characterized (15). The encoded enzyme also displays a prominent invertase activity, allowing the internal release of d-fructose from sucrose (15). Interestingly, the same species was reported to possess a second sucrose-hydrolyzing enzyme (sucrose phosphorylase; EC 2.4.1.7) that supplies the bacterial cell with α-d-glucose-1 phosphate and d-fructose (56). Although analyses of fructose fermentation patterns revealed that B. longum can use d-fructose as a sole carbon source (reference 36 and this work), the events following the uptake of fructose into the cell remain to be defined.
To address this, we wanted to identify the catabolic route allowing d-fructose fermentation. For this purpose, the first product resulting from d-fructose degradation by B. longum A10C crude extracts was revealed by HPAEC-PAD profiling. This resulted in the detection of F6P, glucose-6-phosphate, and sedoheptulose-7-phosphate, but neither fructose-1-phosphate nor fructose-1,6-diphosphate could be detected by this technique (Fig. 1C). The same intermediary products were identified when the d-glucose fermentation pattern was analyzed (Fig. 1B). These results led to two preliminary conclusions: (i) the presence of three intermediates of the bifid shunt, corroborated with the absence of fructose-1,6-diphosphate, indicates that fructose is assimilated via the F6PPK pathway; and (ii) fructose is converted to F6P by one or several enzymatic reactions.
To address the hypothesis that d-fructose is incorporated into the bifid shunt through a one-step transformation catalyzed by a fructokinase, we cloned a putative fructokinase gene from B. longum A10C and characterized the activity of the expressed protein. Thus, the two sequenced genomes of B. longum (DJO10A and NCC2705) were screened for the presence of genes that putatively encode a fructokinase, leading to the identification of one ORF of interest. The amino acid sequence of the fructokinase characterized in this work, as deduced from the DNA sequence, differs at eight amino acids from that predicted for B. longum DJO10A. Among these amino acid changes, three have no effect on the global protein charge (L69 for I69, V168 for A168, and L201 for I201), while three other residue modifications correspond to an increase in the global negative charge due to the replacement of three neutral amino acids (N61, G205, and Q284) with acidic residues (D61, D205, and E284). One amino acid change (Y273 to F273) results in the loss of one potential hydrogen bond. The last substitution concerns a histidine residue (H234) which is replaced with another positively charged amino acid (R234). Although a nucleotide replacement rate of 2.7% (i.e., 97.3% nucleotide identity) within the same species could be considered nonnegligible, this variation is not superior to the global polymorphism displayed by the investigated A10C strain, which shares 85% DNA identity with the B. longum type strain DSM20219 (by DNA-DNA hybridization [F. Gavini, personal communication]).
In order to investigate the Frk involvement in B. longum d-fructose catabolism, we consequently synthesized the protein encoded by the frk ORF. The purified Frk protein was assayed for substrate specificity, and its kinetic parameters were determined. Frk displayed phosphotransferase activity only toward d-fructose, and none of the other investigated hexoses (d-glucose and d-mannose), pentoses (d- and l-arabinose and d-xylose), or the disaccharide sucrose could serve as an acceptor substrate. Moreover, Frk showed a preference for nucleotides derived from purines (ATP, ITP, and to a lesser extent, GTP) as γ-phosphate group donors. Our work also demonstrated that Frk displays a higher affinity for ATP than for other nucleoside triphosphates. Interestingly, an inhibition of Frk activity was noted when the ATP concentration exceeded 12 mM. This result led to the assumption that the enzyme and ATP could form an abortive ternary complex blocking the enzymatic reaction by modification of either the active catalytic conformation or the affinity of the enzyme for d-fructose (4, 25). Enzymatic parameter analyses performed with respect to d- fructose and ATP indicated that Frk has the same affinity for both substrates (Km values of 0.739 ± 0.18 and 0.756 ± 0.08 mM for fructose and ATP, respectively), suggesting a possible BiBi random mechanism. Thus, B. longum A10C Frk is an ATP-dependent phosphotransferase that is highly specific for fructose.
Bacterial fructokinases are usually mannofructokinases (45, 49, 52, 53, 54, 65). Indeed, these enzymes convert both d-fructose and d-mannose to F6P and mannose-6-P, respectively. Therefore, these organisms employ a single kinase to integrate two different hexoses into the same catabolic route. The enzyme purified from a crude extract of Streptomyces violaceoruber constitutes the only example until now of a bacterial fructokinase that is highly specific for d-fructose (39). Consequently, this work presents the first characterization at the molecular and biochemical level of a fructokinase from a gram-positive bacterium that is highly specific for d-fructose.
Interestingly, B. longum A10C is also capable of metabolizing mannose (data not shown), which suggests that this hexose must be either isomerized to d-fructose (by a mannose isomerase; EC 5.3.1.8) or phosphorylated by an as yet unidentified carbohydrate kinase prior to integration into a degradation pathway. Schell and coworkers made the observation that B. longum possesses separate genetic modules dedicated to specific sugar catabolism (44). From an evolutionary point of view, one can speculate that possessing kinases displaying high specificities for different carbohydrate substrates could be a selective advantage for bifidobacteria to survive in a competitive environment containing various carbon sources.
The possible involvement of additional enzymes in the assimilation of fructose by the F6PPK pathway was also assessed in this study by use of an frk mutant. As expected, this mutant displayed no peaks related to d-fructose degradation (Fig. 7A), confirming that a deficiency of the fructokinase Frk leads to the absence of fructose degradation. Consequently, B. longum requires a functional fructokinase in order to metabolize this monosaccharide. Furthermore, enzymatic complementation of the mutant crude extract with purified Frk provided an HPAEC-PAD profile identical to that obtained for fructose-positive strains (Fig. 1C). Thus, the fructokinase Frk characterized in this work is sufficient to directly integrate fructose into the F6PPK pathway.
Genes involved in carbohydrate utilization are rarely constitutively expressed in bacteria (47). Generally, in terms of the regulation of gene expression, two mechanisms direct carbohydrate utilization, i.e., catabolite repression (suggesting the existence of a preferential carbon source) and/or specific induction by the gene product's substrate(s) (36, 37, 61, 62). Therefore, in the present study, we also examined the involvement of these two regulatory mechanisms in Frk synthesis.
Analyses of frk expression as a function of the carbohydrate source were performed by two complementary approaches, specifically semiquantitative RT-PCR and the quantification of enzymatic activity by HPAEC-PAD. The data presented here show differences in response to the carbon source at the levels of both frk expression (RT-PCR) and Frk synthesis (HPAEC-PAD), suggesting that frk is subject to regulation.
The induction of frk by fructose was visible after 8 h of growth in GF medium. Indeed, at this point, there was a sufficient amount of glucose remaining in the medium to exert catabolite repression. Thus, the level of frk mRNA, as estimated by semiquantitative RT-PCR, can be explained only by the induction of frk expression by fructose. Consequently, fructose is an inducer of fructokinase synthesis. Interestingly, d-fructose was also reported to be the best inducer of β-fructofuranosidase biosynthesis in Bifidobacterium infantis (36). Therefore, in addition to the role of d-fructose as an inducer of enzymes involved in the hydrolysis of d-fructose-containing polymers, it also acts as an inducer of d-fructose catabolism. Schell and coworkers (44) proposed a binary model for the regulation of genes involved in carbohydrate utilization in B. longum. Based on bioinformatic analysis, these authors predicted the existence of genetic modules, with each one including enzyme-encoding genes together with their own regulator (44). Interestingly, an ORF encoding a putative transcriptional regulator of the ROK family is located directly upstream of the frk locus (Fig. 6A) and is possibly cotranscribed with frk.
In support of the involvement of catabolite repression in frk expression, we provide evidence that after 8 h of culture, the level of frk expression differed in relation to the carbohydrate source offered in the medium (GF or fructose only) (Fig. 5). As discussed above, since glucose is still present in the GF medium after 8 h, the amount of frk transcript must reflect fructose induction. Hence, the highest level of frk transcription, as estimated for cells grown in fructose, reflected the absence of catabolite repression. This interpretation was confirmed by the results obtained for the 20-h culture, as the cells grown in GF medium displayed an increased level of frk transcription as soon as glucose was no longer available in the culture. Consequently, this work also demonstrates the d-glucose-mediated catabolite repression of frk gene expression in B. longum A10C.
The catabolite repression of genes involved in carbohydrate metabolism has already been reported for Bifidobacterium spp. (56). Nevertheless, the mechanism by which this catabolite repression occurs remains to be determined. In low-GC-content gram-positive bacteria, catabolite control protein A (CcpA) is the central regulator of catabolite repression (19, 34, 57). CcpA has been shown to bind to specific sequences named catabolite-responsive elements (CRE) located near the promoter regions of genes affected by catabolite repression. However, an analysis of the two available B. longum genome sequences revealed neither a CcpA-encoding gene nor a CRE sequence within 2 kb upstream of the frk gene. Another important factor in the catabolite control mechanism is the PTS phosphocarrier HPr (13, 18, 21, 23). However, recent works conducted on high-GC-content bacteria (e.g., Streptomyces coelicolor) showed that the presence of HPr is not necessary for general carbon regulation (10, 35). In both high-GC (S. coelicolor) and low-GC-content (Staphylococcus xylosus and Bacillus megaterium) gram-positive bacteria, a central function in catabolic repression was attributed to a ROK family glucose kinase, namely GlkA (2, 11, 27, 55, 61, 62). Interestingly, in B. longum (strains DJ010A and NCC2705), a gene putatively encoding a glucokinase (EC 2.7.1.2) is located 1.5 kb upstream of the frk locus. In fact, the two genes (glk and frk) are simply separated by the gene encoding a putative regulator of the ROK family (Fig. 6A). Based on this particular genomic context and the importance of both F6P and glucose-6-P in bifidobacterial metabolism, it would be valuable to investigate the glk involvement in catabolite repression by an examination of this phenomenon in a fructose-negative strain (B. longum DSM20219) lacking both the frk and glk genes.
Acknowledgments
This work was supported in part by the Centre National de la Recherche Scientifique (Unité de Glycobiologie Structurale et Fonctionnelle, UMR CNRS-USTL 8576), by the Université des Sciences et Technologies de Lille, and by the Region Nord-Pas de Calais (CPER 2000-2006).
We are indebted to F. Gavini for providing B. longum strains and helpful information. We are grateful to S. Foley for critically reading the manuscript. We thank A. Dupont and E. Stolarczyk for their contributions to frk expression analysis and Frk biochemical characterization, respectively. We also thank E. Aissi and C. Brassart for useful discussions.
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schäffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Angell, S., E. Schwarz, and M. J. Bibb. 1992. The glucose kinase gene of Streptomyces coelicolor A3(2): its nucleotide sequence, transcriptional analysis and role in glucose repression. Mol. Microbiol. 6:2833-2844. [DOI] [PubMed] [Google Scholar]
- 3.Aulkemeyer, P., R. Ebner, G. Heilenmann, K. Jahreis, K. Schmid, S. Wrieden, and J. W. Lengeler. 1991. Molecular analysis of two fructokinases involved in sucrose metabolism of enteric bacteria. Mol. Microbiol. 5:2913-2922. [DOI] [PubMed] [Google Scholar]
- 4.Berghauser, J. 1975. A reactive arginine in adenylate kinase. Biochim. Biophys. Acta 397:370-376. [DOI] [PubMed] [Google Scholar]
- 5.Bockmann, J., H. Heuel, and J. W. Lengeler. 1992. Characterization of a chromosomally encoded, non-PTS metabolic pathway for sucrose utilization in Escherichia coli EC3132. Mol. Gen. Genet. 235:22-32. [DOI] [PubMed] [Google Scholar]
- 6.Bogs, J., and K. Geider. 2000. Molecular analysis of sucrose metabolism of Erwinia amylovora and influence on bacterial virulence. J. Bacteriol. 182:5351-5358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bornet, F. R., F. Brouns, Y. Tashiro, and V. Duvillier. 2002. Nutritional aspects of short-chain fructooligosaccharides: natural occurrence, chemistry, physiology and health implications. Dig. Liver Dis. 34:S111-S120. [DOI] [PubMed] [Google Scholar]
- 8.Bradford, M. M. 1976. A rapid and sensitive method for quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 9.Brann, A., M. Tcherpakov, I. M. Williams, A. H. Futerman, and M. Fainzilber. 2002. Nerve growth factor-induced p75-mediated death of cultured hippocampal neurons is age-dependent and transduced through ceramide generated by neutral sphingomyelinase. J. Biol. Chem. 277:9812-9818. [DOI] [PubMed] [Google Scholar]
- 10.Bruckner, R., and F. Titgemeyer. 2002. Carbon catabolite repression in bacteria: choice of the carbon source and autoregulatory limitation of sugar utilization. FEMS Microbiol. Lett. 209:141-148. [DOI] [PubMed] [Google Scholar]
- 11.Butler, M. J., J. Deutscher, P. W. Postma, T. J. Wilson, A. Galinier, and M. J. Bibb. 1999. Analysis of a ptsH homologue from Streptomyces coelicolor A3(2). FEMS Microbiol. Lett. 177:279-288. [DOI] [PubMed] [Google Scholar]
- 12.Desjardin, L. E., L. G. Hayes, C. D. Sohaskey, L. G. Wayne, and K. D. Eisenach. 2001. Microaerophilic induction of the alpha-crystallin chaperone protein homologue (hspX) mRNA of Mycobacterium tuberculosis. J. Bacteriol. 183:5311-5316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Deutscher, J., E. Kuster, U. Bergstedt, V. Charrier, and W. Hillen. 1995. Protein kinase-dependent HPr/CcpA interaction links glycolytic activity to carbon catabolite repression in gram-positive bacteria. Mol. Microbiol. 15:1049-1053. [DOI] [PubMed] [Google Scholar]
- 14.de Vries, W., S. J. Gerbrandy, and A. H. Stouthamer. 1967. Carbohydrate metabolism in Bifidobacterium bifidum. Biochim. Biophys. Acta 136:415-425. [DOI] [PubMed] [Google Scholar]
- 15.Ehrmann, M. A., M. Korakli, and R. F. Vogel. 2003. Identification of the gene for beta-fructofuranosidase of Bifidobacterium lactis DSM10140 (T) and characterization of the enzyme expressed in Escherichia coli. Curr. Microbiol. 46:391-397. [DOI] [PubMed] [Google Scholar]
- 16.Feldheim, D. A., A. M. Chin, C. T. Nierva, B. U. Feucht, Y. W. Cao, Y. F. Xu, S. L. Sutrina, and M. H. Saier, Jr. 1990. Physiological consequences of the complete loss of phosphoryl-transfer proteins HPr and FPr of the phosphoenolpyruvate:sugar phosphotransferase system and analysis of fructose (fru) operon expression in Salmonella typhimurium. J. Bacteriol. 172:5459-5469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fennington, G. J., Jr., and T. A. Hughes. 1996. The fructokinase from Rhizobium leguminosarum biovar trifolii belongs to group I fructokinase enzymes and is encoded separately from other carbohydrate metabolism enzymes. Microbiology 142:321-330. [DOI] [PubMed] [Google Scholar]
- 18.Fujita, Y., Y. Miwa, A. Galinier, and J. Deutscher. 1995. Specific recognition of the Bacillus subtilis gnt cis-acting catabolite-responsive element by a protein complex formed by CcpA and seryl-phosphorylated HPr. Mol. Microbiol. 17:953-960. [DOI] [PubMed] [Google Scholar]
- 19.Henkin, T. M., F. J. Grundy, W. L. Nicholson, and G. H. Chambliss. 1991. Catabolite repression of alpha-amylase gene expression in Bacillus subtilis involves a trans-acting gene product homologous to the Escherichia coli lacI and galR repressors. Mol. Microbiol. 5:575-584. [DOI] [PubMed] [Google Scholar]
- 20.Hurtado, C. A. R., and R. A. Rachubinski. 2002. Isolation and characterization of YlBEM1, a gene required for cell polarization and differentiation in the dimorphic yeast Yarrowia lipolytica. Eukaryot. Cell 1:526-537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huynh, P. L., I. Jankovic, N. F. Schnell, and R. Bruckner. 2000. Characterization of an HPr kinase mutant of Staphylococcus xylosus. J. Bacteriol. 182:1895-1902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Imamura, L., K. Hisamitsu, and K. Kobashi. 1994. Purification and characterization of beta-fructofuranosidase from Bifidobacterium infantis. Biol. Pharm. Bull. 17:596-602. [DOI] [PubMed] [Google Scholar]
- 23.Jones, B. E., V. Dossonnet, E. Kuster, J. Deutscher, and R. E. Klevit. 1997. Binding of the catabolite repressor protein CcpA to its DNA target is regulated by phosphorylation of its corepressor HPr. J. Biol. Chem. 272:26530-26535. [DOI] [PubMed] [Google Scholar]
- 24.Kaplan, H., and R. W. Hutkins. 2000. Fermentation of fructooligosaccharides by lactic acid bacteria and bifidobacteria. Appl. Environ. Microbiol. 66:2682-2684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kriebardis, T., D. Meng, and S. Aktipis. 1987. Inhibition of the RNA polymerase-catalyzed synthesis of RNA by daunomycin. Effect of the inhibitor on the late steps of RNA chain initiation. J. Biol. Chem. 262:12632-12640. [PubMed] [Google Scholar]
- 26.Krzewinski, F., C. Brassart, F. Gavini, and S. Bouquelet. 1997. Glucose and galactose transport in Bifidobacterium bifidum DSM 20082. Curr. Microbiol. 35:175-179. [DOI] [PubMed] [Google Scholar]
- 27.Kwakman, J. H. J. M., and P. W. Postma. 1994. Glucose kinase has a regulatory role in carbon catabolite repression in Streptomyces coelicolor. J. Bacteriol. 176:2694-2698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Luesink, E. J., J. D. Marugg, O. P. Kuipers, and W. M. de Vos. 1999. Characterization of the divergent sacBK and sacAR operons, involved in sucrose utilization by Lactococcus lactis. J. Bacteriol. 181:1924-1926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Margolles, A., and C. G. de los Reyes-Gavilan. 2003. Purification and functional characterization of a novel alpha-l-arabinofuranosidase from Bifidobacterium longum B667. Appl. Environ. Microbiol. 69:5096-5103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Marx, S. P., S. Winkler, and W. Hartmeier. 2000. Metabolization of beta-(2,6)-linked fructose-oligosaccharides by different bifidobacteria. FEMS Microbiol. Lett. 182:163-169. [DOI] [PubMed] [Google Scholar]
- 31.Matsuki, T., K. Watanabe, R. Tanaka, M. Fukuda, and H. Oyaizu. 1999. Distribution of bifidobacterial species in human intestinal microflora examined with 16S rRNA-gene-targeted species-specific primers. Appl. Environ. Microbiol. 65:4506-4512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mitchell, W. J., J. Reizer, C. Herring, C. Hoischen, and M. H. Saier, Jr. 1993. Identification of a phosphoenolpyruvate:fructose phosphotransferase system (fructose-1-phosphate forming) in Listeria monocytogenes. J. Bacteriol. 175:2758-2761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Moller, P. L., F. Jorgensen, O. C. Hansen, S. M. Madsen, and P. Stougaard. 2001. Intra- and extracellular beta-galactosidases from Bifidobacterium bifidum and B. infantis: molecular cloning, heterologous expression, and comparative characterization. Appl. Environ. Microbiol. 67:2276-2283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mondero, V., M. J. Gosalbes, and J. Perez-Martinez. 1997. Catabolite repression in Lactobacillus casei ATCC 393 is mediated by CcpA. J. Bacteriol. 179:6657-6667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nothaft, H., S. Parche, A. Kamionka, and F. Titgemeyer. 2003. In vivo analysis of HPr reveals a fructose-specific phosphotransferase system that confers high-affinity uptake in Streptomyces coelicolor. J. Bacteriol. 185:929-937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Perrin, S., M. Warchol, J. P. Grill, and F. Schneider. 2001. Fermentations of fructo-oligosaccharides and their components by Bifidobacterium infantis ATCC 15697 on batch culture in semi-synthetic medium. J. Appl. Microbiol. 90:859-865. [DOI] [PubMed] [Google Scholar]
- 37.Perrin, S., C. Fougnies, J. P. Grill, H. Jacobs, and F. Schneider. 2002. Fermentation of chicory fructo-oligosaccharides in mixtures of different degrees of polymerization by three strains of bifidobacteria. Can. J. Microbiol. 48:759-763. [DOI] [PubMed] [Google Scholar]
- 38.Reid, S. J., M. S. Rafudeen, and N. G. Leat. 1999. The genes controlling sucrose utilization in Clostridium beijerinckii NCIMB 8052 constitute an operon. Microbiology 145:1461-1472. [DOI] [PubMed] [Google Scholar]
- 39.Sabater, B., J. Sebastian, and C. Asensio. 1972. Identification and properties of an inducible and highly specific fructokinase from Streptomyces violaceoruber. Biochim. Biophys. Acta 284:414-420. [DOI] [PubMed] [Google Scholar]
- 40.Sabater, B., and G. Delafuente. 1975. Kinetic properties and related changes of molecular weight in a fructokinase from Streptomyces violaceoruber. Biochim. Biophys. Acta 377:258-270. [DOI] [PubMed] [Google Scholar]
- 41.Salyers, A. A., J. K. Palmer, and T. D. Wilkins. 1978. Degradation of polysaccharides by intestinal bacterial enzymes. Am. J. Clin. Nutr. 31:S128-S130. [DOI] [PubMed] [Google Scholar]
- 42.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 43.Scardovi, V. 1986. Genus Bifidobacterium, p. 1418-1434. In P. H. A. Sneath, N. S. Mair, M. E. Sharpe, and J. G. Holt (ed.), Bergey's manual of systematic bacteriology, vol. 2. Williams & Wilkins, Baltimore, Md. [Google Scholar]
- 44.Schell, M. A., M. Karmirantzou, B. Snel, D. Vilanova, B. Berger, G. Pessi, M. C. Zwahlen, F. Desiere, P. Bork, M. Delley, R. D. Pridmore, and F. Arigoni. 2002. The genome sequence of Bifidobacterium longum reflects its adaptation to the human gastrointestinal tract. Proc. Natl. Acad. Sci. USA 99:14422-14427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sebastian, J., and C. Asensio. 1972. Purification and properties of the mannokinase from Escherichia coli. Arch. Biochem. Biophys. 151:227-233. [DOI] [PubMed] [Google Scholar]
- 46.Skerman, V. B. D., V. McGowan, and P. H. A. Sneath. 1980. Approved lists of bacterial names. Int. J. Syst. Bacteriol. 30:225-420. [Google Scholar]
- 47.Spath, C., A. Kraus, and W. Hillen. 1997. Contribution of glucose kinase to glucose repression of xylose utilization in Bacillus megaterium. J. Bacteriol. 179:7603-7605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sprogoe, D., L. A. Van Den Broek, O. Mirza, J. S. Kastrup, A. G. Voragen, M. Gajhede, and L. K. Skov. 2004. Crystal structure of sucrose phosphorylase from Bifidobacterium adolescentis. Biochemistry 43:1156-1162. [DOI] [PubMed] [Google Scholar]
- 49.Sproul, A. A., L. T. Lambourne, D. J. Jean-Jacques, and H. L. Kornberg. 2001. Genetic control of manno(fructo)kinase activity in Escherichia coli. Proc. Natl. Acad. Sci. USA 98:15257-15259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Studier, F. W., A. H. Rosenberg, and J. J. Dunn. 1990. Use of T7 RNA polymerase to direct the expression of cloned genes. Methods Enzymol. 185:60-89. [DOI] [PubMed] [Google Scholar]
- 51.Tanaka, H., H. Hashiba, J. Kok, and I. Mierau. 2000. Bile salt hydrolase of Bifidobacterium longum: biochemical and genetic characterization. Appl. Environ. Microbiol. 66:2502-2512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Thompson, J., and B. M. Chassy. 1981. Uptake and metabolism of sucrose by Streptococcus lactis. J. Bacteriol. 147:543-551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Thompson, J., D. L. Sackett, and J. A. Donkersloot. 1991. Purification and properties of fructokinase I from Lactococcus lactis. Localization of scrK on the sucrose-nisin transposon Tn5306. J. Biol. Chem. 266:22626-22633. [PubMed] [Google Scholar]
- 54.Thompson, J., N. Y. Nguyen, and S. A. Robrish. 1992. Sucrose fermentation by Fusobacterium mortiferum ATCC 25557: transport, catabolism, and products. J. Bacteriol. 174:3227-3235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Titgemeyer, F., J. Reizer, A. Reizer, and M. H. Jr., Saier. 1994. Evolutionary relationships between sugar kinases and transcriptional repressors in bacteria. Microbiology 140:2349-2354. [DOI] [PubMed] [Google Scholar]
- 56.Trindade, M. I., V. R. Abratt, and S. J. Reid. 2003. Induction of sucrose utilization genes from Bifidobacterium lactis by sucrose and raffinose. Appl. Environ. Microbiol. 69:24-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Van Den Bogaard, P. T. C., M. Kleerebezem, O. Kuipers, and W. M. de Vos. 2000. Control of lactose transport, β-galactosidase activity, and glycolysis by CcpA in Streptococcus thermophilus: evidence for carbon catabolite repression by a non-phosphoenolpyruvate-dependent phosphotransferase system sugar. J. Bacteriol. 182:5982-5989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Van Den Broek, L. A., E. L. Van Boxtel, R. P. Kievit, R. Verhoef, G. Beldman, and A. G. Voragen. 2004. Physico-chemical and transglucosylation properties of recombinant sucrose phosphorylase from Bifidobacterium adolescentis DSM20083. Appl. Microbiol. Biotechnol. 61:55-60. [DOI] [PubMed] [Google Scholar]
- 59.Veerkamp, J. H. 1969. Catabolism of glucose and derivatives of 2-deoxy-2-amino-glucose in Bifidobacterium bifidum var. pennsylvanicus. Arch. Biochem. Biophys. 129:257-263. [DOI] [PubMed] [Google Scholar]
- 60.Wade, H. E., and D. M. Morgan. 1953. Detection of phosphate esters on paper chromatograms. Nature 171:529-530. [DOI] [PubMed] [Google Scholar]
- 61.Wagner, E., S. Marcandier, O. Egeter, J. Deutscher, F. Götz, and R. Brückner. 1995. Glucose kinase-dependent catabolite repression in Staphylococcus xylosus. J. Bacteriol. 177:6144-6152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wagner, E., E. Küster-Schöck, and W. Hillen. 2000. Sugar uptake and carbon catabolite repression in Bacillus megaterium strains with inactivated ptsHI. J. Mol. Microbiol. Biotechnol. 2:587-592. [PubMed] [Google Scholar]
- 63.Warchol, M., S. Perrin, J. P. Grill, and F. Schneider. 2002. Characterization of a purified beta-fructofuranosidase from Bifidobacterium infantis ATCC 15697. Lett. Appl. Microbiol. 35:462-467. [DOI] [PubMed] [Google Scholar]
- 64.Weisser, P., R. Kramer, H. Sahm, and G. A. Sprenger. 1995. Functional expression of the glucose transporter of Zymomonas mobilis leads to restoration of glucose and fructose uptake in Escherichia coli mutants. J. Bacteriol. 177:3351-3354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Weisser, P., R. Kramer, and G. A. Sprenger. 1996. Expression of the Escherichia coli pmi gene, encoding phosphomannose-isomerase in Zymomonas mobilis, leads to utilization of mannose as a novel growth substrate, which can be used as a selective marker. Appl. Environ. Microbiol. 62:4155-4161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zembrzuski, B., P. Chilco, X. L. Liu, J. Liu, T. Conway, and R. Scopes. 1992. Cloning, sequencing, and expression of the Zymomonas mobilis fructokinase gene and structural comparison of the enzyme with other hexose kinases. J. Bacteriol. 174:3455-3460. [DOI] [PMC free article] [PubMed] [Google Scholar]