Abstract
Bacterial chemoreceptors are embedded in the inner cell membrane in tight clusters. We show that changes in receptor methylation that generate large changes in kinase activity have relatively little effect on cluster morphology. Thus, changes in receptor activity do not appear to be mediated by changes in receptor-kinase assembly.
The sensory complex mediating chemotaxis in Escherichia coli consists of a mixture of transmembrane receptors, primarily Tar and Tsr (receptors for aspartate and serine), a coupling protein, CheW, and a histidine kinase, CheA (7, 10, 11). These complexes form clusters, primarily at the cell poles, to which other chemotaxis proteins bind (8, 18, 21, 25) (Fig. 1). In E. coli, CheA appears in two forms: a full-length form, CheAL, and a truncated form, CheAS, missing the first 97 amino acids, including the site of autophosphorylation. Both forms localize to receptor clusters (8, 12, 25). CheAL autophosphorylates and subsequently phosphorylates the response regulator CheY. Phosphorylated CheY diffuses through the cytoplasm and binds to flagellar motors to modulate bacterial swimming behavior; it is dephosphorylated by a phosphatase, CheZ.
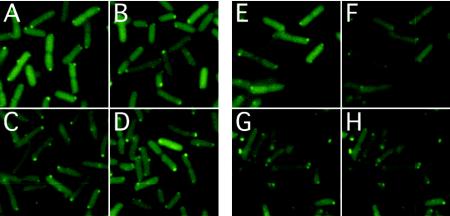
FIG. 1.
CheZ-YFP localization in cells with receptors in different modification states and in cells stimulated by attractant. (A to D) Localization in cells suspended in buffer (10 mM potassium phosphate, 0.1 mM EDTA, 1 μM l-methionine, 10 mM sodium lactate [pH 7]) expressing receptors in states TarEEEE (A), TarQEQE (B), mostly TsrEEEE (C), and TsrQEQE (D). (E to H) Localization in cells expressing TarQEQE in buffer (E), TarQEQE in buffer with 5 mM α-methylaspartate (F), TsrQEQE in buffer (G), or TsrQEQE in buffer with 10 mM serine (H). CheZ-YFP expression in motile cells was induced with 0.01% arabinose (A, B, E, and F) or 0.05 mM IPTG (isopropyl-β-D-thiogalactopyranoside) (C, D, G, and H), and fluorescence microscopy was performed as previously described (19, 21, 22).
Regulation of CheA activity plays a key role in chemotactic signal transduction. Activity is greatly enhanced by association with receptors that are free of chemoattractant, while inhibited by chemoattractant binding (4, 5, 19). Methylation of receptors on four specific glutamate (E) residues by a methyltransferase, CheR, increases CheA activity and is involved in adaptation (3, 14, 15, 19). Receptors are initially expressed in a half-modified state, QEQE, with two glutamines (Q) that are functionally similar to methylated glutamates. Receptors are demethylated and deamidated from glutamines to glutamates by a methylesterase, CheB.
The mechanism of kinase activity regulation by receptors is not well understood. The conventional model assumes that a conformational change in a receptor dimer caused by ligand binding or the addition of methyl groups affects activity of bound CheA (10). Most in vitro studies of intact receptors embedded in membranes have shown that changes in kinase activity with receptors in different modification states are not accompanied by differences in the state of assembly of the receptor complex (6, 11, 13, 14). However, two recent in vitro studies done with soluble receptor constructs (16, 22) have suggested that association of CheA with receptors might be strongly affected by receptor modification, whereas specific kinase activity (i.e., activity per kinase molecule associated with receptors) remains unchanged. In these studies, cytoplasmic receptor fragments were either assembled into complexes through fused leucine zippers (16) or histidine tagged and assembled on nickel-chelating lipid vesicles (22). Although it was argued that such in vitro assemblies might be more sensitive to the changes in electrostatic charge, due to modification than native receptor clusters (13), the relation of these results to receptor complex stability in vivo remained unclear. A recently published computer model of chemotaxis proposed variation in receptor complex stability with modification and ligand binding as a possible mechanism of kinase activity regulation (1). The aim of the present study was to study effects of chemoeffector binding and receptor modification on formation and activity of the receptor-kinase complex in intact cells by fluorescence microscopy.
Effect of receptor modification on receptor complex formation.
To assess the extent of formation of receptor-kinase complex, we used yellow fluorescent protein (YFP) fusions to CheZ and CheA, CheZ-YFP and YFP-CheAΔ258. CheAΔ258 has domains of CheA responsible for catalytic activity and for receptor and CheW binding, but it lacks the first 258 amino acids responsible for binding of CheY, CheB, CheZ, and the autophosphorylation site. CheZ is known to localize to receptor clusters through binding to CheAS (8) and has been used previously by others to study clustering (2). We monitored localization of CheZ-YFP in E. coli strains expressing either Tar or Tsr in different modification states (Fig. 1A to D). We used background strains deleted for the dipeptide receptor, Tap; the methylation and demethylation enzymes, CheR and CheB; and CheY and CheZ. The modification state of Tar was varied by directed substitutions of glutamate for glutamine or vice versa, whereas the modification state of Tsr was decreased by overexpression of CheB from a plasmid, pVS91 (Table 1) in the absence of CheR. Localization of CheZ-YFP was similar in strains with different modification states of the aspartate receptor, TarEEEE (VS144) (Fig. 1A) and TarQEQE (VS148) (Fig. 1B). In strains with different modification levels of the serine receptor, one expected to have mostly unmodified Tsr (pVS91/RP5135) (Fig. 1C) and another with half-modified TsrQEQE (RP5135) (Fig. 1D). Additionally, CheZ localization did not appear to be affected by stimulation with saturating concentrations of attractant added 2 min prior to image acquisition (Fig. 1E to H). Thus, these results show that neither receptor-kinase complex stability nor CheZ localization to the cluster depends strongly on the level of receptor modification or attractant stimulation. Similarly, localization of YFP-CheAΔ258 was not markedly affected (see below). This is consistent with an immunoelectron microscopy study that observed no difference in clustering of receptors in different modification states (17).
TABLE 1.
Strains and plasmids used in this study
| Strains or plasmidsa | Relevant genotype and characteristics | Reference or source |
|---|---|---|
| Strain | ||
| VS131 | tarQ309E Δ(tap-cheZ)2206 | 26 |
| VS147 | tarE491Q Δ(tap-cheZ)2206 | 26 |
| VS134 | tarQ295E Q309E Δ(tap-cheZ)2206 | 26 |
| VS141b | VS131 tsr::Tn5-1a | This work |
| VS144b | VS134 tsr::Tn5-1a | This work |
| VS148b | HCB42 tsr::Tn5-1a | 24 |
| VS149c | Δ(cheR-cheZ) | 24 |
| VS150b | VS147 tsr::Tn5-1a | 24 |
| VS172c | Δ(tar-cheZ) | 24 |
| HCB42 | Δ(tap-cheZ)2206 | J. S. Parkinson; 26 |
| RP5135 | Δ(tar-cheZ)2286 | J. S. Parkinson; 24 |
| RP5723 | tsr::Tn5-1a | J. S. Parkinson |
| Plasmid | ||
| pVS52d | CheZ-YFP expression plasmid, arabinose induction, Cmr | This work |
| pVS64 | CheZ-YFP expression plasmid, IPTG induction, Apr | This work |
| pVS88 | CheY-YFP/CheZ-CFP expression plasmid, IPTG induction, Apr | 24 |
| pVS91 | CheB expression plasmid, arabinose induction, Cmr | This work |
| PVS110de | YFP-CheAΔ258 expression plasmid, arabinose induction, Cmr | This work |
All strains are derivatives of RP437 (20).
Strains were constructed by P1 transduction from RP5723.
Strains were constructed by in-frame deletion of corresponding genes as described previously (25).
YFP was fused by a 5× Gly linker as described previously (25).
This plasmid expresses N-terminal fusion of YFP to CheA deleted for the first 258 amino acids, including P1 (phosphorylation) and P2 (CheY-binding domains).
To make a quantitative comparison of complex formation between strains expressing unmodified and half-modified Tar, we measured the ratio of maximal fluorescence intensity of CheZ-YFP or YFP-CheAΔ258 in the cluster to the mean fluorescence intensity of the cell for several cheR cheB strains with different receptor composition (Table 2). Localization intensities in all strains showed broad distributions with standard deviations being about 40% of the mean. We believe that this reflects natural cell-to-cell variation in cluster intensity. However, means of the distributions could be determined with much higher precision of about 1 to 2%, as characterized by standard errors. Our localization assay was clearly capable of detecting differences in clustering in strains expressing either both or only one of the major receptors. Tsr is more abundant than Tar (9), and degree of localization of both fusion proteins was proportional to the number of receptors in the cluster. In contrast, there was only a small decrease in localization in cells with unmodified receptors compared to cells with half-modified receptors. This decrease was significant for YFP-CheAΔ258, from 5.00 ± 0.11 to 4.41 ± 0.10, but at the limit of significance for CheZ-YFP, from 5.17 ± 0.08 to 5.10 ± 0.09. This was confirmed by Student's t test, which gave P values (the probability that distributions are not significantly different) of 0.0001 and 0.62 for CheA and CheZ fusions, respectively. It has to be noted, however, that our localization assay follows the equilibrium complex formation, and the possibility remains open that receptor modification might affect rates of complex assembly and disassembly to an equal extent without affecting binding equilibrium (15).
TABLE 2.
Localization of fusion proteins in strains with different receptor compositions
| Fusion/receptor modification | Mean localization ratioa | SD | SE | No. of cells with clustersb |
|---|---|---|---|---|
| CheZ-YFP/TarEEEE | 5.10 | 2.19 | 0.09 | 545 |
| CheZ-YFP/TarQEQE | 5.17 | 2.23 | 0.08 | 801 |
| CheZ-YFP/TsrQEQE | 5.63 | 2.56 | 0.10 | 701 |
| CheZ-YFP/TsrQEQE TarQEQE | 5.76 | 2.32 | 0.08 | 935 |
| YFP-CheAΔ258/TarEEEE | 4.41 | 1.74 | 0.10 | 310 |
| YFP-CheAΔ258/TarQEQE | 5.00 | 2.16 | 0.11 | 406 |
| YFP-CheAΔ258/TsrQEQE | 6.10 | 2.67 | 0.11 | 558 |
| YFP-CheAΔ258/TsrQEQE TarQEQE | 7.14 | 2.48 | 0.11 | 507 |
Localization ratio was determined as the maximum intensity (at the cluster) divided by the mean intensity of the cell. Note that this value depends on imaging resolution.
More than 85% of cells in all strains showed clustering. For cells without clusters, mean localization ratio, SD, and SE values were 2.20, 0.37, and 0.02, respectively.
Effect of receptor modification on kinase activity.
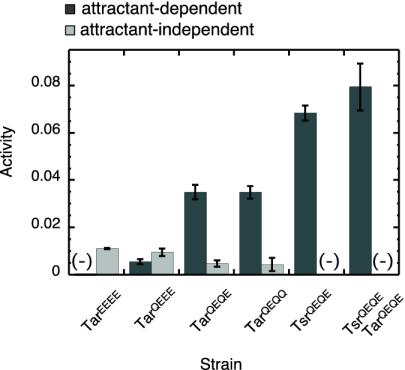
Kinase activity was assayed by a method based on fluorescence resonance energy transfer (FRET) that relies on phosphorylation-dependent interactions of CheZ-CFP (for cyan fluorescent protein) with CheY-YFP (23, 24, 26). The dependence of kinase activity on the state of Tar modification and on receptor composition is shown in Fig. 2. We measured both attractant-dependent activity (i.e., kinase activity associated with receptors, determined by measuring the changes in FRET generated by addition of saturating amounts of α-methylaspartate and serine) and attractant-independent activity (i.e., the difference between total activity, determined by YFP bleaching, and attractant-dependent activity). Changes in attractant-dependent kinase activity due to changes in receptor composition (levels of expression of Tar or Tsr) paralleled changes in intensities of clusters (Table 2). In contrast, changes in attractant-dependent kinase activity due to changes in receptor modification were much more dramatic than changes in intensities of clusters (Table 2). Virtually no attractant-dependent kinase activity was detected in complexes with TarEEEE compared to those with TarQEQE (Fig. 2), whereas their localization ratios decreased only moderately (Table 2). In addition to attractant-dependent kinase activity, FRET also revealed a small amount of attractant-independent kinase activity in the tsr tap strains. As this activity was not observed in the strains with a full complement of receptors (26), it is presumably explained by the activity of CheA that is not associated with receptors. Strains with lower receptor modification showed higher levels of attractant-independent FRET, which can be interpreted as a higher fraction of CheA that is not bound to receptors, consistent with our localization study (Fig. 1).
FIG. 2.
CheY phosphorylation activity in cells with different receptor compositions. Activity is expressed as the fractional change in CFP fluorescence due to FRET. Attractant-dependent kinase activity (dark-gray bars) was determined by adding saturating amounts of α-methylaspartate and serine in 0.1 and 10 mM concentrations, respectively. Total activity was determined by bleaching YFP (19, 20), and attractant-independent activity (light-gray bars) was calculated as the difference between total and attractant-dependent activity. (−), no activity could be measured. Error bars show standard errors.
In conclusion, while there appears to be some correlation with formation of the receptor-kinase complex and the state of receptor modification, the difference in CheZ and CheA binding to TarEEEE or TarQEQE clusters was much smaller than the difference in corresponding receptor kinase activity. So, consistent with the conventional view, changes in kinase activity in the cell do not appear to be due to major changes in the equilibrium of receptor complex assembly. Evidently, the activity must be regulated through conformational changes occurring within the receptor-kinase complex.
Acknowledgments
This work was supported by ZMBH funding to V.S. and by an NIH grant to H.C.B.
REFERENCES
- 1.Albert, R., Y. W. Chiu, and H. G. Othmer. 2004. Dynamic receptor team formation can explain the high signal transduction gain in Escherichia coli. Biophys. J. 86:2650-2659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ames, P., C. A. Studdert, R. H. Reiser, and J. S. Parkinson. 2002. Collaborative signaling by mixed chemoreceptor teams in Escherichia coli. Proc. Natl. Acad. Sci. USA 99:7060-7065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Borkovich, K. A., L. A. Alex, and M. I. Simon. 1992. Attenuation of sensory receptor signaling by covalent modification. Proc. Natl. Acad. Sci. USA 89:6756-6760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Borkovich, K. A., N. Kaplan, J. F. Hess, and M. I. Simon. 1989. Transmembrane signal transduction in bacterial chemotaxis involves ligand-dependent activation of phosphate group transfer. Proc. Natl. Acad. Sci. USA 86:1208-1212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bornhorst, J. A., and J. J. Falke. 2000. Attractant regulation of the aspartate receptor-kinase complex: limited cooperative interactions between receptors and effects of the receptor modification state. Biochemistry 39:9486-9493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bornhorst, J. A., and J. J. Falke. 2003. Quantitative analysis of aspartate receptor signaling complex reveals that the homogeneous two-state model is inadequate: development of a heterogeneous two-state model. J. Mol. Biol. 326:1597-1614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Boukhvalova, M. S., F. W. Dahlquist, and R. C. Stewart. 2002. CheW binding interactions with CheA and Tar. Importance for chemotaxis signaling in Escherichia coli. J. Biol. Chem. 277:22251-22259. [DOI] [PubMed] [Google Scholar]
- 8.Cantwell, B. J., R. R. Draheim, R. B. Weart, C. Nguyen, R. C. Stewart, and M. D. Manson. 2003. CheZ phosphatase localizes to chemoreceptor patches via CheA-short. J. Bacteriol. 185:2354-2361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Clarke, S., and D. E. Koshland, Jr. 1979. Membrane receptors for aspartate and serine in bacterial chemotaxis. J. Biol. Chem. 254:9695-9702. [PubMed] [Google Scholar]
- 10.Falke, J. J., and G. L. Hazelbauer. 2001. Transmembrane signaling in bacterial chemoreceptors. Trends Biochem. Sci. 26:257-265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gegner, J. A., D. R. Graham, A. F. Roth, and F. W. Dahlquist. 1992. Assembly of an MCP receptor, CheW, and kinase CheA complex in the bacterial chemotaxis signal transduction pathway. Cell 70:975-982. [DOI] [PubMed] [Google Scholar]
- 12.Kofoid, E. C., and J. S. Parkinson. 1991. Tandem translation starts in the cheA locus of Escherichia coli. J. Bacteriol. 173:2116-2119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Levit, M. N., T. W. Grebe, and J. B. Stock. 2002. Organization of the receptor-kinase signaling array that regulates Escherichia coli chemotaxis. J. Biol. Chem. 277:36748-36754. [DOI] [PubMed] [Google Scholar]
- 14.Levit, M. N., and J. B. Stock. 2002. Receptor methylation controls the magnitude of stimulus-response coupling in bacterial chemotaxis. J. Biol. Chem. 277:36760-36765. [DOI] [PubMed] [Google Scholar]
- 15.Li, G., and R. M. Weis. 2000. Covalent modification regulates ligand binding to receptor complexes in the chemosensory system of Escherichia coli. Cell 100:357-365. [DOI] [PubMed] [Google Scholar]
- 16.Liu, Y., M. Levit, R. Lurz, M. G. Surette, and J. B. Stock. 1997. Receptor-mediated protein kinase activation and the mechanism of transmembrane signaling in bacterial chemotaxis. EMBO J. 16:7231-7240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lybarger, S. R., and J. R. Maddock. 1999. Clustering of the chemoreceptor complex in Escherichia coli is independent of the methyltransferase CheR and the methylesterase CheB. J. Bacteriol. 181:5527-5529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Maddock, J. R., and L. Shapiro. 1993. Polar location of the chemoreceptor complex in the Escherichia coli cell. Science 259:1717-1723. [DOI] [PubMed] [Google Scholar]
- 19.Ninfa, E. G., A. Stock, S. Mowbray, and J. B. Stock. 1991. Reconstitution of the bacterial chemotaxis signal transduction system from purified components. J. Biol. Chem. 266:9764-9770. [PubMed] [Google Scholar]
- 20.Parkinson, J. S., and S. E. Houts. 1982. Isolation and behavior of Escherichia coli deletion mutants lacking chemotaxis functions. J. Bacteriol. 151:106-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shiomi, D., I. B. Zhulin, M. Homma, and I. Kawagishi. 2002. Dual recognition of the bacterial chemoreceptor by chemotaxis-specific domains of the CheR methyltransferase. J. Biol. Chem. 277:42325-42333. [DOI] [PubMed] [Google Scholar]
- 22.Shrout, A. L., D. J. Montefusco, and R. M. Weis. 2003. Template-directed assembly of receptor signaling complexes. Biochemistry 42:13379-13385. [DOI] [PubMed] [Google Scholar]
- 23.Sourjik, V., and H. C. Berg. 2002. Binding of the Escherichia coli response regulator CheY to its target measured in vivo by fluorescence resonance energy transfer. Proc. Natl. Acad. Sci. USA 99:12669-12674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sourjik, V., and H. C. Berg. 2004. Functional interactions between receptors in bacterial chemotaxis. Nature 428:437-441. [DOI] [PubMed] [Google Scholar]
- 25.Sourjik, V., and H. C. Berg. 2000. Localization of components of the chemotaxis machinery of Esherichia coli using fluorescent protein fusions. Mol. Microbiol. 37:740-751. [DOI] [PubMed] [Google Scholar]
- 26.Sourjik, V., and H. C. Berg. 2002. Receptor sensitivity in bacterial chemotaxis. Proc. Natl. Acad. Sci. USA 99:123-127. [DOI] [PMC free article] [PubMed] [Google Scholar]