Abstract
Several sequencing projects unexpectedly uncovered the presence of genes that encode ribulose 1,5-bisphosphate (RuBP) carboxylase/oxygenase (RubisCO) in anaerobic archaea. RubisCO is the key enzyme of the Calvin-Benson-Bassham (CBB) reductive pentose phosphate pathway, a scheme that does not appear to contribute greatly, if at all, to net CO2 assimilation in these organisms. Recombinant forms of the archaeal enzymes do, however, catalyze a bona fide RuBP-dependent CO2 fixation reaction, and it was recently shown that Methanocaldococcus (Methanococcus) jannaschii and other anaerobic archaea synthesize catalytically active RubisCO in vivo. To complete the CBB pathway, there is a need for an enzyme, i.e., phosphoribulokinase (PRK), to catalyze the formation of RuBP, the substrate for the RubisCO reaction. Homology searches, as well as direct enzymatic assays with M. jannaschii, failed to reveal the presence of PRK. The apparent lack of PRK raised the possibility that either there is an alternative pathway to generate RuBP or RubisCO might use an alternative substrate in vivo. In the present study, direct enzymatic assays performed with alternative substrates and extracts of M. jannsachii provided evidence for a previously uncharacterized pathway for RuBP synthesis from 5-phospho-d-ribose-1-pyrophosphate (PRPP) in M. jannaschii and other methanogenic archaea. Proteins and genes involved in the catalytic conversion of PRPP to RuBP were identified in M. jannaschii (Mj0601) and Methanosarcina acetivorans (Ma2851), and recombinant Ma2851 was active in extracts of Escherichia coli. Thus, in this work we identified a novel means to synthesize the CO2 acceptor and substrate for RubisCO in the absence of a detectable kinase, such as PRK. We suggest that the conversion of PRPP to RuBP might be an evolutional link between purine recycling pathways and the CBB scheme.
Ribulose 1,5-bisphosphate (RuBP) carboxylase/oxygenase (RubisCO) is one of the two unique enzymes of the Calvin-Benson-Bassham (CBB) reductive pentose phosphate pathway, along with phosphoribulokinase (PRK), the enzyme responsible for generating RuBP, the substrate for RubisCO. Archaeal genome sequencing projects for several organisms, including Methanocaldococcus (Methanococcus) jannaschii, Archaeoglobus fulgidus, Thermococcus kodakaraensis, and Methanosarcina acetivorans, unexpectedly revealed genes encoding RubisCO (3, 6, 12, 14), and recombinant enzymes encoded by genes derived from each of these organisms were shown to catalyze authentic RubisCO reactions (5, 14, 28). Subsequently, M. jannaschii, A. fulgidus, and M. acetivorans were found to synthesize catalytically active RubisCO under normal growth conditions (5). Based on sequence homologies, these archaeal RubisCO proteins have been placed in a new class, designated form III, that is distinct from the previously characterized form I and form II RubisCO proteins from plants, algae, and bacteria that require the CBB pathway for autotrophic growth (8, 9, 22, 27). Despite the presence of RubisCO, it is clear from previous whole-cell CO2 incorporation studies that methanogenic archaea do not employ the CBB pathway for primary CO2 assimilation (21). In addition, no evidence for the presence of PRK, the second enzyme unique to the CBB cycle, was obtained from either sequence comparisons or extensive in vitro assays (5). It appears to be unlikely that an organism would invest energy in producing a protein like RubisCO in vivo if it was unable to synthesize its substrate. The lack of demonstrable PRK activity led to the proposition that archaeal RubisCO might utilize a substrate different from RuBP or that there is an alternative pathway for producing RuBP. Certainly, it is clear that homologs of RubisCO, the RubisCO-like proteins (RLP) (8), catalyze non-RubisCO-type reactions (1, 8, 9). Thus, perhaps the archaeal protein, despite being a bona fide RubisCO, does catalyze a physiologically significant alternative reaction with a substrate(s) other than RuBP. Previously, however, we showed that the archaeal RubisCO gene complements RubisCO deletion strains of nonsulfur purple photosynthetic bacteria to autotrophic CO2-dependent growth (5). The archaeal enzyme thus obviously uses RuBP as a physiologically significant substrate. Although by no means conclusive, the latter results appear to argue for the possibility that there may be some alternative means to synthesize RuBP in methanogenic archaea as archaeal RubisCO, unlike RLP, does function to assimilate CO2 in a background in which this capability is absolutely essential (5).
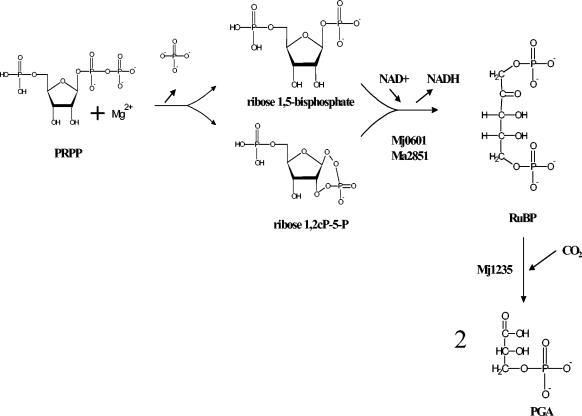
The present study provided strong evidence that an alternative RuBP-generating pathway is employed, at least in M. jannaschii, as well as other methanogenic archaea. The key precursor metabolite used for RuBP synthesis in this scheme is 5-phospho-d-ribose-1-pyrophosphate (PRPP). We propose that dephosphorylation of the β-phosphate at the C-1 position of PRPP produces ribose 1,5-bisphosphate and that this is followed by enzyme-catalyzed conversion of ribose 1,5-bisphosphate to RuBP. This pathway sidesteps the need for a specific kinase similar to PRK in the CBB pathway because PRPP already contains both phosphates; instead, the issue becomes one of converting ribose 1,5-bisphosphate to the keto form of the sugar, namely, RuBP.
MATERIALS AND METHODS
Growth of M. jannaschii and preparation of cell extracts.
M. jannaschii was cultured as previously described (17). The cell material was frozen in liquid nitrogen and stored at −80°C. Frozen cell material was chipped off inside an anaerobic chamber and suspended in TEMMB buffer (pH 7.5) (25 mM Tris-Cl, 1 mM EDTA, 10 mM MgCl2, 10 mM 2-mecaptoethanol, 50 mM sodium bicarbonate) for enzyme assays or TEMMB buffer (pH 8.5) for purifying the enzyme that catalyzes RuBP formation. The cells were disrupted in a French pressure cell under an argon atmosphere, and the lysate was subsequently decanted inside the chamber into an anaerobic centrifuge tube with a sealable O ring. The cell lysate was centrifuged at 30,000 × g for 20 min at 4°C. The supernatant was filtered with a 0.45-μm-pore-size filter. The protein concentration was determined by the Lowry method by using bovine serum albumin as a standard (15).
Immunoprecipitation of M. jannaschii RubisCO in cell extracts.
It was empirically determined that 25 μl of crude polyclonal RubisCO antisera could efficiently inhibit the RubisCO activity found in 1.5 ml of desalted M. jannaschii crude extract (1.44 mg). Desalting was accomplished by passing the extract through a G-25 superfine gel filtration column inside an anaerobic chamber. The extract and antisera were incubated on ice for 1 h. During the incubation, 100 μl of Staphylococcus aureus Cowan I was resuspended and then mixed with the lysate after the 1-h incubation. The S. aureus Cowan I-lysate slurry was incubated for 30 min on ice and then centrifuged at 10,000 × g for 15 min at 4°C. The supernatant was transferred to a fresh tube and used for subsequent assays.
DNA manipulations, genetic techniques, gene expression, and recombinant protein preparation.
The Mj0601 gene was amplified from genomic M. jannaschii DNA by using Unipol (PGC Scientific) and incorporating an NdeI site at the N terminus (5′AAGATACATATGGTGAATCTAATGAATATAAAGGAT3′) and a BamHI site at the C terminus (5′AAAAGCGGATCCTTATTCTTTATTTTTCAACTTTTCAGT3′). The amplified product was cloned into pCRTopo 2.1 (Invitrogen) and sequenced to ensure that no point mutation occurred after amplification of the gene. For expression in Escherichia coli, the gene was subcloned into the unique NdeI and BamHI sites in pET11A (Stratagene) and transformed into the E. coli BL21 overexpression strain. The strategy used to clone the homologue of Mj0601 from M. acetivorans (Ma2851) was very similar; the primers used contained an NdeI site at the N terminus (5′AAGATACATATGGAACTTGACGAAGTCATAATCACA3′) and a BamHI site at the C terminus (5′AAAAGCGGATCCTTAGATTGTTTTAAGCCTGTCAAGGGC3′). The amplified product was ligated into pCRTopo 2.1 (Invitrogen) and sequenced. The gene was then subcloned into pET11A as described above for the Mj0601 DNA sequence. For expression, E. coli BL21 containing the Ma2851 gene in the pET vector was grown in 1 liter of Luria-Bertani medium (10 g of tryptone per liter, 5 g of yeast extract per liter, 10 g of NaCl per liter; pH 7.5) supplemented with 100 μg of ampicillin per liter in a 2-liter flask. Cultures were grown with shaking at 120 rpm at 37°C until an optical density at 600 nm of 0.4 was reached. The culture was shifted to a temperature of 42°C for 30 min and then cooled to 25°C. At this time, isopropyl-β-d-thiogalactopyranoside (IPTG) was added to a final concentration of 0.1 mM, and induction was performed for 4 h at 25°C with shaking at 85 rpm. After induction, cells were centrifuged aerobically and stored at −80°C. For recombinant Ma2851 protein preparation, approximately 4 g (wet weight) of E. coli BL21 cells was resuspended in 50 ml of anaerobic TEMB buffer (TEMMB lacking 2-mercaptoethanol) and disrupted by two passes through a French pressure cell under anaerobic conditions. The extract was decanted into an anaerobic centrifuge tube sealed with a cap containing an O ring. The disrupted cell material was centrifuged at 20,000 × g for 30 min at 4°C. The soluble protein was then filtered through a 0.45-μm-pore-size filter in an anaerobic chamber.
RubisCO assay.
The radiometric RubisCO assay was modified to allow for a strict anaerobic environment (5, 23). The assay buffer for M. jannaschii extracts contained 80 mM HEPES-KOH (pH 7.2). All manipulations were performed by using gas-tight Hamilton syringes with beveled needles. All assay vials contained H14CO3− at a final concentration of 2 μCi, and the temperature used for the assays was 83°C. The assay mixtures typically contained 0.8 mM RuBP. PRPP was substituted at the same concentration where indicated below. The assays were quenched by injection of propionic acid, and the serum vials were opened to the air, treated, and then counted for radioactivity as previously described (5). To verify the authenticity of any RuBP-dependent CO2 fixation observed, the RubisCO transition state analog 2-carboxyarabinitol-1,5-bisphosphate (CABP) was used at a final concentration of 0.24 mM (18).
PRPP utilization and stoichiometry.
To determine the stoichiometry of PRPP utilization, PRPP conversion was coupled to RubisCO catalysis, and the amount of the product formed, 3-phosphoglyceric acid (PGA), was determined by standard spectrophotometric assays. PRPP disappearance was coupled to RubisCO catalysis in 1.0-ml cuvettes (9SOG-10-GL14-S Starna cells) with sealable caps with rubber septa that had been equilibrated anaerobically. The assay mixtures (1.0 ml) contained 800 μg of M. jannaschii extract, 0.6 mM NAD, 100 μg of purified recombinant A. fulgidus RubisCO (as coupling enzyme), 20 mM bicarbonate, 25 mM MgCl2, and 2.4 mM PRPP. The assays were carried out in a heated spectrophotometric block (Cary 100 UV/Spec) at 83°C for 10 min. The cuvettes were then exposed to air and placed on ice. Samples were removed from the cuvettes and prepared for PRPP and PGA determinations.
To determine the levels of PGA formed, RubisCO assays were performed anaerobically at 83°C with unlabeled bicarbonate. The reaction was terminated by exposure to air and heating the reaction mixture to 100°C for 5 min. A sample was centrifuged for 60 min at 4.5 rcf (1.18 × 10−6 × radius [in millimeters] × [revolutions per minute]2) at 4°C in an Amicon Ultrafree-MC 5,000 NMWL centrifugal filter device (Millipore). Heating and filtering were necessary to reduce the high background levels of NADH oxidation found in untreated M. jannaschii extracts. The filtrate was injected into an aerobic cuvette (Starna cell [see above]), and PGA was measured exactly as previously described (23).
PRPP utilization was determined by using a coupled assay with orotic acid and commercially available ortidine-5-phosphate pyrophosphorylase and ortidine-5-phosphate decarboxylase, as modified from the method of Micheli et al. (16). M. jannaschii extracts were filtered as described above but not heated to 100°C. The assays were performed aerobically at room temperature, and PRPP utilization was monitored at a wavelength of 295 nm in quartz cuvettes (Varian). The assay mixture contained 20 mM Tris-Cl (pH 8.0), 200 μM orotic acid, and 600 μM MgCl2. The linear range for the assay was between 50 and 150 μM PRPP. After a baseline absorbance was reached, 2.5 U of ortidine-5-phosphate pyrophosphorylase and 2.5 U of ortidine-5-phosphate decarboxylase were injected into the cuvette and mixed with a plumper. An extinction coefficient of 3,950 M−1 cm−1 at 295 nm (16) was used to detect the amount of orotic acid utilized, and this value was used to calculate the amount of PRPP remaining.
Enzymatic conversion of ribose 1,5-bisphosphate to RuBP.
Radiometric RubisCO-coupled assays were employed to determine the ability of recombinant Ma2851 to catalyze the formation of RuBP at 37°C. Prior to the assay, 8.3 mM PRPP, 50 mM NaHCO3, 25 mM MgCl2, and 2 μCi H14CO3− in 80 mM HEPES-KOH (pH 7.2) was incubated in a sealed vial at 83°C for 1 h. This high-temperature preincubation is known to stimulate conversion of PRPP to ribose 1,5-bisphosphate in the presence of Mg2+ ions (2, 4, 19). After cooling for a minimum of 5 min, the solution was injected into either open or closed serum vials equilibrated at 37°C containing NAD, Ma2851 protein, and purified recombinant Rhodosprillum rubrum RubisCO. Controls containing the RubisCO transition state analog CABP were assayed in parallel. The final concentration of PRPP was 4.0 mM, and 0.6 mM NAD was added to each serum vial whether the assay was performed aerobically or anaerobically. The assays were quenched, and acid-stable labeled PGA was counted like it was in RubisCO assays.
Synthesis of ribose 1,5-bisphosphate.
Ribose 1,5-bisphosphate was enzymatically synthesized by incubating glucose 1,6-phosphate with ribose 1-phosphate in the presence of commercially available glucose-6-phosphate dehydrogenase and phosphoglucomutase (11, 20). Ribose 1,5-bisphosphate was purified by using a DEAE-Sepharose Fast Flow column (7). For quantification of ribose 1,5-bisphosphate we used established protocols (7) after mild acidic hydrolysis of ribose 1,5-bisphosphate to ribose 5-phosphate.
Purification of Mj0601 from extracts of M. jannaschii.
All purification procedures and assays of column fractions were performed under strict anaerobic conditions. Isolation of Mj0601 from M. jannaschii was accomplished by resuspending 16 g (wet weight) of M. jannaschii cells in 40 ml of TEMMB buffer (pH 8.5); these cells were disrupted with a French pressure cell under an argon atmosphere. The extract was decanted inside an anaerobic chamber into an anaerobic centrifuge bottle with an O-ring seal and centrifuged at 30,000 × g for 20 min at 4°C. The supernatant was decanted inside the anaerobic chamber and filtered through a 0.45-μm-pore-size filter. The filtered extract was loaded onto a DEAE-Sepharose Fast Flow column, and active fractions eluted between 17 and 20 mS/cm after a KCl gradient was applied. The sample was then loaded onto a Q-Sepharose high-performance (QSHP) column, and active fractions eluted between 22 and 29 mS/cm after a KCl gradient was applied. Anaerobic ammonium sulfate was added to the active QSHP fraction to a final concentration of 1.7 M, and the sample was then loaded onto a Phenyl-Sepharose high-performance (PSHP) column. The column was subjected to a reverse gradient of ammonium sulfate, and the protein eluted at 17 mS/cm. The protein was concentrated by centrifugation by using a Millipore Biomax 50K NMWL membrane filter (Sigma) inside the chamber and was loaded onto a 110-ml Superose-12 gel filtration column. Active fractions eluted from the column at a position corresponding to a molecular mass of 168 kDa and appeared as a single band on a sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE) gel. The protein was then subjected to N-terminal sequencing by using protocols provided by the Molecular Structure Facility at the University of California, Davis (http://msf.ucdavis.edu).
RESULTS
Potential precursors of RuBP formation and subsequent CO2 fixation.
Previous work indicated that extracts of M. jannaschii possess RubisCO activity (5); however, all attempts to demonstrate PRK activity failed. These negative results suggested that this organism does not possess a specific kinase to phosphorylate ribulose 5-phosphate to RuBP. In the present study, a variety of different compounds were tested and assayed anaerobically at 83°C to determine if radiolabeled 14CO2 could be incorporated into an acid-stable product (PGA) by the endogenous RubisCO activity found in extracts of M. jannaschii. The compounds tested included RuBP, PRPP, ribose 1,5-bisphosphate, ADP plus ribose 5-phosphate, ribose 5-phosphate, AMP, ADP plus ribose 1-phosphate, ribose 1-phosphate, ATP, GTP, CTP, TTP, glucose 1,6-bisphosphate, and fructose 1,6-bisphosphate. Besides RuBP, the only other substrates that supported significant 14CO2 fixation into an acid-stable product were ribose 1,5-bisphosphate and PRPP. In the presence of the RubisCO-specific transition state inhibitor CABP, little incorporation of 14CO2 into an acid-stable product was obtained with either of these substrates (Fig. 1A). In assay mixtures containing ribose 1,5-bisphosphate, a compound prepared at low yields, the activity was low compared to the activity with PRPP and RuBP (added at a concentration of 800 μM) because the available levels of ribose 1,5-bisphosphate (51.8 μM) were limiting in the assay. Neither PRPP nor ribose 1,5-bisphosphate served as a substrate for RubisCO since purified M. jannaschii RubisCO failed to use either compound as a CO2 acceptor (data not shown).
FIG. 1.
PRPP- and ribose 1,5-bisphosphate-dependent 14CO2 fixation into an acid-stable product by extracts of M. jannaschii. (A) PRPP- and ribose 1,5-bisphosphate (R1,5-P)-dependent 14CO2 fixation in the presence (grey bars) and in the absence (open bars) of the RubisCO transition state inhibitor CABP. (B) RubisCO dependence of acid-stable 14CO2 incorporation in extracts of M. jannaschii. The open bars show the results obtained under normal assay conditions. The gray bars show the results of assays performed in the presence of antibody generated against recombinant M. jannaschii RubisCO with mixtures incubated as described in Materials and Methods. All assays were performed for 60 min.
Specificity of PRPP-dependent CO2 fixation.
In extracts of M. jannaschii, PRPP-dependent CO2 fixation was inhibited by both CABP and antibodies to M. jannaschii RubisCO (Fig. 1B). Control experiments with preimmune serum indicated that the serum itself did not inhibit RubisCO activity (data not shown). To further prove the specificity of the M. jannaschii antibodies for inhibition of PRPP-dependent CO2 fixation, assays were performed with mixtures containing the M. jannaschii antibodies as described above; however, purified R. rubrum RubisCO was injected into the anaerobic reaction mixture (at 30°C). When form II R. rubrum RubisCO, which shows absolutely no cross-reactivity with the M. jannaschii RubisCO, was added, significant amounts of acid-stable product were again obtained. In summary, inhibition of CO2 fixation either by the RubisCO-specific transition state inhibitor CABP or by M. jannaschii RubisCO antibodies proved that PRPP-dependent 14CO2 incorporation was dependent on RubisCO catalysis, which requires RuBP as the only demonstrable CO2 acceptor.
To determine if the novel PRPP-dependent RuBP formation reaction might occur in organisms other than archaea, experiments were performed with extracts from photoautotrophically grown Rhodobacter capsulatus strain SB1003. In the presence of PRPP or ribose 1,5 bisphosphate, no 14CO2 incorporation or acid-stable product was detected (data not shown). Thus, a M. jannaschii-type RuBP formation enzyme may not be present in this organism, which contains an established CBB pathway, in which PRK is expected to catalyze RuBP formation.
The incorporation of 14CO2 into an acid-stable product followed a typical time course when either RuBP or PRPP was used as the substrate in RubisCO-coupled assays (Fig. 2A). 14CO2 fixation with M. jannaschii extracts exhibited saturation kinetics with increasing concentrations of PRPP (Fig. 2B). When assays were optimized for saturating amounts of each substrate, the specific activity for RuBP-dependent CO2 fixation was 13.3 nmol/min/mg, which was about twice the specific activity of the PRPP-dependent CO2 fixation obtained (6.8 nmol/min/mg). Similar to the findings obtained with RuBP, not only was PRPP-dependent activity sensitive to CABP, but the activity was also abolished in the presence of air.
FIG. 2.
Evidence that PRPP is converted to RuBP. (A) Time-dependent radiometric assay of RuBP- and PRPP-dependent 14CO2 incorporation into an acid-stable product. The assay mixtures contained M. jannaschii extract, excess purified recombinant A. fulgidus RubisCO, NAD, and RuBP (⧫), RuBP plus CABP (▪), PRPP (▴), or PRPP plus CABP (×). (B) RubisCO activity as a function of increasing concentration of PRPP (average values from 5- and 10-min assays).
Because 14CO2 fixation was dependent on RubisCO, the acid-stable product was assumed to be PGA. An established and absolutely specific enzymatic method, in which commercial enzymes that are conveniently coupled to NADH oxidation, was used to prove that the product was indeed PGA (23). In addition, this assay allowed convenient quantitation of PGA levels during the enzymatic consumption of PRPP. Prior to determination of substrate consumption and product formation, the stabilities of PRPP, RuBP, and PGA were tested at 83°C. The results showed that PGA is a thermostable enzymatic product of the conversion of PRPP to RuBP and that RuBP is slightly more stable than PRPP at 83°C (Fig. 3). Of particular note, PRPP was previously shown to be converted to ribose 1,5-bisphosphate nonenzymatically at elevated temperatures in the presence of magnesium in a time-dependent fashion (4, 24, 25). This finding was reproduced in our studies; i.e., when PRPP was incubated at 83°C for more than 30 min, a white precipitate formed, which was indicative of the formation of magnesium phosphate and ribose 1,5-bisphosphate (data not shown). The conditions under which subsequent assays were performed took into consideration both the nonenzymatic conversion and the enzymatic conversion of PRPP to ribose 1,5-bisphosphate and the subsequent enzymatic conversion to PGA. For any study in which the physiological role of RubisCO in thermophilic archaea is considered, it is important that the stability of PGA and RuBP at 83°C and higher temperatures be considered.
FIG. 3.
Time-dependent thermostability of PRPP (⧫), PGA (▪), and RuBP (⧫) at 83°C in the absence of M. jannaschii extract. PGA and PRPP concentrations were determined as described in Materials and Methods.
Experiments performed with M. jannaschii extracts showed that for every molecule of PRPP consumed enzymatically, two molecules of PGA were formed. The average values for triplicate experiments indicated that after a 10-min assay, 45.1 nmol of PRPP was enzymatically consumed, leading to the formation of 88.7 nmol of PGA. The standard deviations for detecting PRPP disappearance and RuBP formation were ±0.5 and ±0.3, respectively. Enzymatically consumed PRPP refers to the decrease in PRPP obtained in the presence of extract after 10 min of incubation at 83°C minus the nonenzymatic breakdown that occurred under these conditions. The ratio of the amount of substrate PRPP consumed to the amount of PGA produced is 1:2. As previously shown for the radiometric assay, the presence of air and the RubisCO inhibitor CABP completely inhibited the conversion of PRPP to PGA.
Enzyme that catalyzes RuBP formation.
A protocol was designed to purify the enzyme or complex responsible for the conversion of PRPP to RuBP. By using 16 g of M. jannaschii cells, a stepwise purification protocol was designed with DEAE, QSHP, PSHP (Fig. 4A), and Superose 12 columns in an anaerobic environment. The calibrated Superose 12 column, in the final stage of the purification, yielded a sharp peak of activity, which eluted at a molecular mass of 168 kDa (data not shown). This corresponded to a single 27-kDa protein on an SDS-PAGE gel (Fig. 4B). The N-terminal sequence obtained for the 27-kDa protein corresponded to the open reading frame Mj0601. Mj0601 is annotated as a protein responsible for synthesizing thiamine phosphate and has a calculated subunit molecular mass of 28,655.1 Da. Based on the gel filtration studies described above, the protein appeared to be hexamer in solution with an estimated molecular mass of 168 kDa. The specific activity of Mj0601 at this stage was 305 nmol/min/mg of protein. Alignments of the protein indicated there is an NAD or flavin adenine dinucleotide binding site. Radiometric assays in the presence of NAD increased the specific activity of the protein twofold, while flavin adenine dinucleotide did not have any stimulatory effect; in addition, separate assays indicated that NAD did not stimulate RubisCO activity (data not shown). In future studies we will address whether significant amounts of NAD are bound to the isolated, active enzyme; perhaps recombinant Mj0601 was inactive because of issues related to incorporation of a needed cofactor (see below).
FIG. 4.
(A) Purification of the Mj0601 protein: nondenaturing gel electrophoresis (8% acrylamide) of concentrated PSHP column eluate fractions. Each gel contained 20 μl per well. Lane 1, molecular mass standards (660, 440, 232, and 144 kDa); lanes 2, 3, and 4, consecutive active fractions eluted from the column; lane 5, A. fulgidus RubisCO, with a predicted molecular mass of 97 kDa. (B) SDS-PAGE gel containing the extracted bands from the 8% acrylamide nondenaturing gel. Each of the three protein bands in panel A, lane 3, was individually extracted from the nondenaturing gel, electrophoresed in an SDS-PAGE gel, and then extracted for N-terminal sequencing. The three bands of this gel corresponded to the three major bands in panel A, lane 3. Each of the three bands in lanes 2, 3, and 4 was then sequenced and identified. Lane 1 contained molecular mass standards (66, 45, 31, and 21 kDa); the bands in lanes 2 to 4 were identified as Mj0216 (lane 2), Mj1025 or Mj0720 (lane 3), and Mj0601 (lane 4), the target protein.
Homologous proteins are present in several other organisms (Table 1). Subsequently, genes encoding the Mj0601 enzyme and its homolog from M. acetivorans, Ma2851, were amplified from genomic DNA and cloned into expression vectors for the production of recombinant protein in E. coli. For Mj0601, despite the fact that high levels of recombinant protein were made, no activity was obtained when cells were grown under low-aeration or anaerobic conditions. However, the mesophilic Ma2851 protein was active in extracts of E. coli strain BL21 as PRPP supported 14CO2 fixation when the reaction was coupled to R. rubrum RubisCO activity (Fig. 5). Interestingly, radiolabel incorporation was not inhibited by oxygen; instead, an aerobic environment enhanced activity (Fig. 5).
TABLE 1.
Homologs of Mj0601, the protein thought to catalyze conversion of PRPP or ribose 1,5-bisphosphate to RuBP in M. jannaschii
| Organism | % Identity |
|---|---|
| Methanococcus maripaludisa | 69 |
| Sulfolobus tokodaii | 49 |
| Sulfolobus solfataricus | 47 |
| Archaeoglobus fulgidus | 47b |
| Methanothermobacter thermautotrophicus strain Delta | 46 |
| Pyrococcus abyssi | 46 |
| Pyrococcus horikoshii | 46b |
| Pyrococcus furiosus DSM 3638 | 45b |
| Methanopyrus kandleri AV19 | 45 |
| Methanosarcina barkeri | 43b |
| Pyrobaculum aerophilum | 43 |
| Methanosarcina acetivorans C2A | 42b |
| Methanosarcina mazei Goel | 41b |
| Thermotoga maritima | 40 |
| Aeropyrum pernix | 34 |
| Halobacterium sp. strain NRC-1 | 32 |
| Schizosaccharomyces pombe | 32 |
| Arabidopsis thaliana | 31 |
| Saccharomyces cerevisiae | 27 |
http://www.genome.washington.edu/UWGC/methanococus/.
The organisms are known to contain archaeal form III RubisCO.
FIG. 5.
Time-dependent recombinant Ma2851 activity in soluble extracts of E. coli. The assay conditions are explained in Materials and Methods. Radiolabel incorporation was RubisCO dependent as determined by assays performed in the presence of the RubisCO inhibitor CABP. Assays were performed in an air atmosphere (▪) or under strict anaerobic conditions (⧫).
DISCUSSION
The present study provided an alternative means by which RuBP may be synthesized that circumvents the need for a classical kinase, such as PRK, in the CBB pathway. The first indication that there might be an alternative way to generate RuBP in extracts of M. jannaschii involved the use of AMP, ADP, and ribose monophosphates as potential substrates (data not shown). These compounds, however, were not nearly as effective as PRPP. The initial results led to the possibility that over the 1.5-h assay time the AMP and other compounds were converted to PRPP through PRPP synthetase, which could then be converted to RuBP by some unknown pathway. Indeed, additional experiments in which PRPP was used as the sole substrate resolved the need for a kinase dedicated to RuBP generation, because PRPP already contains the relevant phosphates at both the C-1 and C-5 positions. It is believed that either there is a selective enzymatic dephosphorylation step at the C-1 position or, as previously shown, nonenzymatic dephosphorylation occurs at the pyrophosphate at both moderate and high temperatures in the presence of magnesium at neutral pH (4, 19). In either case the product would be ribose 1,5-bisphophosphate, a compound known to be synthesized in many other biological systems, including macrophages and red blood cells under conditions of hypoxia (10, 26). The other possibility is that 5-ribose-1,2-cyclic phosphate is formed, which is extremely thermostable (13, 19), unlike RuBP (Fig. 3). Since RubisCO is classically a sluggish enzyme and any diminution of RuBP levels would further exacerbate this issue, it is feasible that the precursor compound that leads to the formation of its substrate (RuBP) is maintained as the heat-stable cyclic form of ribose in extreme thermophiles that grow at or near the boiling point of water. In addition, storing the ribose as a cyclic bisphosphate rather than as a triphosphate prevents the possible diversion of the compound to purine recycling.
Neither ribose 1,5-bisphosphate nor PRPP nor presumably 5-ribose-1,2-cyclic phosphate is a direct substrate for archaeal RubisCO because purified recombinant M. jannaschii RubisCO did not catalyze RubisCO-dependent CO2 fixation in the presence of ribose 1,5-bisphosphate or PRPP. Only when ribose 1-5 bisphosphate or PRPP was assayed in the presence of soluble extracts from M. jannaschii was an acid-stable product formed. This product was determined to be PGA. These results indicated that another enzyme(s) in the M. jannaschii extract was responsible for catalyzing the conversion of PRPP and/or the different forms of ribose bisphosphate to the keto sugar RuBP. A further indication of a novel and specific enzymatic reaction(s) was the stoichiometric conversion of PRPP to RuBP with extracts of M. jannaschii, such that that one molecule of PRPP was converted to two molecules of PGA. These results provided experimental verification for the proposed pathway (Fig. 6) and, along with the CABP and antibody studies, reinforced the idea that RubisCO catalysis is essential to convert PRPP to PGA. The proposed unique enzymatic step of this pathway is the conversion of ribose 1,5-bisphosphate or 5-ribose-1,2-cyclic phosphate to RuBP encoded by open reading frames Mj0601 and Ma2851 in M. jannaschii and M. acetivorans, respectively.
FIG. 6.
Proposed pathway for conversion of PRPP to PGA in extracts of M. jannaschii. It is not clear whether PRPP is converted strictly to ribose 1,5-bisphosphate or, if the reaction is enabled by elevated temperatures in this extreme thermophile, PRPP is converted to a cyclic bisphosphate intermediate in the presence of magnesium (19, 26). Mj0601 (or Ma2851 in M. acetivorans), with NAD as a cofactor, catalyzed the conversion of ribose 1,5-bisphosphate or the cyclic bisphosphate intermediate to RuBP, with subsequent production of two molecules of PGA via RubisCO (Mj1235) catalysis.
Sequence alignments and motif searches indicated that there is an NAD binding site in the Mj0601 or Ma2851 protein; the addition of NAD increased the conversion of PRPP to PGA, but an absolute requirement for NAD could not be demonstrated, perhaps because NAD is already tightly bound to the protein. This cofactor most likely facilitates oxidation at the C-3 position to a keto form, followed by elimination of the C-4 hydroxyl with the hydrogen coming from C-2. An enol-ketol conversion and reduction at C-3 would most likely produce the product RuBP. Oxidation of the aldo sugar to the keto form must occur because ribose 1,5-bisphosphate and PRPP were not viable substrates for recombinant M. jannaschii RubisCO. Thus, the enzymes Mj0601 and Ma2851, although annotated as enzymes necessary for thiamine monophosphate formation (3), could have a dual catalytic activity, which we suggest may be a ribose 1,5-bisphosphate dehydrogenase-like activity. At this time, there is no biochemical evidence to indicate that Mj0601 and Ma2851 catalyze any reaction of thiamine metabolism as this assignment is tentative and merely reflects a putative function based on sequence alignments. In future work we will address whether these enzymes can catalyze such reactions in vitro, but it should be noted that there is ample precedent for dual specificity of enzymes of sugar metabolism since R. rubrum RubisCO was recently shown to function in a methionine salvage pathway in Bacillus subtilis (1). The production of active recombinant Ma2851 protein (Fig. 5) and its subsequent purification with a high yield will direct further investigations in our laboratory on this novel enzyme.
In conclusion, the evidence presented in this paper appears to resolve the paradox that anaerobic archaea contain catalytically competent RubisCO but have no visible means (i.e., no PRK) to make RuBP, which appears to be the sole CO2 acceptor for this form III RubisCO. It is completely unnecessary for an organism to require a specific kinase to catalyze ribulose 5-phosphate phosphorylation if the organism contains an alternative enzyme that catalyzes the oxidization of ribose 1,5-bisphosphate or 5-ribose-1,2-cyclic phosphate to RuBP. This novel pathway may point to unique evolutionary links between purine recycling pathways and the CBB cycle because PRPP is a central metabolite linking the two pathways.
Acknowledgments
We are extremely grateful to Biswarup Mukhopadhyay for M. jannaschii cell material, technical advice, and much encouragement. We also thank Janet Gibson for comments on the manuscript.
This study was supported by National Institutes of Health grants GM24497 and GM45404.
REFERENCES
- 1.Ashida, H., Y. Saito, C. Kojima, K. Kobayashi, N. Ogasawara, and A. Yokota. 2003. A functional link between RubisCO-like protein of Bacillus and photosynthetic RubisCO. Science 302:286-290. [DOI] [PubMed] [Google Scholar]
- 2.Bhatia, M. B., and C. Grubmeyer. 1993. The role of divalent magnesium in activating the reaction catalyzed by orotate phosphoribosyltransferase. Arch. Biochem. Biophys. 303:321-325. [DOI] [PubMed] [Google Scholar]
- 3.Bult, C. J., O. White, G. J. Olsen, L. Zhou, R. D. Fleischmann, G. G. Sutton, J. A. Blake, L. M. FitzGerald, R. A. Clayton, J. D. Gocayne, A. R. Kerlavage, B. A. Dougherty, J. F. Tomb, M. D. Adams, C. I. Reich, R. Overbeek, E. F. Kirkness, K. G. Weinstock, J. M. Merrick, A. Glodek, J. L. Scott, N. S. Geoghagen, and J. C. Venter. 1996. Complete genome sequence of the methanogenic archaeon, Methanococcus jannaschii. Science 273:1058-1073. [DOI] [PubMed] [Google Scholar]
- 4.Dennis, A. L., M. Puskas, S. Stasaitis, and R. K. Sandwick. 2000. The formation of a 1-5 phosphodiester linkage in the spontaneous breakdown of 5-phosphoribosyl-α-1-pyrophosphate. J. Inorg. Biochem. 81:73-80. [DOI] [PubMed] [Google Scholar]
- 5.Finn, M., and W. F. R. Tabita. 2003. Synthesis of catalytically active form III ribulose 1,5-bisphosphate carboxylase/oxygenase in archaea. J. Bacteriol. 185:3049-3059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Galagan, J. E., C. Nusbaum, A. Roy, M. G. Endrizzi, P. Macdonald, W. FitzHugh, S. Calvo, R. Engels, S. Smirnov, D. Atnoor, A. Brown, N. Allen, J. Naylor, N. Stange-Thomann, K. DeArellano, R. Johnson, L. Linton, P. McEwan, K. McKernan, J. Talamas, A. Tirrell, W. Ye, A. Zimmer, R. D. Barber, I. Cann, D. E. Graham, D. A. Grahame, A. M. Guss, R. Hedderich, C. Ingram-Smith, H. C. Kuettner, J. A. Krzycki, J. A. Leigh, W. Li, J. Liu, B. Mukhopadhyay, J. N. Reeve, K. Smith, T. A. Springer, L. A. Umayam, O. White, R. H. White, E. Conway de Macario, J. G. Ferry, K. F. Jarrell, H. Jing, A. J. Macario, I. Paulsen, M. Pritchett, K. R. Sowers, R. V. Swanson, S. H. Zinder, E. Lander, W. W. Metcalf, and B. Birren. 2002. The genome of M. acetivorans reveals extensive metabolic and physiological diversity. Genome Res. 12:532-542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Guha, S. K., and Z. B. Rose. 1986. The enzymatic synthesis of ribose 1,5-bisphosphate: studies of its role in metabolism. Arch. Biochem. Biophys. 250:513-518. [DOI] [PubMed] [Google Scholar]
- 8.Hanson, T. E., and F. R. Tabita. 2001. A ribulose-1,5-bisphosphate carboxylase/oxygenase (RubisCO)-like protein from Chlorobium tepidum that is involved with sulfur metabolism and the response to oxidative stress. Proc. Natl. Acad. Sci. USA 98:4397-4402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hanson, T. E., and F. R. Tabita. 2003. Insights into the stress response and sulfur metabolism revealed by proteome analysis of a Chlorobium tepidum mutant lacking the RubisCO-like protein. Photosynth. Res. 78:231-248. [DOI] [PubMed] [Google Scholar]
- 10.Kawaguchi, T., R. L. Veech, and K. Uyeda. 2001. Regulation of energy metabolism in macrophages during hypoxia. J. Biol. Chem. 276:28554-28561. [DOI] [PubMed] [Google Scholar]
- 11.Khorana, H. G., J. F. Fernandes, and A. Kornberg. 1958. Pyrophosphorylation of ribose 5-phosphate in the enzymatic synthesis of 5-phosphorylribose 1-pyrophosphate. J. Biol. Chem. 230:941-948. [PubMed] [Google Scholar]
- 12.Klenk, H. P., R. A. Clayton, J. F. Tomb, O. White, K. E. Nelson, K. A. Ketchum, R. J. Dodson, M. Gwinn, E. K. Hickey, J. D. Peterson, D. L. Richardson, A. R. Kerlavage, D. E. Graham, N. C. Kyrpides, R. D. Fleischmann, J. Quackenbush, N. H. Lee, G. G. Sutton, S. Gill, E. F. Kirkness, B. A. Dougherty, K. McKenney, M. D. Adams, B. Loftus, J. C. Venter, et al. 1997. The complete sequence of the hyperthermophilic sulphate-reducing archaeon Archaeoglobus fulgidus. Nature 390:364-370. [DOI] [PubMed] [Google Scholar]
- 13.Kornberg, A., I. Lieberman, and E. S. Simms. 1954. Enzymatic synthesis and properties of 5-phosphoribosylpyrophosphate. J. Biol. Chem. 215:389-402. [PubMed] [Google Scholar]
- 14.Maeda, N., K. Kitano, T. Fukui, S. Ezaki, H. Atomi, K. Miki, and T. Imanaka. 1999. Ribulose bisphosphate carboxylase/oxygenase from the hyperthermophilic archaeon Pyrococcus kodakaraensis KOD1 is composed solely of large subunits and forms a pentagonal structure. J. Mol. Biol. 293:57-66. [DOI] [PubMed] [Google Scholar]
- 15.Markwell, M. K., S. M. Hass, L. L. Bieber, and N. E. Tolbert. 1978. A modification of the Lowry method to simplify protein determination in membrane and lipoprotein samples. Anal. Biochem. 87:206-210. [DOI] [PubMed] [Google Scholar]
- 16.Micheli, V., G. Pompucci, and R. Marcolongo. 1975. An improved method for the determintation of 5-phosphoribosyl 1-pyrophosphate. Clin. Chim. Acta 65:181-185. [DOI] [PubMed] [Google Scholar]
- 17.Mukhopadhyay, B., E. F. Johnson, and R. S. Wolfe. 1999. Reactor-scale cultivation of the hyperthermophilic methanarchaeon Methanococcus jannaschii to high cell densities. Appl. Environ. Microbiol. 65:5059-5065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pierce, J., N. E. Tolbert, and R. E. Barker. 1980. Interaction of ribulose-bisphosphate carboxylase/oxygenase with transition-state analogues. Biochemistry 19:934-942. [DOI] [PubMed] [Google Scholar]
- 19.Remy, C. N., W. T. Remy, and J. M. Buchanan. 1955. Biosynthesis of the purines. enzymatic synthesis and utilization of α-5-phosphoribosylpyrophosphate. J. Biol. Chem. 215:885-895. [PubMed] [Google Scholar]
- 20.Rose, I. A., and J. V. B. Warms. 1974. Glucose and mannose-1,6P as activators of phosphofructokinase in red blood cells. Biochem. Biophys. Res. Commun. 59:1333-1340. [DOI] [PubMed] [Google Scholar]
- 21.Sprott, G. D., I. Ekiel, and G. Patel. 1993. Metabolic pathways in Methanococcus jannaschii and other methanogenic bacteria. Appl. Environ. Microbiol. 59:2-1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tabita, F. R. 1999. Microbial ribulose 1,5-bisphosphate carboxylase/oxygenase: adifferent perspective. Photosyn. Res. 60:28. [Google Scholar]
- 23.Tabita, F. R., P. Caruso, and W. Whitman. 1978. Facile assay of enzyme unique to the Calvin cycle in intact cells, with special reference to ribulose 1,5 bisphosphate carboxylase. Anal. Biochem. 84:462-472. [DOI] [PubMed] [Google Scholar]
- 24.Thompson, R. E., E. L. F. Li, H. O. Spivey, J. P. Chandler, A. J. Katz, and J. R. Appleman. 1978. Apparent stability constants of H+ and Mg2+ complexes of 5-phosphoribosyl alpha-1-pyrophosphate. Bioinorg Chem. 9:35-45. [DOI] [PubMed] [Google Scholar]
- 25.Trembacz, H., and M. M. Jezewska. 1990. The route of non-enzymic and enzymic breakdown of 5-phosphoribosyl 1-pyrophosphate to ribose 1-phosphate. Biochem. J. 271:621-625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vanderheiden, B. S. 1970. Phosphate esters in human erythrocytes. VII. Further evidence for ribose 1,5-diphosphate as a natural metabolite. Biochim. Biophys. Acta 215:242-248. [DOI] [PubMed] [Google Scholar]
- 27.Watson, G. M. F., and F. R. Tabita. 1997. Microbial ribulose 1,5-bisphosphate carboxylase/oxygenase: a molecule for phylogenetic and enzymological investigation. FEMS Lett. 146:13-22. [DOI] [PubMed] [Google Scholar]
- 28.Watson, G. M. F., J. P. Yu, and F. R. Tabita. 1999. Unusual ribulose1,5-bisphosphate carboxylase/oxygenase of anoxic archaea. J. Bacteriol. 181:1569-1575. [DOI] [PMC free article] [PubMed] [Google Scholar]