Abstract
Streptomyces coelicolor A3(2) strain 2106 carries a 1.85-Mb linear plasmid, SCP1′-cysD, in addition to a 7.2-Mb linear chromosome. Macrorestriction analysis indicated that both linear DNAs are hybrids of the wild-type chromosome and the linear plasmid SCP1 on each side. Nucleotide sequencing of the fusion junctions revealed no homology between the recombination regions. SCP1′-cysD contains an SCP1 telomere and a chromosomal telomere at each end and therefore does not have terminal inverted repeats. In addition, SCP1′-cysD could not be eliminated from strain 2106 by various mutagenic treatments. Thus, we concluded that both the 7.2-Mb chromosome and SCP1′-cysD are chimeric chromosomes generated by a single crossover of the wild-type chromosome and SCP1. This may be regarded as a model of chromosomal duplication in genome evolution.
Streptomyces species are gram-positive filamentous soil bacteria with a high G+C composition (70 to 74%) and are well known for the production of a large number of secondary metabolites. Moreover, this genus is unusual among bacteria because its members have a linear chromosome (7, 38, 39, 52) and a linear plasmid(s) (16, 30, 33). Streptomyces linear chromosomes and plasmids have the same principal structural features: terminal inverted repeats (TIRs) are present at both ends and a terminal protein is covalently linked to the 5′ ends (1, 55).
Streptomyces coelicolor A3(2), the genetically best studied Streptomyces species, contains a linear chromosome (45) as well as two sex plasmids, the linear plasmid SCP1 (18, 50) and the circular plasmid SCP2 (4, 47). Recently, genome projects have been completed for all three of these replicons, namely, the S. coelicolor A3(2) chromosome (8,668 kb) (2), SCP1 (356 kb) (3), and SCP2*, a derivative of SCP2 (31 kb) (15).
Considerable attention has been given to SCP1 because of its interaction with the host chromosome. Hopwood and colleagues isolated variants carrying a hybrid SCP1-chromosome structure (18, 19, 21). Namely, SCP1 is integrated into the central region or other regions of the S. coelicolor A3(2) chromosome. Free SCP1-prime plasmids containing a certain chromosomal DNA stretch were also found. In matings with SCP1-free partners, SCP1-integrated strains showed either a unidirectional or bidirectional gradient of transfer of genetic markers with respect to the SCP1 integration site.
Previous studies reported the integrated structures of SCP1 in the normal fertility (NF) strain 2612 and the NF-like strain A634 (14, 54). These strains show similar types of bidirectional DNA transfer, although the directions of SCP1 integration are opposite and the deletion sizes at each end are totally different. Therefore, the molecular reason for directional DNA transfer is still unknown. On the other hand, genetic studies of the cysB donor strain 1984 and the cysD donor strain 2106 suggested that they contained an SCP1-prime plasmid, either SCP1′-cysB or SCP1′-cysD (20). A preliminary physical analysis revealed that SCP1′-cysB and SCP1′-cysD are giant linear plasmids, of 550 and 1,700 kb, respectively (35). However, their structural details and interaction with the host chromosome have not been clarified.
For this paper, we precisely analyzed the structures of SCP1′-cysD and the chromosome of S. coelicolor A3(2) strain 2106 and revealed the interaction between SCP1 and the wild-type chromosome in these linear DNA elements. Based on our results, we discuss the universality and function of TIRs of Streptomyces linear replicons and the origin and evolution of linear chromosomes.
MATERIALS AND METHODS
Bacterial strains, cosmids, plasmids, and media.
S. coelicolor A3(2) strain 1147 is the wild-type strain (17), and strains M145, M138, and 2106 are derivatives of strain 1147. The genotype and plasmid status of each strain are as follows (29): 1147, wild type/SCP1+ SCP2+; M145, prototroph/SCP1− SCP2−; M138, argA1 proA1 cysD1/SCP1+ SCP2−; and 2106, cysD donor pheA1/SCP1′-cysD SCP2+. Escherichia coli SURE2 was used as a host for a cosmid library, and E. coli XL1-Blue was used for cloning and nucleotide sequencing. A cosmid library of strain 2106 was constructed by a previously described method (44, 45) using the vector Supercos1 (10) and a Gigapack III Gold packaging kit (Stratagene, La Jolla, Calif.). pSCP201 contains the 3.9-kb SpeI end fragment of SCP1 (34), and pSUL221 contains the 1.3-kb BamHI end fragment of the M145 chromosome (22). YEME medium (29) was used for liquid cultures of S. coelicolor A3(2) strains. MYM agar, which was used for methylenomycin sensitivity assays, contains 0.4% maltose, 0.4% yeast extract, 1.0% malt extract, and 2.0% agar.
DNA preparation and pulsed-field gel electrophoresis.
Total DNAs of S. coelicolor A3(2) strains were prepared as described by Kieser et al. (29). DNA gel samples were prepared by the mycelium method (31, 38), and SCP1′-cysD and SCP1 were separated by contour-clamped homogeneous electric fields (CHEF) (9). CHEF assays were performed with 0.5× Tris-borate-EDTA buffer and 1.0% agarose gels at 14°C. E. coli strains were cultured in Luria-Bertani medium, and cosmid and plasmid DNAs were isolated as described by Sambrook et al. (46).
Southern hybridization experiments.
DNA fragments were separated by CHEF or conventional agarose gel electrophoresis and then transferred to nylon membrane filters as described by Kieser et al. (29). For colony hybridization, cosmid clones were inoculated directly onto nylon membrane filters, cultured on Luria-Bertani ampicillin plates, and treated as described by Sambrook et al. (46). For the preparation of large hybridization probes, such as the 146-kb AseI fragment of SCP1 and the AseI-B fragment of the M145 chromosome (1,450 kb), a CHEF assay was performed with low-melting-point agarose, and excised bands were melted at 70°C and extracted with phenol and then phenol-chloroform (1:1). DNA fragments were labeled with digoxigenin-11-dUTP (Roche Diagnostics, Mannheim, Germany), and hybridization and detection were carried out as described by the supplier.
Telomere cloning and nucleotide sequencing.
The telomere fragments of SCP1′-cysD were separated first by CHEF electrophoresis and then by conventional agarose gel electrophoresis and were cloned into pUC19 by force cloning. For force cloning (12, 34, 37), the telomere fragment, which should still retain a small peptide covalently bound to the 5′ end after a protease treatment, was directly ligated to the vector and subjected to transformation. Nucleotide sequences were determined by the dideoxy termination method with a Dye Terminator cycle sequencing kit (Amersham Pharmacia Biotech, Uppsala, Sweden) and an ABI-373S sequencer (PE Biosystems, Foster City, Calif.). The Genetyx-Mac 10.1 program (Software Development, Tokyo, Japan) was used for analyses of sequence data.
PCR.
Primers for PCR amplification of the junction fragment of SCP1′-cysD were designed based on the junction sequence of the 7.2-Mb chromosome, and their sequences were as follows: primer A, 5′-GCGTCCTTCAGaAGCTtGACAAG-3′ (complementary to nucleotides [nt] 125780 to 125758 of the SCP1 sequence [accession no. AL590463]); and primer B, 5′-AAGAGCCGAatTCCCGTTGGGGTG-3′ (complementary to nt 176767 to 176744 of the M145 chromosome sequence [accession no. AL023861]). Lowercase letters indicate the bases that were introduced to create recognition sites for HindIII and EcoRI, respectively. PCRs were done with the total DNA of strain 2106 and with KOD-Plus DNA polymerase (Toyobo, Tokyo, Japan).
Curing experiments and bioassay.
To eliminate SCP1′-cysD from strain 2106, we used the following curing methods. Strain 2106 was grown in liquid YEME medium at 28°C for 2 days and then cultured for an additional 24 h in the presence of acriflavin (20 to 60 μg/ml) or ethidium bromide (20 to 60 μg/ml) (26) or at a high temperature (42°C). The mycelia exposed to mutagens were washed with 20% glycerol and separated by use of a homogenizer. Suitable dilutions of each mycelium suspension were spread and grown on MYM agar, and 500 single colonies were isolated from each group, with each group showing <1% survival. All 1,500 single colonies were grown adjacent to strain M138 (SCP1+) on MYM agar, and their sensitivity to methylenomycin was tested.
RESULTS
Macrorestriction analysis of SCP1′-cysD and the 7.2-Mb chromosome of strain 2106.
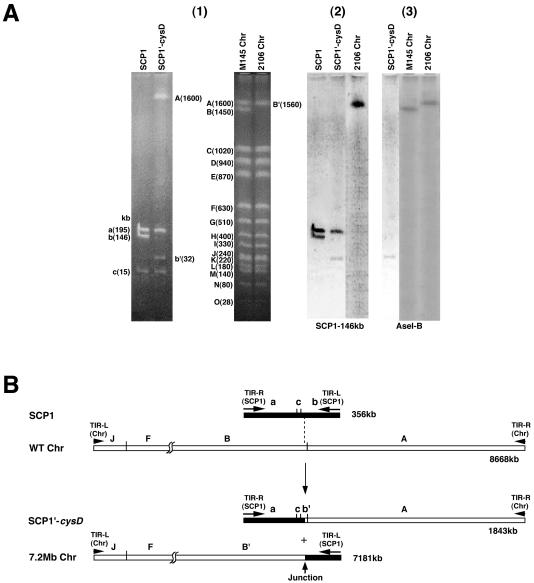
The structures of the SCP1-prime plasmid SCP1′-cysD and the chromosome of S. coelicolor A3(2) strain 2106 were analyzed by comparisons with SCP1 isolated from strain M138 and the total DNA of strain M145, with the latter being used as a wild-type chromosome. As shown in Fig. 1A (panel 1), three fragments (195, 146, and 15 kb; see Fig. 1B for a restriction map) were observed for an AseI digest of SCP1 (32). In an SCP1′-cysD digest, the 195- and 15-kb fragments were conserved, but two new fragments, of 1,600 and 32 kb, appeared in place of the 146-kb fragment. Since the 32-kb fragment gave positive hybridization with both the 146-kb SCP1 fragment and the AseI B fragment of the M145 chromosome (Fig. 1A, panels 2 and 3), this fragment was suggested to be a hybrid of SCP1 and the wild-type chromosome. Hybridization of the 146-kb SCP1 probe to the 195-kb fragment of both SCP1 and SCP1′-cysD (Fig. 1A, panel 2) was due to the presence of the 79-kb TIRs at both ends of SCP1. Hybridization experiments using an ordered cosmid library of the M145 chromosome (45) revealed that the 1,600-kb fragment of SCP1′-cysD was the AseI A fragment of the wild-type chromosome itself (data not shown).
FIG. 1.
Comparison of AseI fragments of the linear DNA elements in S. coelicolor A3(2) strains by CHEF assay and Southern hybridization (A) and generation model of SCP1′-cysD and the 7.2-Mb chromosome in strain 2106 (B). (A) (1) CHEF electrophoresis. CHEF electrophoresis was done at 150 V with 90-s pulses for 36 h. Fragment sizes were based on data from genome projects. (2 and 3) Southern hybridization. The 146-kb AseI fragment of SCP1 and the AseI B fragment of the M145 chromosome were used as probes for panels 2 and 3, respectively. (B) Generation model. Black and white bars indicate the SCP1 and chromosomal regions, respectively, with the former enlarged to twice its size. AseI recognition sites, fragment names, and TIRs are also included. The two recombination points are connected by a dashed line.
We next digested the chromosomal DNA of strain 2106, which remained in the wells of CHEF gels, and the M145 chromosome with AseI and compared the results. As shown in Fig. 1A, panel 1, all of the AseI fragments of the M145 chromosome, except for the AseI B fragment, seemed to be present in strain 2106. However, the AseI A fragment of the M145 chromosome did not hybridize to the 1,560-kb AseI B′ fragment (data not shown), while both the 146-kb SCP1 fragment and the M145 AseI B fragment hybridized to this fragment (Fig. 1A, panels 2 and 3). These results indicated that the 1,560-kb AseI B′ fragment was a hybrid of the 146-kb fragment of SCP1 and the AseI B fragment of the wild-type chromosome. Based on the total sizes of the AseI fragments of strain 2106 together with genome project data for the M145 chromosome (2), the chromosomal size of strain 2106 was calculated to be 7.2 Mb. Thus, it was suggested that in strain 2106, SCP1′-cysD and the 7.2-Mb chromosome were generated by a single crossover between the 8,668-kb wild-type chromosome and the 356-kb linear plasmid SCP1 (Fig. 1B; the direction of SCP1 is shown opposite to that reported in reference 32).
Nucleotide sequences of SCP1-chromosome junctions.
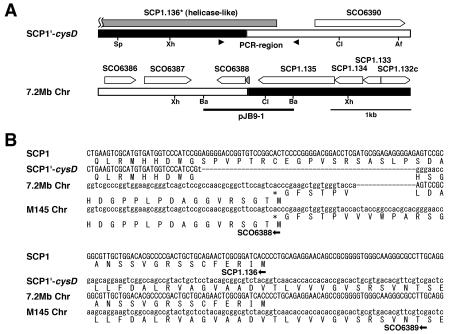
To confirm that there was a single crossover between SCP1 and the wild-type chromosome in strain 2106, we tried to clone the SCP1-chromosome junctions in SCP1′-cysD and the 7.2-Mb chromosome. First, a cosmid library was constructed for the total DNA of strain 2106 by use of a partial digest with Sau3AI. Of 2,000 cosmids obtained, 83 clones were selected by positive hybridization to SCP1 DNA. An additional round of hybridization using the chromosomal AseI B fragment identified four candidate cosmids (6B3, 7G1, 8E8, and 11B2). Hybridization with SCP1′-cysD and the 7.2-Mb chromosome revealed that these cosmids were not derived from SCP1′-cysD but from the 7.2-Mb chromosome.
The four junction clones hybridized to both SCP1 cosmid 32 (44) and the chromosomal cosmid 3C8 (45), with the latter being located in the AseI B fragment. Restriction and hybridization analysis with these cosmids located the junction of the 7.2-Mb chromosome in a 1.1-kb BamHI fragment (pJB9-1) (Fig. 2A). This fragment was subcloned from cosmid 8E8 and then sequenced (Fig. 2B). The results, together with genome project data for SCP1 and the M145 chromosome, determined the SCP1-chromosome junction site in the 7.2-Mb chromosome and also defined another junction in SCP1′-cysD. The deduced 1.0-kb DNA fragment containing the latter junction was amplified by a PCR with the two primers indicated in Fig. 2A (arrowheads) (see Materials and Methods for primer sequences), cloned into pUC19, and sequenced.
FIG. 2.
Gene organization (A) and nucleotide sequences (B) around the junctions of SCP1′-cysD and the 7.2-Mb chromosome in strain 2106. (A) pJB9-1 contains the junction of the 7.2-Mb chromosome, and the two arrowheads indicate the primers for PCR amplification of the SCP1′-cysD junction. The names of open reading frames are according to genome projects for S. coelicolor A3(2) and SCP1. SCP1.136* codes for the mutated helicase in SCP1′-cysD. Af, AflII; Ba, BamHI; Cl, ClaI; Sp, SphI; Xh, XhoI. (B) Nucleotide sequences of the corresponding regions of SCP1 and the M145 chromosome. It was revealed that a 55-bp SCP1 DNA and a 15-bp chromosomal DNA were deleted and a T residue was inserted during the recombination process.
As shown in Fig. 2B, a sequence comparison of the two junctions and their corresponding regions in SCP1 (cosmid 32; accession no. AL590463) and the wild-type chromosome (cosmid 3C8; accession no. AL023861) revealed no homology between the two recombination regions. Thus, the two junctions were generated by nonhomologous recombination between SCP1 and the wild-type chromosome. During this event, a 55-bp DNA from SCP1 and a 15-bp DNA from the wild-type chromosome, shown with dashed lines in the figure, were deleted, and one T residue (in lowercase) was added. The crossover points were located on a putative helicase gene (SCP1.136) in SCP1 and on an unknown gene (SCO6388) in the chromosome. Two fusion genes were generated by recombination: they were a mutated helicase gene in SCP1′-cysD coding for an elongated N-terminal region and a hypothetical gene in the 7.2-Mb chromosome coding for a small protein of only 24 amino acids, which therefore may not function.
Nucleotide sequences of the telomeres of SCP1′-cysD.
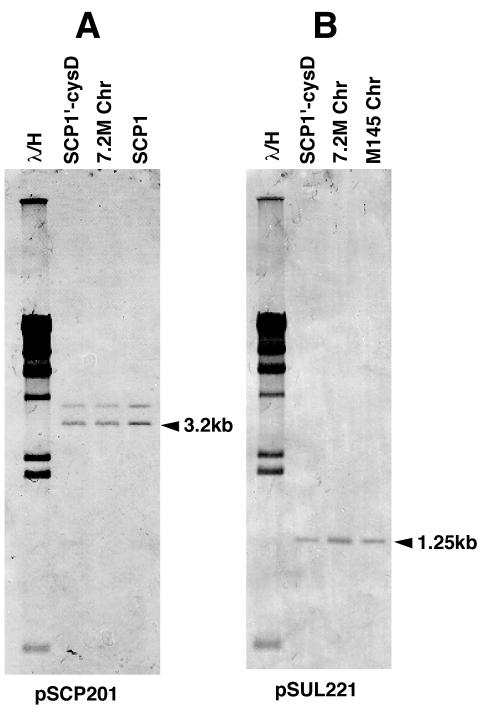
Most Streptomyces linear chromosomes and plasmids that have been analyzed thus far have TIRs at both ends. In this respect, SCP1′-cysD and the 7.2-Mb chromosome were quite interesting, because they seemed to not have TIRs; rather, the telomere sequences of SCP1 (34) and the M145 chromosome (22) are totally different. When it was probed by the SCP1 end clone pSCP201 (34) and the chromosomal end clone pSUL221 (22), an NcoI digest of SCP1′-cysD showed hybridizing signals at the same positions as SCP1 (3.2 kb) and the M145 chromosome (1.25 kb) (Fig. 3; we used NcoI because BamHI gave terminal fragments of similar sizes [1.25 and 1.33 kb] for both SCP1 and the chromosome). In addition, the 7.2-Mb chromosome also showed the same two signals (Fig. 3).
FIG. 3.
Southern hybridization analysis of the ends of SCP1′-cysD and the 7.2-Mb chromosome of strain 2106. SCP1, SCP1′-cysD, the M145 chromosome, and the 7.2-Mb chromosome were digested with NcoI, separated by conventional agarose gel electrophoresis, and analyzed by Southern hybridization. pSCP201 (A) and pSUL221 (B) were used as probes for the SCP1 end and the chromosomal end, respectively.
To clone both ends of SCP1′-cysD, we digested it with SpeI and separated the fragments by CHEF electrophoresis. The 3.9- and 50-kb fragments, which contain the SCP1 (left) and chromosome (right) ends, respectively, were isolated. The former was cloned into pUC19 which had been digested with BamHI and HincII. The 50-kb SpeI fragment was further digested with BamHI, and the resultant 1.33-kb end fragment was cloned. The nucleotide sequences of the two right-end clones were identical to that of the S. coelicolor A3(2) chromosome telomere (…TGTGGTACCCGCTCCGCGGG-3′). On the other hand, the three left-end clones showed minor heterogeneity. Specifically, two clones contained four G residues at the 5′ end and one clone contained three G residues [5′-(G)GGGCGGAGAGGCCTAACGGC…]. A similar heterogeneity (four to six G residues) was previously reported for the 5′ end of SCP1 (34). It is not known whether these differences represent real heterogeneity in SCP1′-cysD or if they were generated during the cloning process. In either case, the terminal sequences of the chimeric molecule are different and therefore are not repeats of each other.
Failure of curing of SCP1′-cysD from strain 2106.
It was easily speculated that SCP1′-cysD is indispensable for the survival of strain 2106 because it contains a 1.6-Mb DNA fragment from the right end of the wild-type chromosome. To study this further, we tried to cure SCP1′-cysD from strain 2106 by the following three mutagenic treatments: incubation with acriflavin or ethidium bromide (26) and incubation at a high temperature (42°C). Since SCP1′-cysD carries the methylenomycin biosynthetic and resistance genes (6), mutants that lost SCP1′-cysD would become sensitive to methylenomycin. Thus, 500 colonies isolated after each mutagenic treatment were grown adjacent to strain M138, which carries SCP1 and produces methylenomycin. Eleven, 9, and 3 colonies of the acriflavin, ethidium bromide, and high-temperature groups showed some growth inhibition. All of these colonies, plus 15 well-grown colonies randomly selected from each group, were further analyzed by CHEF electrophoresis for the presence of SCP1′-cysD. All of the tested colonies were confirmed to still carry SCP1′-cysD.
DISCUSSION
For this study, we analyzed the structures of the two linear DNA elements, SCP1′-cysD and the 7.2-Mb chromosome, in S. coelicolor A3(2) strain 2106. Macrorestriction analysis and nucleotide sequencing revealed that both DNA elements were formed by a single crossover between nonhomologous regions of the wild-type chromosome and SCP1, with deletions of a 15-bp chromosomal DNA and a 55-bp SCP1 DNA. Therefore, the sizes of SCP1′-cysD and the chromosome of strain 2106 were calculated to be 1,843 and 7,181 kb, respectively. The right and left telomeres of SCP1′-cysD were also cloned and sequenced, and the results showed that the telomeres of the wild-type chromosome and SCP1 are conserved at each end. This may also be true for the 7.2-Mb chromosome, as its NcoI digest gave hybridizing signals at the same positions as the end fragments of the chromosome (1.25 kb) and SCP1 (3.2 kb). We were not able to cure SCP1′-cysD by various mutagenic treatments, which indicated that SCP1′-cysD is indispensable for survival. Thus, both the 7.2-Mb chromosome and SCP1′-cysD are chimeric chromosomes, and we named them chromosomes I and II, respectively.
Strain 2106 transferred a cysD phenotype (cysD donor) in a mating with an SCP1-negative strain (19). Consequently, it was believed that the cysD gene is located on the giant linear plasmid SCP1′-cysD. However, from our sequence data, the cysD gene was deduced to be present on the AseI B′ fragment of the 7.2-Mb chromosome at a spot that is 356 kb from the junction. Therefore, the 1,843-kb linear DNA element should be called chromosome II instead of SCP1′-cysD, and the gene-transferring property of strain 2106 should be analyzed physically based on the present results. Apart from this problem, the chimeric chromosomes of strain 2106 are noteworthy in two aspects, i.e., the universality and function of the TIRs of Streptomyces linear replicons and the origin and evolution of linear chromosomes, which are discussed below in detail.
Most of the linear chromosomes and plasmids isolated so far from Streptomyces have TIRs at both ends. The sizes of the TIRs of linear plasmids are quite different, ranging from 44 bp for SLP2 in S. lividans (8) to 95 kb for pPZG101 in S. rimosus (13). The sizes of Streptomyces linear chromosomes are also different, ranging from 168 bp for S. avermitilis (24) to about 6,500 kb for the end-to-end fused chromosome of the S. ambofaciens mutant NSA65 (53). On the other hand, Kalkus et al. (27) reported that plasmid pHG201 in Rhodococcus opacus has TIRs of only 3 bp, if they can be called TIRs. Pandza et al. (41) showed that recombination between plasmid pPZG101 and the chromosome of S. rimosus led to an exchange of their ends. Based on its restriction map, the generated hybrid plasmid pPZG103 seemed not to have TIRs. Recently, the linear plasmid SLP2 was shown to be a composite plasmid comprising the chromosome and a linear plasmid on each side (23). In addition, this study revealed that neither of the two chimeric chromosomes in strain 2106 has TIRs but that they contain an SCP1 telomere and a chromosomal telomere at each end.
Streptomyces linear chromosomes are unstable and frequently cause deletions at one or both ends (7, 36, 51). The deleted chromosomes subsequently display dynamic rearrangements such as circularization, arm replacement, and amplification. We proposed the following hypothesis for the function of TIRs in Streptomyces linear replicons (48). When one of the two telomeres is deleted inside the TIR region, recombinational DNA repair may function between the intact and deleted TIR sequences on the same chromosome and recover an intact telomere. This event always repairs telomere damage inside the TIR region. However, we cannot observe it because an identical TIR structure is reproduced. Thus, the TIRs may guarantee homologous sequences for recombination, and this is the reason that most of the Streptomyces linear replicons have TIRs at both ends.
When a deletion in one telomere extends beyond the TIR region, the intact TIR structure cannot be recovered in this way. However, if nonallelic similar sequences are present on the right and left arms, homologous recombination between them causes chromosomal arm replacement and reproduces a telomere at the end (11, 48). In this case, some chromosomal sequences are lost and new, longer TIRs are formed. This may be the reason that the sizes of TIRs vary greatly in Streptomyces strains and that homologous genes or insertion elements are frequently found at the inside ends of TIRs, and this possibility was pointed out previously (34). When both telomeres are deleted, the extreme ends cannot be recovered, and therefore the chromosome will be circularized by nonhomologous recombination to survive (25, 28). Long TIRs, which are formed by chromosomal arm replacement, also suffer telomere deletions. If a second recombination occurs at the deletion ends inside the long TIRs, a circular chromosome with a large palindromic structure is generated (49). Qin and Cohen (43) analyzed similar structural changes of pSLA2 derivatives and discussed the strategies of Streptomyces linear replicons after telomere damage.
Without TIRs at both ends, how can Streptomyces linear replicons recover an intact telomere after terminal deletion? Qin and Cohen (43) demonstrated that when a linear plasmid which contained a pSLA2 telomere at one end and a damaged chromosomal telomere at another end was introduced into S. lividans ZX7, the damaged telomere was repaired by intermolecular recombination between the plasmid and the chromosome. This result, together with the presence of linear replicons without TIRs, such as pHG210, pPZG103, SLP2, and the two chimeric chromosomes in strain 2106, suggests that TIRs are not essential for Streptomyces linear replicons under exceptional conditions. Specifically, if identical telomere sequences are present on two different linear replicons in the same cell, they may compensate for each other intermolecularly in recombinational DNA repair.
Streptomyces linear chromosomes contain two replication mechanisms, bidirectional replication from a centrally located replication origin (40) and terminal replication primed by a terminal protein (42). The replication origins of Streptomyces chromosomes are quite similar to those of typical bacterial circular chromosomes (5, 56), while their telomere sequences can make a secondary fold-back structure like those of adenoviruses and parvoviruses (22, 42). Thus, it was proposed that Streptomyces linear chromosomes may have been generated in the evolutionary past by a single crossover between a bacterium-type circular chromosome and a linear plasmid or phage which contained TIRs at both ends and terminal proteins bound to the 5′ ends (7, 52). In addition, the results obtained in this study suggested another evolutionary possibility, i.e., that duplication and multiplication of a linear chromosome may have occurred in a similar way to the mechanism for the generation of chromosomes I and II in S. coelicolor A3(2) strain 2106.
Acknowledgments
We are indebted to D. A. Hopwood for S. coelicolor A3(2) strains and to C. W. Chen for plasmid pSUL221.
This work was supported by a grant-in-aid for Scientific Research on Priority Area “Genome Biology” from the Ministry of Education, Culture, Sports, Science and Technology of Japan. M. Yamasaki was supported by a scholarship from the Japan Society for the Promotion of Sciences.
REFERENCES
- 1.Bao, K., and S. N. Cohen. 2001. Terminal proteins essential for the replication of linear plasmids and chromosomes in Streptomyces. Genes Dev. 15:1518-1527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bentley, S. D., K. F. Chater, A. M. Cerdeno-Tarraga, G. L. Challis, N. R. Thomson, K. D. James, D. E. Harris, M. A. Quail, H. Kieser, D. Harper, A. Bateman, S. Brown, G. Chandra, C. W. Chen, M. Collins, A. Cronin, A. Fraser, A. Goble, J. Hidalgo, T. Hornsby, S. Howarth, C. H. Huang, T. Kieser, L. Larke, L. Murphy, K. Oliver, S. O'Neil, E. Rabbinowitsch, M. A. Ralandream, K. Rutherford, S. Rutter, K. Seeger, D. Saunders, S. Sharp, R. Squares, S. Squares, K. Taylor, T. Warren, A. Wietzorrek, J. Woodward, B. G. Barrell, J. Parkhill, and D. A. Hopwood. 2002. Complete genome sequence of the model actinomycete Streptomyces coelicolor A3(2). Nature 417:141-147. [DOI] [PubMed] [Google Scholar]
- 3.Bentley, S. D., S. Brown, L. D. Murphy, D. E. Harris, M. A. Quail, J. Parkhill, B. G. Barrell, J. R. McCormick, R. I. Santamaria, R. Losick, M. Yamasaki, H. Kinashi, C. W. Chen, G. Chandra, D. Jakimowicz, H. M. Kieser, T. Kieser, and K. F. Chater. 2004. SCP1, a 356,023 bp linear plasmid adapted to the ecology and developmental biology of its host, Streptomyces coelicolor A3(2). Mol. Microbiol. 51:1615-1628. [DOI] [PubMed] [Google Scholar]
- 4.Bibb, M. J., R. F. Freeman, and D. A. Hopwood. 1977. Physical and genetic characterization of a second sex factor, SCP2 for Streptomyces coelicolor A3(2). Mol. Gen. Genet. 154:155-166. [Google Scholar]
- 5.Calcutt, M., and F. J. Schmidt. 1992. Conserved gene arrangement in the origin region of the Streptomyces lividans chromosome. J. Bacteriol. 174:3220-3225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chater, K. F., and C. J. Bruton. 1984. Resistance, regulatory and production genes for the antibiotic methylenomycin are clustered. EMBO J. 4:1893-1897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen, C. W., C. H. Huang, H. H. Lee, H. H. Tsai, and R. Kirby. 2002. Once the circle has been broken: dynamics and evolution of Streptomyces chromosomes. Trends Genet. 18:522-529. [DOI] [PubMed] [Google Scholar]
- 8.Chen, C. W., T. W. Yu, Y. S. Lin, H. M. Kieser, and D. A. Hopwood. 1993. The conjugative plasmid SLP2 of Streptomyces lividans is a 50 kb linear molecule. Mol. Microbiol. 7:925-932. [DOI] [PubMed] [Google Scholar]
- 9.Chu, G., D. Vollrath, and R. W. Davis. 1986. Separation of large DNA molecules by contour-clamped homogeneous electric fields. Science 234:1582-1585. [DOI] [PubMed] [Google Scholar]
- 10.Evans, G. A., K. Lewis, and B. E. Rothenberg. 1989. High efficiency vectors for cosmid microcloning and genomic analysis. Gene 79:9-20. [DOI] [PubMed] [Google Scholar]
- 11.Fischer, G., T. Wenner, B. Decaris, and P. Leblond. 1998. Chromosomal arm replacement generates a high level of intraspecific polymorphism in the terminal inverted repeats of the linear chromosomal DNA of Streptomyces ambofaciens. Proc. Natl. Acad. Sci. USA 95:14296-14301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Goshi, K., T. Uchida, A. Lezhava, M. Yamasaki, K. Hiratsu, H. Shinkawa, and H. Kinashi. 2002. Cloning and analysis of the telomere and terminal inverted repeat of the linear chromosome of Streptomyces griseus. J. Bacteriol. 184:3411-3415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gravius, B., D. Glocker, J. Pigac, K. Pandza, D. Hranueli, and J. Cullum. 1994. The 387 kb linear plasmid pPZG101 of Streptomyces rimosus and its interactions with the chromosome. Microbiology 140:2271-2277. [DOI] [PubMed] [Google Scholar]
- 14.Hanafusa, T., and H. Kinashi. 1992. The structure of an integrated copy of the giant linear plasmid SCP1 in the chromosome of Streptomyces coelicolor 2612. Mol. Gen. Genet. 231:363-368. [DOI] [PubMed] [Google Scholar]
- 15.Haug, I., A. Weissenborn, D. Brolle, S. Bentley, T. Kieser, and J. Altenbuchner. 2003. Streptomyces coelicolor A3(2) plasmid SCP2*: deductions from the complete sequence. Microbiology 149:505-513. [DOI] [PubMed] [Google Scholar]
- 16.Hayakawa, T., T. Tanaka, K. Sakaguchi, N. Otake, and H. Yonehara. 1979. A linear plasmid-like DNA in Streptomyces sp. producing lankacidin group antibiotics. J. Gen. Appl. Microbiol. 25:255-260. [Google Scholar]
- 17.Hopwood, D. A. 1959. Linkage and mechanism of recombination in Streptomyces coelicolor. Ann. N. Y. Acad. Sci. 81:887-898. [DOI] [PubMed] [Google Scholar]
- 18.Hopwood, D. A., and T. Kieser. 1993. Conjugative plasmids of Streptomyces, p. 293-311. In D. B. Clewell (ed.), Bacterial conjugation. Plenum Press, New York, N.Y.
- 19.Hopwood, D. A., and H. M. Wright. 1976. Interaction of the plasmid SCP1 with the chromosome of Streptomyces coelicolor A3(2), p. 607-619. In K. D. MacDonald (ed.), Second International Symposium on the Genetics of Industrial Microorganisms. Academic Press, London, United Kingdom.
- 20.Hopwood, D. A., and H. M. Wright. 1976. Genetic studies on SCP1-prime strains of Streptomyces coelicolor A3(2). J. Gen. Microbiol. 95:107-120. [DOI] [PubMed] [Google Scholar]
- 21.Hopwood, D. A., R. J. Harold, A. Vivian, and H. M. Ferguson. 1969. A new kind of fertility variant in Streptomyces coelicolor. Genetics 62:461-477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huang, C. H., Y. S. Lin, Y. L. Yang, S. W. Huang, and C. W. Chen. 1998. The telomeres of Streptomyces chromosomes contain conserved palindromic sequences with potential to form complex secondary structures. Mol. Microbiol. 28:905-916. [DOI] [PubMed] [Google Scholar]
- 23.Huang, C. H., C. Y. Chen, H. H. Tsai, C. Chen, Y. S. Lin, and C. W. Chen. 2003. Linear plasmid SLP2 of Streptomyces lividans is a composite replicon. Mol. Microbiol. 47:1563-1576. [DOI] [PubMed] [Google Scholar]
- 24.Ikeda, H., J. Ishikawa, A. Hanamoto, M. Shinose, H. Kikuchi, T. Shiba, Y. Sakaki, M. Hattori, and S. Omura. 2003. Complete genome sequence and comparative analysis of the industrial microorganism Streptomyces avermitilis. Nat. Biotechnol. 21:526-531. [DOI] [PubMed] [Google Scholar]
- 25.Inoue, S., K. Higashiyama, T. Uchida, K. Hiratsu, and H. Kinashi. 2003. Chromosomal circularization in Streptomyces griseus by nonhomologous recombination of deletion ends. Biosci. Biotechnol. Biochem. 67:1101-1108. [DOI] [PubMed] [Google Scholar]
- 26.Kahler, R., and D. Noack. 1974. Action of acridine orange and ethidium bromide on growth and antibiotic activity of Streptomyces hygroscopicus JA 6599. Z. Allg. Mikrobiol. 14:529-533. [DOI] [PubMed] [Google Scholar]
- 27.Kalkus, J., R. Menne, M. Reh, and H. G. Schlegel. 1998. The terminal structures of linear plasmids from Rhodococcus opacus. Microbiology 144:1271-1279. [DOI] [PubMed] [Google Scholar]
- 28.Kameoka, D., A. Lezhava, H. Zenitani, K. Hiratsu, M. Kawamoto, K. Goshi, K. Inada, H. Shinkawa, and H. Kinashi. 1999. Analysis of fusion junctions of circularized chromosomes in Streptomyces griseus. J. Bacteriol. 181:5711-5717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kieser, T., M. J. Bibb, M. J. Buttner, K. F. Chater, and D. A. Hopwood. 2000. Practical Streptomyces genetics. The John Innes Foundation, Norwich, England.
- 30.Kinashi, H. 1994. Linear plasmids from actinomycetes. Actinomycetologica 8:87-96. [Google Scholar]
- 31.Kinashi, H. 1994. Pulsed field gel electrophoresis: isolation and analysis of large linear plasmids, p. 227-240. In J. W. Dale and P. G. Sanders (ed.), Methods in gene technology, vol. 2. JAJ Press, London, United Kingdom.
- 32.Kinashi, H., and M. Shimaji-Murayama. 1991. Physical characterization of SCP1, a giant linear plasmid from Streptomyces coelicolor. J. Bacteriol. 173:1523-1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kinashi, H., M. Shimaji, and A. Sakai. 1987. Giant linear plasmids in Streptomyces which code for antibiotic biosynthesis genes. Nature 328:454-456. [DOI] [PubMed] [Google Scholar]
- 34.Kinashi, H., M. Shimaji-Murayama, and T. Hanafusa. 1991. Nucleotide sequence analysis of the unusually long terminal inverted repeats of giant linear plasmid, SCP1. Plasmid 26:123-130. [DOI] [PubMed] [Google Scholar]
- 35.Kinashi, H., M. Murayama, H. Matsushita, and O. Nimi. 1993. Structural analysis of the giant linear plasmid SCP1 in various Streptomyces coelicolor strains. J. Gen. Microbiol. 139:1261-1269. [Google Scholar]
- 36.Leblond, P., and B. Decaris. 1994. New insights into the genetic instability of Streptomyces. FEMS Microbiol. Lett. 123:225-232. [DOI] [PubMed] [Google Scholar]
- 37.Levings, C. S., III, and R. R. Sederoff. 1983. Nucleotide sequence of the S-2 mitochondrial DNA from the S cytoplasm of maize. Proc. Natl. Acad. Aci. USA 80:4055-4059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lezhava, A., T. Mizukami, T. Kajitani, D. Kameoka, M. Redenbach, H. Shinkawa, O. Nimi, and H. Kinashi. 1995. Physical map of the linear chromosome of Streptomyces griseus. J. Bacteriol. 177:6492-6498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lin, Y. S., H. M. Kieser, D. A. Hopwood, and C. W. Chen. 1993. The chromosomal DNA of Streptomyces lividans 66 is linear. Mol. Microbiol. 10:923-933. [DOI] [PubMed] [Google Scholar]
- 40.Musialoski, M. S., F. Flett, G. B. Scott, G. Hobes, C. P. Smith, and S. G. Oliver. 1994. Functional evidence that the principal DNA replication origin of the Streptomyces coelicolor chromosome is close to the dnaA-gyrB region. J. Bacteriol. 176:5123-5125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pandza, S., G. Biukovic, A. Paravic, A. J. Dadbin, J. Cullum, and D. Hranueli. 1998. Recombination between the linear plasmid pPZG101 and the linear chromosome of Streptomyces rimosus can lead to exchange of ends. Mol. Microbiol. 28:1165-1176. [DOI] [PubMed] [Google Scholar]
- 42.Qin, Z., and S. N. Cohen. 1998. Replication at the telomeres of the Streptomyces linear plasmid pSLA2. Mol. Microbiol. 28:893-903. [DOI] [PubMed] [Google Scholar]
- 43.Qin, Z., and S. N. Cohen. 2002. Survival mechanism for Streptomyces linear replicons after telomere damage. Mol. Microbiol. 45:785-794. [DOI] [PubMed] [Google Scholar]
- 44.Redenbach, M., K. Ikeda, M. Yamasaki, and H. Kinashi. 1998. Cloning and physical mapping of the EcoRI fragments of the giant linear plasmid SCP1. J. Bacteriol. 180:2796-2799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Redenbach, M., H. M. Kieser, D. Denapalte, A. Eichner, J. Cullum, H. Kinashi, and D. A. Hopwood. 1996. A set of ordered cosmids and a detailed genetic and physical map for the 8 Mb Streptomyces coelicolor A3(2) chromosome. Mol. Microbiol. 21:77-96. [DOI] [PubMed] [Google Scholar]
- 46.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 47.Schrempf, H., H. Bujard, and D. A. Hopwood. 1975. Isolation of covalently closed circular deoxyribonucleic acid from Streptomyces coelicolor. J. Bacteriol. 121:416-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Uchida, T., M. Miyawaki, and H. Kinashi. 2003. Chromosomal arm replacement in Streptomyces griseus. J. Bacteriol. 185:1120-1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Uchida, T., N. Ishihara, H. Zenitani, K. Hiratsu, and H. Kinashi. 2004. Circularized chromosome with a large palindromic structure in Streptomyces griseus mutants. J. Bacteriol. 186:3313-3320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Vivian, A., and D. A. Hopwood. 1973. Genetic control of fertility in Streptomyces coelicolor A3(2): new kinds of donor strains. J. Gen. Microbiol. 76:147-162. [DOI] [PubMed] [Google Scholar]
- 51.Volff, J. N., and J. Altenbuchner. 1998. Genetic instability of the Streptomyces chromosome. Mol. Microbiol. 27:239-246. [DOI] [PubMed] [Google Scholar]
- 52.Volff, J. N., and J. Altenbuchner. 2000. A new beginning with new ends: linearisation of circular chromosomes during bacterial evolution. FEMS Microbiol. Lett. 186:143-150. [DOI] [PubMed] [Google Scholar]
- 53.Wenner, T., V. Roth, G. Fischer, C. Fourrier, B. Aigle, B. Decaris, and P. Leblond. 2003. End-to-end fusion of linear deleted chromosomes initiates a cycle of genome instability in Streptomyces ambofaciens. Mol. Microbiol. 50:411-425. [DOI] [PubMed] [Google Scholar]
- 54.Yamasaki, M., M. Redenbach, and H. Kinashi. 2001. Integrated structures of the linear plasmid SCP1 in two bidirectional donor strains of Streptomyces coelicolor A3(2). Mol. Gen. Genet. 264:634-642. [DOI] [PubMed] [Google Scholar]
- 55.Yang, C. C., C. H. Huang, C. Y. Li, Y. G. Tsay, S. C. Lee, and C. W. Chen. 2002. The terminal proteins of linear Streptomyces chromosomes and plasmids: a novel class of replication priming proteins. Mol. Microbiol. 43:297-305. [PubMed] [Google Scholar]
- 56.Zakrzewska-Czerwinska, J., and H. Schrempf. 1992. Characterization of an autonomously replicating region from the Streptomyces lividans chromosome. J. Bacteriol. 147:2688-2693. [DOI] [PMC free article] [PubMed] [Google Scholar]