Abstract
The man5K gene of Clostridium cellulolyticum was cloned and overexpressed in Escherichia coli. This gene encodes a 424-amino-acid preprotein composed of an N-terminal leader peptide, followed by a dockerin module and a C-terminal catalytic module belonging to family 5 of the glycosyl hydrolases. Mature Man5K displays 62% identity with ManA from Clostridium cellulovorans. Two forms of the protein were purified from E. coli; one form corresponds to the full-length enzyme (45 kDa), and a truncated form (39 kDa) lacks the N-terminal dockerin module. Both forms exhibit the same typical family 5 mannanase substrate preference; they are very active with the galactomannan locust bean gum, and the more galacto-substituted guar gum molecules are degraded less. The truncated form, however, displays fourfold-higher activity with galactomannans than the full-length enzyme. Man5K was successfully overproduced in C. cellulolyticum by using expression vectors. The trans-produced protein was found to be incorporated into the cellulosomes and became one of the major enzymatic components. Modified cellulosomes displayed 20-fold-higher specific activities than control fractions on galactomannan substrates, whereas the specific activity on crystalline cellulose was reduced by 20%. This work clearly showed that the composition of the cellulosomes is obviously regulated by the relative amounts of the enzymes produced and that this composition can be engineered in clostridia by structural gene cloning.
The plant cell wall is composed mainly of a complex aggregation of polysaccharides, such as cellulose, hemicellulose [xylan, mannan, glucomannan, β(1,3-1,4) d-glucans, laminarin], and pectin. This composite structure makes it a robust material that is recalcitrant to degradation. A number of microorganisms are able to degrade the plant cell wall components, essentially through the action of various glycosyl hydrolases; the oligomers and/or monomers produced are used as carbon and energy sources. Cellulolytic microorganisms secrete several cellulases, which cleave the β-1,4-glycosidic bonds that link the glucose units in the cellulose chains; they often also produce specialized enzymes (mannanases, xylanases, lichenase, pectate lyase) that hydrolyze the associated noncellulosic polysaccharides (2, 13). The actions of these enzymes allow cellulases to access the cellulose.
The cellulolytic mesophilic anaerobic bacterium Clostridium cellulolyticum secretes multienzyme complexes called cellulosomes. In the cellulosomes, several different enzymes displaying various substrate specificities and modes of activity are associated with a scaffolding protein designated CipC (9). CipC mediates attachment of the cellulosomes to cellulose through a carbohydrate binding module and anchors the dockerin-containing enzymes on its eight hydrophobic cohesin domains (18, 20). The enzymatic composition of the cellulosomes is probably regulated by the amounts of the different available dockerin-containing enzymes since the cohesin-dockerin interaction has been shown not to be enzyme specific (7, 8, 19). In C. cellulolyticum, most of the known genes coding for cellulosomal components are in a 26-kb cluster, the cipC-cel48F-cel8C-cel9G-cel9E-orfX-cel9H-cel9J-man5K-cel9M-rgl11Y-cel9N clus-ter (1, 3, 21). These genes mainly encode endocellulases and processive cellulases, including the major processive cellulase Cel48F (9, 26). The cel cluster also encodes two helper enzymes, the rhamnogalacturonan lyase Rgl11Y (21) and the mannanase Man5K (GenBank accession number AF316823).
C. cellulolyticum is a cellulosome-producing bacterium for which genetic tools are available (12, 14, 16, 24, 28). It has been shown in three different studies that the cellulolytic capacity of this organism might be genetically engineered. An increase in its capacity to degrade cellulose has been achieved by cloning an additional fermentation pathway (12). On the other hand, in vivo modification of the cellulosome enzyme composition has been accomplished by using antisense RNAs targeted against cel48F mRNA (24). Furthermore, an engineered expression vector, pSOS952 (23), has been successfully used to perform cipC transcomplementation of a cipC insertional mutant (16).
The aim of the present work was to test the possibility of increasing the amount of one defined enzyme in the cellulosomes by overexpressing the corresponding gene in C. cellulolyticum. We first purified and characterized the recombinant form of the cellulosomal mannanase Man5K, which was overproduced in Escherichia coli. Then we transformed C. cellulolyticum with expression vectors harboring the man5K gene and analyzed the modified cellulosomes that were purified from the transformants. We thus explored the possibility of designing, by genetic engineering of C. cellulolyticum, functionally tailor-made cellulosomes displaying a preferential and/or new substrate specificity.
MATERIALS AND METHODS
Bacterial strains, plasmids, and media.
E. coli DH5α (Life Technologies), E. coli BL21(DE3) (Life Technologies), and E. coli SG13009(pREP4) (QIAGEN) (Table 1) were grown at 37°C in Luria-Bertani medium supplemented with appropriate antibiotics (100 μg of ampicillin per ml, 50 μg of kanamycin per ml). C. cellulolyticum ATCC 35319 (25) was grown anaerobically at 32°C on basal medium (10) supplemented with either cellobiose (2 g/liter; Sigma-Aldrich) or MN300 cellulose (5 g/liter; Serva). To select and maintain recombinant C. cellulolyticum strains, erythromycin (10 μg/ml) was added to the medium. Colonies of recombinant C. cellulolyticum strains were isolated on solid medium (basal medium supplemented with 2 g of cellobiose per liter, 15 g of agar per liter, and 10 μg of erythromycin per ml) under the anaerobic atmosphere of a glove box (N2-H2, 95:5 [vol/vol]). Plates were incubated in anaerobic jars under 2 × 105 Pa of an N2-CO2 (80:20 [vol/vol]) atmosphere.
TABLE 1.
Bacterial strains and plasmids
| Strain or plasmid | Relevant characteristicsa | Source and/or reference |
|---|---|---|
| Escherichia coli strains | ||
| DH5α | F−endA1 hsdR17(rK− mK+) supE44 thi-1 λ−gyrA96 relA1 Δ(lacZYA argF) U169 (φ80 lacZΔM15) recA | Roche Diagnostics |
| SG13009(pREP4) | F−his pyrD Δlon-100 rpsL (pREP4) | Qiagen (11) |
| BL21(DE3) | F−ompT hsdS (rB− mB−) gal dcm (DE3) | Novagen |
| Clostridium cellulolyticum strains | ||
| ATCC 35319 | Wild type | 25 |
| D3K | ATCC 35319(pD3K) | This study |
| 952K | ATCC 35319(p952K) | This study |
| 954K | ATCC 35319(p954K) | This study |
| Plasmids | ||
| pGEMT-Easy | E. coli cloning vector; Apr | Invitrogen |
| pET22b(+) | E. coli expression vector; Apr | Novagen |
| pET-K | pET22b(+) derivative carrying the 1,202-bp NdeI-XhoI fragment encoding mature Man5K | This study |
| pSOS95 | E. coli-Clostridium shuttle vector (ColEI and plM13 replicons); Apr Emr carrying the adc-ctfA-ctfB operon transcribed from the thl promoter | 29 |
| pSOSzero | pSOS95 derivative with the entire expression cassette (Pthl-adc-ctfA-ctfB) deleted | 24 |
| pSOS952 | pSOS95 derivative carrying two lac operators upstream and downstream of the thl promoter | 23 |
| pSOS954 | pSOS952 derivative carrying a thl −35 mutated box | This study |
| pD3 | pSOS95 derivative with the 377-bp fragment carrying the upstream region of the cipC open reading frame in place of the thl promoter | This study |
| p952K | pSOS952 derivative carrying the 1,313-bp BamHI-NarI cipC′-′man5K gene | This study |
| p954K | pSOS954 derivative carrying the 1,313-bp BamHI-NarI cipC′-′man5K gene | This study |
| pD3K | pD3 derivative carrying the 1,313-bp BamHI-NarI cipC′-′man5K gene | This study |
Abbreviations: adc, acetoacetate decarboxylase gene; ctfA, coenzyme A transferase subunit gene; ctfB, CoAT subunit B gene; thl promoter, thl promoter region for the thiolase gene (thl) of C. acetobutylicum ATCC 824; Apr, ampicillin resistant; Emr, erythromycin resistant.
The plasmid vectors used were pGEMT-Easy (Invitrogen) for cloning a PCR fragment and pET22b+ (Novagen) for overexpressing man5K in E. coli. pSOS95 (kindly provided by P. Soucaille, INSA, Toulouse, France) and the derivatives pSOS952 (23), pSOS954, and pD3 (this study) were used for overexpressing man5K in C. cellulolyticum. pSOSzero (24) was used as the reference empty expression vector in C. cellulolyticum.
Production of Man5K in E. coli.
The region of the man5K gene that encodes the mature protein was amplified by PCR by using oligonucleotides manKdir (5′-GATTCCATATGGCGACATACAAACTT-3′) and manKrev1 (5′-GGGCTCGAGTCTGACGTCAGGATG-3′). These primers introduced NdeI and XhoI sites (underlined) upstream and downstream of the coding sequence, respectively. One ATG initiation codon (boldface type) was included in the first primer. The 1.3-kb DNA fragment was amplified by PCR. Since man5K contains an internal NdeI site, two steps were necessary to clone the 1.3-kb DNA fragment into pET22b(+). The PCR fragment was first digested with NdeI and EcoRI. The resulting 0.4-kb NdeI-EcoRI fragment was cloned into NdeI-EcoRI-linearized pET22b(+), resulting in pET-K1. Then the PCR fragment was digested with EcoRI and XhoI, and the resulting 0.9-kb EcoRI-XhoI fragment was cloned in EcoRI-XhoI-linearized pET-K1. The resulting plasmid, pET-K, contained the coding sequence for the Man5K mature protein fused in frame at its C terminus with a sequence encoding six histidine residues (His tag). Plasmid pET-K was used to transform BL21(DE3) cells to produce the recombinant protein.
Expression vectors for C. cellulolyticum.
The expression vectors pSOS952 and pSOS954 were derived from the E. coli-Clostridium shuttle vector pSOS95 (Table 1) (29). These vectors were constructed for cloning genes between the constitutive promoter of the thiolase gene and the transcriptional terminator of the adc gene (encoding acetoacetate decarboxylase) by using BamHI and NarI sites. pSOS952 (23) and pSOS954 contained a regulatory region composed of two lac operators (21 bp each), which allowed negative modulation of the level of expression of the gene which follows the thiolase promoter in E. coli. This regulatory region was added to facilitate cloning of the genes encoding a heterologous secreted protein which may be toxic. We constructed pSOS954 by using the procedure described previously for pSOS952 (23). In the pSOS954 vector, we introduced one additional mutation (A→T) in the −35 box of the thiolase promoter. This mutation was introduced by insertion of the first lac operator into the pSOS95 vector by using primer SosAm (5′-TCCTGCAGGTCGACTTTTTAACAAAATATATTGATTAAAATAATAAT-3′; a SalI site is underlined, the −35 box is indicated by boldface type, and the mutation is indicated by boldface type and underlining) instead of the SosA primer used for construction of pSOS952 (23).
The expression vector pD3 was derived from pSOS95 by replacing the thiolase promoter with the 377-bp fragment located upstream of the cipC coding sequence. The 5′ end of the cipC gene was synthesized by PCR by using forward primer cipPdir (5′-GTCGACTATGTAGCTGTAAGCATTAT-3′), reverse primer cipPrev (5′-GGATCCACATGAAAACCATTTGG-3′), and genomic DNA as the template. The forward and reverse primers were designed to anneal 391 and 25 bases upstream of the ATG initiation codon of cipC, respectively, and to contain one SalI site and one BamHI site, respectively (underlined). The PCR fragment hypothesized to carry the cipC promoter was cloned into the pGEMT-Easy vector. The 377-bp BamHI-SalI fragment of the resulting vector was inserted between the SalI and BamHI sites located upstream of the adc-ctfA-ctfB operon in pSOS95.
Cloning of the man5K gene into C. cellulolyticum.
A chimeric cipC-man5K coding sequence was amplified by two PCR steps by using C. cellulolyticum genomic DNA as the template. The first PCR step generated a 133-bp fragment encoding the 5′ end of the cipC gene, including 25 bp upstream of the ATG start codon and the first 90 bp of the coding sequence. This fragment, synthesized by using primers SScipCdir (5′-AGGGATCCTAAAATAAAAAATAGGAGGTTTACAATG-3′; BamHI site underlined) and SScipCrev (5′-CCAAGTTTGTAAGTACCTGCTGCAAAAGCTGTA-3′), contained the cipC ribosome binding site and encoded the CipC signal sequence (SSCipC). A 1,211-bp PCR fragment was synthesized in a second step by using primers ManKmat (5′-GCAGGTACTTACAAACTTGGTGATGTTGACAAT-3′) and ManKrev2 (5′-TTTCCGGCGCCTTATCTGACTACGTCAGGATGCTCT-3′; NarI site underlined). This fragment contained the sequence coding for mature Man5K. Both fragments were combined by overlap extension PCR performed with the external primers SScipCdir and ManKrev2. The SSCipC-Man5K-encoding DNA (1.3 kb) was subsequently digested with BamHI and NarI and ligated into BamHI-NarI-digested pSOS952, pSOS954, and pD3, resulting in vectors pSOS952K, pSOS954K, and pD3K, respectively. The expression vectors were treated with MspI methylase (New England Biolabs) before C. cellulolyticum electrotransformation (14, 28). Electrotransformed cells were incubated overnight in cellobiose-supplemented basal medium without any antibiotics and then subcultured in fresh medium supplemented with erythromycin to select for transformants. Erythromycin-resistant colonies were isolated on solid medium supplemented with erythromycin.
Purification of rMan5K produced in E. coli.
Recombinant mannanase 5K (rMan5K) was purified as follows. E. coli BL21(DE3)(pET-K) was grown at 37°C with shaking to an optical density at 600 nm of 0.7. Isopropyl-β-d-thiogalactopyranoside (IPTG) was added to a final concentration of 100 μM, and the culture (3 liters) was incubated with shaking at 25°C for 15 h. The cells were then harvested by centrifugation and broken in a French press. The supernatant was loaded onto a 3-ml Ni-nitrilotriacetic acid column, and the His-tagged protein was eluted from the column with imidazole. The eluate was dialyzed and concentrated with an Amicon concentrator in 50 mM phosphate buffer (pH 7.2)-150 mM NaCl. The concentrated solution was then subjected to gel filtration chromatography [Superdex 200pg (26/60); Amersham Pharmacia Biotech]. The fractions obtained for each peak were pooled, dialyzed, and concentrated with an Amicon concentrator in 20 mM Tris-HCl (pH 8)-1 mM CaCl2. The protein concentration was estimated by measuring the absorbance at 280 nm in 6 M guanidinium chloride based on the known amino acid composition of the desired protein.
Purification of the cellulose-adsorbed cellulolytic system from C. cellulolyticum.
The Clostridium strain was grown in 400 ml of cellulose-supplemented basal medium for 6 days. The culture was filtered through a 3-μm-pore-size GF/D glass filter (Whatman). The retained cellulose was washed with 500 ml of ice-cold 50 mM phosphate buffer (pH 7) and then with 500 ml of 12.5 mM phosphate buffer (pH 7). Bound proteins (mainly cellulosomes) were eluted with 100 ml of ice-cold MilliQ water. Cellulose residues were removed by centrifugation of the eluted fraction. The supernatant (Fc fraction) was concentrated in a stirred ultrafiltration cell equipped with a polyethersulfone ultrafiltration membrane (10-kDa cutoff; Amicon) and subsequently stored at −20°C. Protein concentration was determined by the method of Lowry et al. (15).
N-terminal sequencing.
The N-terminal amino acid sequence was obtained with an Applied Biosystems 470A sequence analyzer with the proteins blotted onto polyvinylidene difluoride.
Enzyme assays.
Galactomannans were used as substrates for mannanases and mannanase-enriched cellulosomes. These substrates are composed of a linear homopolymer of mannose linked by β-1,4 bonds substituted with α-1,6-galactose side chains. In locust bean gum (LBG) and guar gum (Fluka), the mannose/galactose ratios were 4:1 and 2:1, respectively. The soluble carboxymethyl cellulose (CMC) (medium viscosity; Sigma) and the insoluble Avicel microcrystalline cellulose (PH101; Fluka) were also tested. All these substrates were prepared in 20 mM Tris-maleate (pH 6.0). Guar gum and LBG were washed three times before use. Enzymatic assays were performed at 37°C. A suitable amount of the Fc fraction was diluted in 1 ml of 20 mM Tris-maleate (pH 6.0) and then mixed with 4 ml of a preincubated substrate preparation at a final concentration of 0.8% (CMC, Avicel) or 0.5% (LBG, guar gum). After various incubation times aliquots were examined to determine the soluble reducing sugar content by the method of Park and Johnson (22); glucose or mannose was used as the standard. One international unit of enzymatic activity corresponded to 1 μmol of reducing sugar released per min.
PAGE and Western blot analysis.
Sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE) (Prosieve 50 gel; TEBU) was performed by using a vertical electrophoresis system apparatus (Amersham Pharmacia Biotech). Native PAGE was performed with precast 4 to 15% polyacrylamide gradient gels by using a Phast-System apparatus (Amersham Pharmacia Biotech). Gels were stained with Coomassie blue or were electrotransferred onto nitrocellulose BA 83 membranes (Schleicher & Schuell). After saturation, membranes were probed with polyclonal rabbit antibodies raised against Man5K (or Cel48F) purified from E. coli. Antibodies were detected by using anti-rabbit horseradish peroxidase conjugate and a chemiluminescent substrate (Amersham Pharmacia Biotech).
RESULTS
Sequence analysis of Man5K.
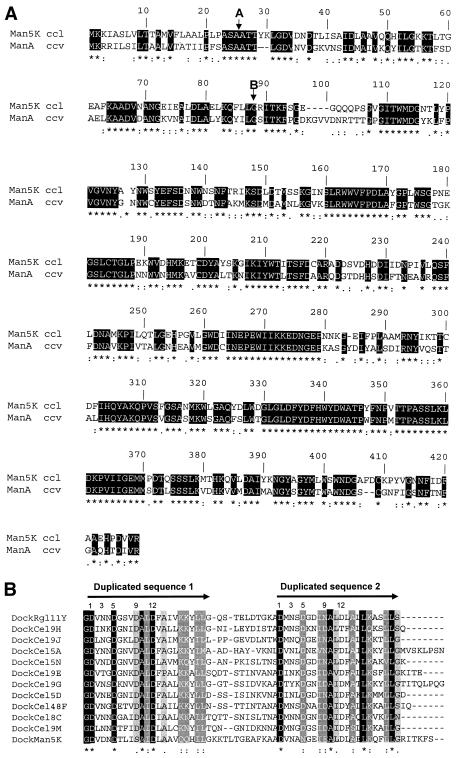
The man5K gene was identified in the large cel cluster between cel9J and cel9 M (GenBank accession number AF316823). This gene encodes a 424-amino-acid protein which includes a putative N-terminal signal peptide. The cleavage site of this signal peptide might be located between Ala residues 25 and 26, at the typical signal peptidase I processing site Ala23-X-Ala25 (Fig. 1A). The mature protein, which contains 399 amino acids, has a calculated molecular mass of 45,072 Da and a predicted pI of 4.6. A typical dockerin domain, composed of a duplicated sequence consisting of 22 amino acid residues, was found at an unusual position (N terminus). Its sequence, although very similar to the sequences of the dockerin domains of the other 11 known cellulosomal enzymes from C. cellulolyticum, has several unique characteristics (Fig. 1B). In the first duplicated sequence, an aspartate residue was found in place of an asparagine at the third position, and a serine was found in place of an aspartate residue at the ninth position. Furthermore, the lysine residues at the 17th and 18th positions in the consensus sequence are replaced in Man5K by two glutamine residues. In the second duplicated sequence, an asparagine residue was found at the fifth position in place of the aspartate residue that is usually found. The catalytic module, which belongs to family 5 of the glycosyl hydrolases, was found at the C terminus. The whole protein exhibits 62% identity with the cellulosomal mannanase ManA from C. cellulovorans (Fig. 1A) (27).
FIG. 1.
(A) Alignment of Man5K of C. cellulolyticum (accession number AF316823) (Man5K ccl) with ManA of C. cellulovorans (accession number AF132735) (ManA ccv). Arrow A indicates the signal sequence putative cleavage site, and arrow B indicates the last amino acid of the dockerin domain. (B) Alignment of the dockerin domain sequences found in cellulosomal C. cellulolyticum proteins.
Production, purification, and catalytic properties of rMan5K.
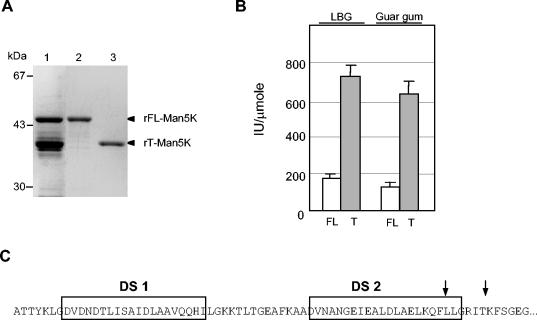
The DNA sequence encoding the full-length mature protein was cloned in frame with a sequence encoding a His tag into the expression vector pET-22b(+). The resulting plasmid, pET-K, was used to transform E. coli expression strain BL21(DE3). A fraction of the recombinant protein was processed when it was produced in E. coli; two proteins were therefore purified from the cellular extract on Ni-nitrilotriacetic acid agarose, and they had apparent masses of 46 and 39 kDa (Fig. 2A, lane 1). The mass of the first protein was in good agreement with the expected size of the full-length mannanase (rFL-Man5K). Furthermore, its N-terminal sequence, determined by sequencing, perfectly matched the expected sequence for the entire protein. The analysis of the N terminus of the 39-kDa protein revealed that the truncated form of Man5K had lost most of the N-terminal dockerin domain (Fig. 2C). This form was designated rT-Man5K. rFL-Man5K and rT-Man5K were subsequently separated by gel filtration chromatography (Fig. 2A, lanes 2 and 3).
FIG. 2.
rMan5K produced by E. coli. (A) Purified proteins analyzed by SDS-PAGE and stained with Coomassie blue. Lane 1, fraction of Man5K eluted with imidazole from Ni-nitrilotriacetic acid agarose; lanes 2 and 3, rFL-Man5K and rT-Man5K separated by gel filtration chromatography, respectively. (B) Specific activities of rFL-Man5K (FL) and rT-Man5K (T) with galactomannan substrates (0.5% LBG and 0.5% guar gum). (C) N-terminal sequence of rFL-Man5K. The arrows indicated the two major cleavage sites leading to rT-Man5K. DS 1 and DS 2, duplicated sequences 1 and 2.
The two purified proteins were analyzed to determine their enzymatic properties with CMC and two different galacto-substituted mannan substrates, LBG (mannose/galactose ratio, 4:1) and guar gum (mannose/galactose ratio, 2:1). Both enzymes efficiently degraded the two galactomannan substrates tested (Fig. 2B) and exhibited very low (rT-Man5K; 1.2 IU/μmol) or undetectable (rFL-Man5K) activity with CMC. Both enzymes were found to be about 20% more active with LBG than with guar gum. Remarkably, the truncated mannanase showed fourfold-higher activity than FL-Man5K, whatever the substrate.
Production and purification of Man5K-enriched cellulosomes in C. cellulolyticum.
To investigate the possibility of modifying the enzymatic composition of the cellulosomes secreted by C. cellulolyticum, we overexpressed the cellulosomal man5K gene in C. cellulolyticum by gene cloning in various expression vectors, expecting that the composition of the cellulosomes would depend mainly on the relative amounts of the different dockerin-containing enzymes. Man5K is the only known cellulosomal mannanase in C. cellulolyticum. An increase in the amount of this enzyme in the complexes should therefore be easily detectable by measuring the specific activities of the modified cellulosomes on the mannanase substrates and comparing these activities with those exhibited by the cellulosomes produced by wild-type strain ATCC 35319 and control strain ATCC 35319(pSOSzero). In order to overproduce Man5K at different levels in C. cellulolyticum, we constructed three different vectors, designated pSOS952K, pSOS954K, and pD3K, which were designed to encode Man5K. Each vector differs from the others by the promoter sequence; pSOS952 has the intact promoter of the Clostridium acetobutylicum thiolase gene (Pthl), pSOS954 has a mutated thiolase promoter (the wild-type −35 box TTGATA is replaced by the TTGATT sequence), and pD3 is expected to contain the cipC promoter (Table 1). Plasmid pSOSzero, which does not contain any expression cassette (empty plasmid), was used as the control (24). pSOS952 has been used previously to clone the cipC gene into the mutant strain cipCMut1 (16), leading to a well-produced and secreted recombinant CipC protein. In order to guarantee secretion of large amounts of the mature full-length mannanase in the Clostridium strain, we decided to keep the same signal peptide that was used for CipC overproduction. The DNA coding for the signal sequence of CipC was fused to the mature Man5K-encoding sequence and cloned in the three different vectors. The sequence surrounding the signal peptide cleavage site of CipC was used in the chimeric protein. Consequently, the N-terminal sequence of the recombinant mature protein was expected to differ slightly from the native Man5K sequence (Fig. 3C), allowing its identification.
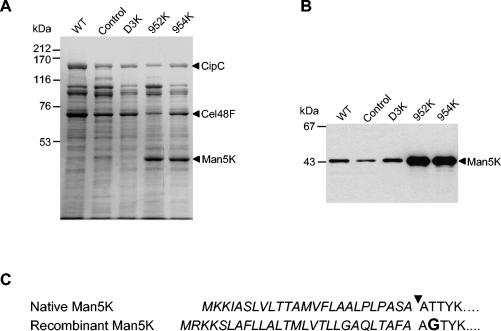
FIG. 3.
Overproduction of Man5K by C. cellulolyticum. (A) SDS-PAGE analysis of the Fc fraction (10 μg) from wild-type (WT), control, D3K, 952K, and 954K strains. (B) Western blot probed with antibodies raised against rMan5K (10 μg loaded for wild-type, control, and D3K Fc fractions and 2.5 μg loaded for 952K and 954K Fc fractions). (C) N-terminal sequences of native Man5K and rMan5K synthesized by C. cellulolyticum. Signal sequences are italicized. The arrowhead indicates the cleavage site. In the mature rMan5K, a G residue (boldface type) replaces the T residue found at the second position in the native mature Man5K.
The C. cellulolyticum transformants were isolated by plating cells on erythromycin-containing solid medium. The plasmids were detected in the cells by PCR and by agarose gel electrophoresis of total DNA, as previously described (14) (data not shown). The amount of Man5K associated with the cellulosomes was evaluated for the five different strains (wild type, control, D3K, 952K, and 954K [Table 1]) cultivated in liquid medium supplemented with MN300 cellulose. The secreted proteins bound to residual cellulose (referred to as the Fc fraction) were purified from a 6-day-old culture of each strain and analyzed by SDS-PAGE (Fig. 3A). A high-intensity band composed of proteinaceous components at an apparent Mr of 44,000 was detected by Coomassie blue staining in the Fc fractions purified from the 952K and 954K strains, but not in the Fc fraction from the D3K strain or in the Fc fractions from the wild-type and control strains. The molecular mass of this protein (44 kDa) was in good agreement with the expected size for the full-length mannanase (45,027 Da). The thickness of the bands revealed by specific polyclonal antibodies raised against Man5K confirmed that the 44-kDa protein overproduced by the 952K and 954K strains was Man5K (Fig. 3B). The trans-produced enzyme comigrated with the natural mannanase synthesized from the chromosomal gene in all strains tested. N-terminal sequence analysis of proteins located in the thick 44-kDa bands identified for the 952K and 954K strains revealed only one protein species. The sequence perfectly matched the expected N-terminal sequence for the mature rMan5K (Fig. 3C), indicating that the native protein produced from the chromosomal gene was a minor component of these bands. This result also showed that the CipC signal peptide grafted on Man5K was processed and that the overproduced mannanase carried its N-terminal dockerin domain.
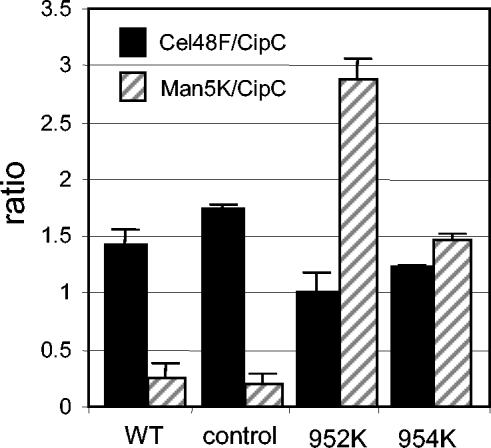
The cellulolytic system compositions of strains 954K and 952K were found to be not identical, as revealed by the SDS-PAGE patterns (Fig. 3A). A larger amount of Man5K appeared to be present in the Fc fraction from the 952K strain than in the Fc fraction purified from the 954K strain. The Man5K/CipC and Cel48F/CipC ratios were calculated from a densitometric analysis of the SDS-PAGE patterns of the Fc fractions purified from the wild-type, control, 952K, and 954K strains. The Man5K/CipC ratio was found to be twofold higher in the 952K Fc fraction than in the 954K Fc fraction and sevenfold higher in the Fc fraction from the 954K strain than in the Fc fraction from the control strain (Fig. 4). On the other hand, we observed that the Cel48F/CipC ratios in the 954K and 952K Fc fractions were only 30 and 40% lower, respectively, than the ratio obtained for the control strain. All these results clearly show that Man5K was overproduced and secreted as a mature full-length mannanase by strains 954K and 952K but not by the D3K strain. Moreover, strain 952K produced twofold more mannanase than strain 954K produced.
FIG. 4.
Densitometry analysis of the Fc fractions from the wild-type (WT), control, 952K, and 954K strains. Two independent SDS-PAGE gels stained with Coomassie blue were scanned and analyzed by using the Quantity One quantitation software from Bio-Rad. The Cel48F/CipC and Man5K/CipC ratios were calculated from the band intensities measured for the Cel48F, Man5K, and CipC proteins.
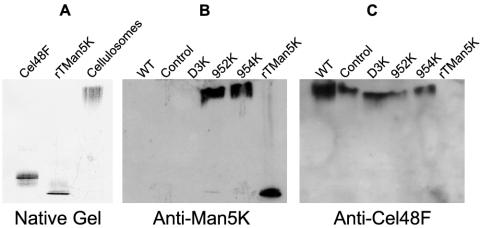
To check the incorporation of Man5K into the cellulosomes and to detect potential free enzymes, we analyzed the Fc fractions of the different strains separated in a native polyacrylamide gel by Western blotting with antibodies raised against Man5K (Fig. 5). In the Fc fraction from strains 954K and 952K, Man5K was found almost exclusively in the enzymatic complexes. Man5K was not detected in the complexes from the wild-type, control, and D3K strains in the experimental conditions used. Conversely, Cel48F, the major cellulosomal enzyme, was detected by specific antibodies in the cellulosomes from all strains studied. Similar to the results for Man5K, no free Cel48F was found in any of the Fc fractions. Thus, Cel48F and Man5K were mainly found to be incorporated into the cellulosomes in the Fc fractions of the 952K and 954K strains.
FIG. 5.
Native PAGE (4 to 15% polyacrylamide) analysis of the Fc fractions from wild-type (WT) (5 μg), control (5 μg), D3K (5 μg), 952K (2.5 μg), and 954K (2.5 μg) strains. (A) Migration of purified Cel48F, rT-Man5K, and cellulosomes revealed by Coomassie blue staining. (B) Western blot probed with antibodies raised against Man5K. (C) Western blot probed with antibodies raised against Cel48F.
Catalytic properties of Man5K-enriched cellulosome fractions.
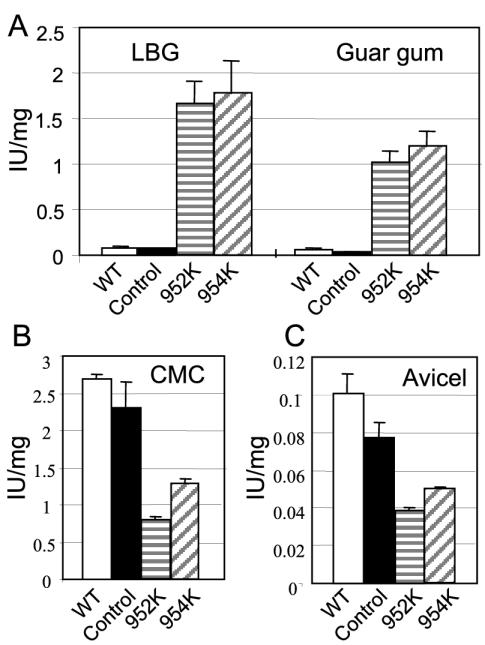
We showed that the Fc fractions from 952K and 954K contained large amounts of cellulosomal Man5K. In order to measure the impact of this mannanase enrichment on the enzymatic properties, the activities of the Fc fractions were tested with various substrates, including the galactomannan substrates guar gum and LBG and the cellulosic substrates CMC and Avicel. On both galactomannan substrates, a basal specific activity of ∼0.09 IU/mg was obtained for the wild-type and control Fc fractions (Fig. 6). The Fc fractions collected from the 952K and 954K strains had identical specific activities on the galactomannan substrates; these specific activities were 20 times higher than the basal specific activity. Both Fc fractions were more active with LBG than with guar gum. The Fc fraction from strain D3K did not show increased activity with the galactomannan substrates compared to the wild-type and control Fc fractions (data not shown). With cellulosic substrates, both modified Fc fractions displayed lower activities than the wild-type and control Fc fractions. On the other hand, the 952K Fc fraction displayed 40 and 20% lower activity than the 954K Fc fraction on CMC and Avicel, respectively. Thus, the Fc fraction which contained the largest amount of mannanase (952K) was the least active Fc fraction against cellulosic substrates.
FIG. 6.
Specific activities of the Fc fractions from the ATCC 35319 (WT), control, 952K, and 954K strains with the galactomannan substrates at a concentration of 0.5% (A), with 0.8% CMC (B), and with 0.8% Avicel (C). For CMC and Avicel, the values shown for ATCC 35319 and the control strain were obtained from previous results (24).
DISCUSSION
In C. cellulolyticum, 12 genes encoding cellulosomal catalytic subunits have been discovered so far. Most of these genes code for cellulases. Nevertheless, it has been shown that C. cellulolyticum cellulosomes (9) can degrade, in addition to cellulose, other plant cell wall polysaccharides. In some cases, these extra activities may be due to the ability of cellulases to degrade related substrates, such as xylan (5, 6), but it has also been shown that cellulosomes contain genuine pectinase (Rgl11Y, 21) and xylanases (17). Recently, a gene likely to encode a family 5 mannanase was found. The enzyme, designated Man5K, is highly homologous to the ManA enzyme produced by C. cellulovorans. Moreover, the two enzymes have the same unusual feature: a dockerin domain located at the N terminus and not at the C terminus, which is the case in all other cellulosomal enzymes secreted by both bacteria (27). ManA was characterized as an endo-acting β-1,4-mannanase active on glucomannan and galactomannans. Endomannanases are synthesized by bacteria and fungi, and their catalytic modules have been classified into glycosyl hydrolase families GH5 and GH26 (http://afmb.cnrs-mrs.fr/∼cazy/CAZY/index.html). While GH26 enzymes exhibit activity only with mannans, several GH5 mannanases display some activity with cellulosic substrates (4). Recombinant C. cellulovorans ManA was purified from E. coli as a truncated form in which the dockerin domain was removed. As observed for recombinant ManA (rManA), rT-Man5K and rFL-Man5K exhibited greater activity with LBG (a less galacto-substituted mannan) than with guar gum. Nevertheless, the C. cellulolyticum enzyme showed some enzymatic characteristics which were different from those established for ManA. rManA is fivefold less active with guar gum (3.0 IU/mg) than rT-Man5K (15.4 IU/mg; 613 IU/μmol). In addition, low but significant activity of rT-Man5K was detected with CMC (0.03 IU/mg; 1.2 IU/μmol), in contrast to rManA, for which carboxymethyl cellulase activity was undetectable. Surprisingly, the full-length Man5K was found to be four to five times less active with LBG and guar gum than the truncated form of the mannanase. Similar observations were made for the endocellulases Cel5A and Cel8C; in the latter cases it has been shown that deletion of the C-terminal dockerin improved the carboxymethyl cellulase activities approximately twofold (5, 6). The lower specific activities of the full-length mannanase on galactomannan substrates might be explained by high flexibility of the dockerin domain; in the free state, the dockerin domain might interact with the catalytic module and diminish its efficiency.
With the aim of overproducing Man5K in C. cellulolyticum, we used three plasmids having different promoters, pSOS952K, pSOS954K, and pD3K. We observed that the pD3K plasmid did not lead to overproduction of Man5K, thus suggesting that the 377-base DNA strand expected to contain the cipC promoter is not functional. In contrast, trans production of Man5K was successful when we used either intact Pthl (strain 952K) or mutated Pthl (strain 954K). Furthermore, the Fc fraction from the 952K strain was found to contain about twice as much mannanase as the Fc fraction from the 954K strain. This suggests that the mutated thiolase promoter directed weaker expression of the cloned gene in strain 954K.
Native gel Western blot analysis of the 952K and 954K Fc fractions (Fig. 5) indicated that Man5K was found almost exclusively in the cellulosomes. The major cellulases Cel48F and Cel9E were also found to be located almost exclusively in these complexes (data not shown). In concentrated culture supernatants, only trace amounts of Man5K and Cel48F were detected in the free state (data not shown). Man5K and Cel48F may therefore coexist as major components in the cellulosomes. These results are in good agreement with the results of previous biochemical studies, which indicated that the cohesin-dockerin interaction is not enzyme specific in a given species (7, 8, 19). Altogether, the data show that the enzymatic composition of cellulosomes is obviously regulated by the relative amounts of the enzymes available to be incorporated into the complexes. In the Man5K-overproducing strains, the mannanase units synthesized in large amounts might replace some natural cellulosomal enzymes or interact with unoccupied cohesins. Surprisingly, no major cellulosomal cellulases were found in the free state, which suggests that the major part of these enzymes was incorporated into the cellulosomes. It is possible that these enzymes are synthesized less in the recombinant strains and/or that these enzymes are degraded when they are not translocated and assembled into the cellulosomes.
Man5K enrichment induced a drastic increase in the cellulosome activity against galactomannan substrates compared to wild-type and control cellulosomes. The free rFL-Man5K and rT-Man5K and the Man5K-enriched cellulosomes preferred mannan with a low level of galacto substitution as the substrate (LBG). Incorporation of large amounts of Man5K concomitantly reduced the specific activity of the Fc fraction with cellulosic substrates. The decrease in cellulolytic activity appeared to be correlated with the amount of incorporated mannanase, since the 952K Fc fraction was 20 and 40% less active than the 954K Fc fraction on Avicel and CMC, respectively. The decrease probably reflected the addition of mannanase molecules in incomplete cellulosomes and/or replacement of cellulases by mannanases within the complexes.
No significant difference was observed between the specific activities exhibited by the two Fc fractions from strains 952K and 954K with galactomanann substrates. However, the cellulosomes from strain 952K have been shown to contain twofold more mannanase than the cellulosomes from strain 954K. The latter content was therefore sufficient to obtain the optimal activity with galactomannans in the experimental conditions used. Incorporation of additional mannanases into the complexes (strain 952K) did not lead to an increase in the rate of hydrolysis. It is hypothesized that only limiting amounts of hydrolyzable bonds are available in the vicinity of the complexes.
In previous work, an antisense RNA strategy was used to reduce the content of Cel48F produced by C. cellulolyticum ATCC 35319(pSOSasrF). A three- to fourfold decrease in the Cel48F content of the cellulosomes led to a 20% reduction in the specific activity with Avicel (24). The present work showed that the composition of the cellulosomes produced by C. cellulolyticum can be efficiently engineered in clostridia by structural gene expression instead of regulatory gene expression. Two strains producing different levels of mannanase were engineered. Other promoters should be examined in order to adapt more precisely the enzymatic composition of the complexes to the substrate to be degraded (pure cellulose or heterogeneous cellulosic substrate). New enzymatic activities might also be added by this method in order to tentatively increase the rate of hydrolysis of complex polysaccharides, like those found in the plant cell wall.
Acknowledgments
We thank R. Lebrun for performing N-terminal sequencing. We are grateful to Philippe Soucaille, Pascale de Philip, Sandrine Pagès, and Anne Galinier for fruitful discussions and to Odile Valette for technical assistance.
We acknowledge the financial support received from the Centre National de la Recherche Scientifique and Université de Provence and from Conseil Général des Bouches du Rhône and Région Provence-Alpes-Côtes d'Azur.
REFERENCES
- 1.Bagnara-Tardif, C., C. Gaudin, A. Belaich, P. Hoest, T. Citard, and J. P. Belaich. 1992. Sequence analysis of a gene cluster encoding cellulases from Clostridium cellulolyticum. Gene 119:17-28. [DOI] [PubMed] [Google Scholar]
- 2.Bayer, E. A., Y. Shoham, and R. Lamed. 2000. Cellulose-decomposing bacteria karyotes and their enzyme systems. In M. Dworkin, S. Falkow, E. Rosenberg, K.-H. Schleifer, and E. Stackebrandt (ed.), The prokaryotes: an evolving electronic resource for the microbiological community, 3rd ed. (release 3.7, February 2001). [Online.] Springer-Verlag, New York, N.Y. http://141.150.157.117:8080/prokPUB/chaprender/jsp/showchap.jsp?chapnum=297.
- 3.Bélaich, A., G. Parsiegla., L. Gal, C. Villard, R. Haser, and J.-P.Belaich. 2002. Cel9M, a new family 9 cellulase of the Clostridium cellulolyticum cellulosome. J Bacteriol. 184:1378-1384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cann, I. K. O., S. Kocherginskaya, M. R. King, B. A. White, and R. I. Mackie. 1999. Molecular cloning, sequencing, and expression of a novel multidomain mannanase gene from Thermoanaerobacterium polysaccharolyticum. J. Bacteriol. 181:1643-1651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fierobe, H. P., C. Gaudin, A. Belaich, M. Loutfi, E. Faure, C. Bagnara, and J. P. Belaich. 1991. Characterization of endoglucanase A from Clostridium cellulolyticum. J. Bacteriol. 173:7956-7962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fierobe, H. P., C. Bagnara-Tardif, C. Gaudin, F. Guerlesquin, P. Sauve, A. Belaich, and J. P. Belaich. 1993. Purification and characterization of endoglucanase C from Clostridium cellulolyticum—catalytic comparison with endoglucanase A. Eur. J. Biochem. 217:557-565. [DOI] [PubMed] [Google Scholar]
- 7.Fierobe, H. P., A. Mechaly, C. Tardif, A. Bélaich, R. Lamed, Y. Shoham, J. P. Bélaich, and E. A. Bayer. 2001. Design and production of active cellulosome chimeras. J. Biol. Chem. 276:21257-21261. [DOI] [PubMed] [Google Scholar]
- 8.Fierobe, H. P., E. A. Bayer, C. Tardif, M. Czjzek, A. Mechaly, A. Bélaich, R. Lamed, Y. Shoham, and J. P. Bélaich. 2002. Degradation of cellulose substrates by cellulosome chimeras-substrate targeting versus proximity of enzyme components. J. Biol. Chem. 277:49621-49630. [DOI] [PubMed] [Google Scholar]
- 9.Gal, L., S. Pagès, C. Gaudin, A. Belaich, C. Reverbel-Leroy, C. Tardif, and J. P. Belaich. 1997. Characterization of the cellulolytic complex (cellulosome) produced by Clostridium cellulolyticum. Appl. Environ. Microbiol. 63:903-909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Giallo, J., C. Gaudin, J. P. Bélaich, E. Petitdemange, and F Caillet-Mangin. 1983. Metabolism of glucose and cellobiose by cellulolytic mesophilic Clostridium sp. strain H10. Appl. Environ. Microbiol. 45:843-849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gottesman, S., E. Halpern, and P. Trisler. 1981. Role of sulA and sulB in filamentation by lon mutants of Escherichia coli K-12. J. Bacteriol. 148:265-273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Guedon, E., M. Desvaux, and H. Petitdemange. 2002. Improvement of cellulolytic properties of Clostridium cellulolyticum by metabolic engineering. Appl. Environ. Microbiol. 68:53-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hogg, D., G. Pell, P. Dupree, F. Goubet, S. M. Martin-Orue, S. Armand, and H. Gilbert. 2003. The modular architecture of Cellvibrio japonicus mannanases in glycoside hydrolase families 5 and 26 points to differences in their role in mannan degradation. Biochem. J. 371:1027-1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jennert, K. C., C. Tardif, D. L. Young, and M. Young. 2000. Gene transfer to Clostridium cellulolyticum ATCC 35319. Microbiology 146:3071-3080. [DOI] [PubMed] [Google Scholar]
- 15.Lowry, O. H., N. Rosebrough, A. L. Farr, and R. J. Randall. 1951. Protein measurement with the Folin phenol reagent. J. Biol. Chem. 193:265-275. [PubMed] [Google Scholar]
- 16.Maamar, H., O. Valette, H.-P. Fierobe, A. Bélaich, J.-P. Bélaich, and C. Tardif. 2004. Cellulolysis is severely affected in Clostridium cellulolyticum strain cipCMut1. Mol. Microbiol. 51:589-598. [DOI] [PubMed] [Google Scholar]
- 17.Mohand-Oussaid, O., S. Payot, E. Guedon, E. Gelhaye, A. Youyou, and H. Petitdemange. 1999. The extracellular xylan degradative system in Clostridium cellulolyticum cultivated on xylan: evidence for cell-free cellulosome production. J. Bacteriol. 181:4035-4040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pagès, S., A. Belaich, C. Tardif, C. Reverbel-Leroy, C. Gaudin, and J. P. Belaich. 1996. Interaction between the endoglucanase CelA and the scaffolding protein CipC of the Clostridium cellulolyticum cellulosome. J. Bacteriol. 178:2279-2286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pagès, S., A. Belaich, J. P. Belaich, E. Morag, R. Lamed, Y. Shoham, and E. A. Bayer. 1997. Species-specificity of the cohesin-dockerin interaction between Clostridium thermocellum and Clostridium cellulolyticum: prediction of specificity determinants of the dockerin domain. Proteins 29:517-527. [PubMed] [Google Scholar]
- 20.Pagès, S., A. Belaich, H. P. Fierobe, C. Tardif, C. Gaudin, and J. P. Belaich. 1999. Sequence analysis of scaffolding protein CipC and ORFXp, a new cohesin-containing protein in Clostridium cellulolyticum: comparison of various cohesin domains and subcellular localization of ORFXp. J. Bacteriol. 181:1801-1810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pagès, S., O. Valette, L. Abdou, A. Bélaich, and J.-P. Bélaich. 2003. A rhamnogalacturonan lyase in the Clostridium cellulolyticum cellulosome. J. Bacteriol. 185:4727-4733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Park, J. T., and M. J. Johnson. 1949. A submicrodetermination of glucose. J. Biol. Chem. 181:149-151. [PubMed] [Google Scholar]
- 23.Perret, S., L. Casalot, H.-P. Fierobe, C. Tardif., F. Sabathe, J.-P. Bélaich, and A. Bélaich. 2004. Production of heterologous and chimeric scaffoldins by Clostridium acetobutylicum ATCC 824. J. Bacteriol. 186:253-257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Perret, S., H. Maamar, J.-P. Bélaich, and C. Tardif. 2004. Use of antisense RNA to modify the composition of cellulosomes produced by Clostridium cellulolyticum. Mol. Microbiol. 51:599-607. [DOI] [PubMed] [Google Scholar]
- 25.Petitdemange, E., F. Caillet, J. Giallo, and C. Gaudin. 1984. Clostridium cellulolyticum sp. nov., a cellulolytic, mesophilic species from decayed grass. Int. J. Syst. Bacteriol. 34:155-159. [Google Scholar]
- 26.Reverbel-Leroy, C., S. Pagès, A. Belaich, J. P. Belaich, and C. Tardif. 1997. The processive endocellulase CelF, a major component of the Clostridium cellulolyticum cellulosome: purification and characterization of the recombinant form. J. Bacteriol. 179:46-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tamaru, Y., and R. H. Doi. 2000. The engL gene cluster of Clostridium cellulovorans contains a gene for cellulosomal ManA. J. Bacteriol. 182:244-247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tardif, C., H. Maamar, M. Balfin, and J.-P. Bélaich. 2001. Electrotransformation studies in Clostridium cellulolyticum. J. Ind. Microbiol. Biotechnol. 27:271-274. [DOI] [PubMed] [Google Scholar]
- 29.Tummala, S. B., N. E. Welker, and E. T. Papoutsakis. 1999. Development and characterisation of a gene expression reporter system for Clostridium acetobutylicum ATCC 824. Appl. Environ. Microbiol. 65:3793-3799. [DOI] [PMC free article] [PubMed] [Google Scholar]