Abstract
An ampicillin enrichment strategy following transposon insertion mutagenesis was employed to obtain NaCl-sensitive mutants of a gltBD (glutamate synthase [GOGAT]-deficient) strain of Escherichia coli. It was reasoned that the gltBD mutation would sensitize the parental strain even to small perturbations affecting osmotolerance. Insertions conferring an osmosensitive phenotype were identified in the proU, argP (formerly iciA), and glnE genes encoding a glycine betaine/proline transporter, a LysR-type transcriptional regulator, and the adenylyltransferase for glutamine synthetase, respectively. The gltBD+ derivatives of the strains were not osmosensitive. The argP mutation, but not the glnE mutation, was associated with reduced glutamate dehydrogenase activity and a concomitant NH4+ assimilation defect in the gltBD strain. Supplementation of the medium with lysine or a lysine-containing dipeptide phenocopied the argP null mutation for both osmosensitivity and NH4+ assimilation deficiency in a gltBD background, and a dominant gain-of-function mutation in argP was associated with suppression of these lysine inhibitory effects. Osmosensitivity in the gltBD strains, elicited either by lysine supplementation or by introduction of the argP or glnE mutations (but not proU mutations), was also correlated with a reduction in cytoplasmic glutamate pools in cultures grown at elevated osmolarity. We propose that an inability to accumulate intracellular glutamate at high osmolarity underlies the osmosensitive phenotype of both the argP gltBD and glnE gltBD mutants, the former because of a reduction in the capacity for NH4+ assimilation into glutamate and the latter because of increased channeling of glutamate into glutamine.
When bacteria are exposed to environments with elevated osmolarity, there is a passive outflow of water from the intracellular compartment, which results in a growth-inhibiting loss of cell turgor and a reduction in the cytoplasmic volume. The set of metabolic changes that occur under these conditions to restore bacterial growth is referred to as osmoregulation. In enterobacteria such as Escherichia coli and Salmonella enterica serovar Typhimurium, the osmoregulatory processes that have been identified so far include cell envelope alterations and the cytoplasmic accumulation (to concentrations that approach several hundred millimolar) of K+ ions, glutamate, trehalose, proline, and glycine betaine (reviewed in references 12 and 13). Of these, the accumulation of glutamate and trehalose is mediated by increased synthesis of these compounds, whereas accumulation of K+, proline, and glycine betaine is achieved by increased uptake from the culture medium. Glutamate serves as a cytoplasmic counterion for K+ in osmotically stressed cells, and the available evidence suggests that perturbations in accumulation of either one of these molecules adversely affects the accumulation of the other (31, 43). Proline and glycine betaine are also referred to as osmoprotectants, because at low concentrations they dramatically enhance the growth rates of enterobacteria in media with elevated osmolarity.
In addition to its perceived role in osmoregulation, glutamate is also a central player in global nitrogen metabolism (reviewed in references 35 to 37); 75 to 90% of all cellular nitrogen is assimilated via glutamate. NH4+ is the preferred nitrogen source for E. coli, and it is assimilated into glutamate through two pathways. One of these pathways is the glutamate dehydrogenase (GDH) pathway, in which 2-oxoglutarate undergoes reductive condensation with NH4+, yielding glutamate. The second pathway is the two-step glutamine synthetase (GS)-glutamate synthase (GOGAT) pathway, in which glutamine that is synthesized in the first step from one molecule of NH4+ and glutamate (in the presence of ATP) is involved in a reductive reaction with 2-oxoglutarate, which yields two molecules of glutamate. GDH, GS, and GOGAT are encoded by the gdhA, glnA, and gltBD genes, respectively.
The GDH pathway is functional for nitrogen assimilation in media containing ≥1 mM NH4+. In media with limiting NH4+ concentrations or with alternative poor nitrogen sources, nitrogen assimilation into glutamate is mediated through the GS-GOGAT pathway, whose regulation in turn is tied to the complex cascade of nitrogen regulation that is referred to as Ntr (35-37). In NH4+-replete media, glnA is expressed only at basal levels; in addition, there is a reduction in the catalytic activity of GS resulting from the adenylylation of its homopolymeric subunits by the glnE-encoded adenylyltransferase. The residual activity of GS is then sufficient to meet the cell's anabolic requirement for glutamine for protein synthesis. On the other hand, in low-NH4+ medium or during growth on poor nitrogen sources, expression of the Ntr regulon (of which glnA is a member) is activated, and in addition GS is deadenylylated by GlnE; the vastly increased activity of GS is now able to catalyze sufficient glutamine synthesis to meet the cell's nitrogen assimilatory requirement.
With high exogenous levels of NH4+, glutamate accumulation at high osmolarity is not affected by single mutations that block either biosynthetic pathway (3, 31), but in low-NH4+ media, GOGAT-deficient mutants are osmosensitive (14, 43). A correlation between osmotolerance and NH4+ assimilation efficiency in GOGAT-deficient strains was established in a previous study (39). The data obtained previously provided indirect genetic evidence that increased glutamate synthesis is necessary for optimal growth under hyperosmotic stress conditions.
In the present study, we performed transposon insertion mutagenesis of a GOGAT-deficient (gltBD) strain to identify mutants that are osmosensitive on NH4+-replete medium. The rationale was that the gltBD strain would be sensitized to even small perturbations that affect osmotolerance (given that multiple additive mechanisms operate for osmoregulation). Furthermore, it was reasoned that transposon insertions would generate null mutations and that the possibility of obtaining conditional-lethal salt-sensitive mutants with mutations in essential housekeeping genes would be avoided (12).
We report here identification of insertions in glnE, as well as in argP, each of which acts synergistically with gltBD in conferring an osmosensitive phenotype. As mentioned above, glnE is involved in covalently modifying and thus modulating GS activity. The argP gene (also called iciA) encodes a LysR-type transcriptional regulator that has previously been implicated in the regulation of arginine transport (6-8, 34) and in the control of chromosomal DNA replication initiation (20, 21, 42) in E. coli.
MATERIALS AND METHODS
Bacterial strains and growth media.
Genotypes of E. coli K-12 strains are shown in Table 1. The routine rich and minimal growth media used were Luria-Bertani medium (33) and 0.2% glucose-minimal A medium (33), respectively, and the incubation temperature was 37°C. Minimal A medium contains 15 mM NH4+ and hence is considered to be NH4+ replete. Growth in liquid cultures was monitored with a Klett-Summerson photoelectric colorimeter. NH4+ assimilation growth experiments were performed by using W salts basal medium (40), to which 0.4% glucose was added as a carbon source and the desired concentration of ammonium sulfate was added as a nitrogen source. The medium was supplemented with lysine (Lys), lysylalanine (Lys-Ala), histidylalanine, or glycine betaine (each at a concentration of 1 mM) and with aspartate (0.2%). Unless otherwise indicated, the concentrations of antibiotics used were the concentrations described previously (39).
TABLE 1.
E. coli K-12 strainsa
| Strain | Genotypeb |
|---|---|
| MC4100 | Δ(argF-lac)U169 rpsL150 relA1 araD139 flbB5301 deoC1 ptsF25 |
| GJ2529 | MC4100 ΔgltBDF500 zha-6::Tn10 |
| GJ2530 | GJ2529 fnr::Ωc |
| GJ2560 | GJ2529 proU610::λplacMu55(Kan)d |
| GJ4534 | MC4100 proU610::λplacMu55(Kan) |
| GJ4536 | MC4100 argP202::λplacMu55(Kan) |
| GJ4540 | MC4100 argP203::λplacMu55(Kan) |
| GJ4544 | MC4100 proU611::λplacMu55(Kan) |
| GJ4545 | MC4100 glnE463::λplacMu55(Kan) |
| GJ4547 | MC4100 glnE464::λplacMu55(Kan) |
| GJ4549 | MC4100 glnE465::λplacMu55(Kan) |
| GJ4652 | MC4100 ΔgltBDF500 |
| GJ4654 | GJ4652 argP202::λplacMu55(Kan) |
| GJ4655 | GJ4652 argP203::λplacMu55(Kan) |
| GJ4661 | GJ4652 apaG611::λplacMu55(Kan) |
| GJ4663 | GJ4652 glnE463::λplacMu55(Kan) |
| GJ4664 | GJ4652 glnE464::λplacMu55(Kan) |
| GJ4735 | GJ4652 apaH::Kan |
| GJ4842 | GJ4652 ΔruvABC::Cm |
| GJ4843 | GJ4652 recA56 srl-300::Tn10 |
| GJ4891 | GJ4663 argP202::λplacMu55(Kan) ΔdsbC::Cm |
| GJ4928 | GJ4652 proU611::λplacMu55(Kan) |
| GJ4929 | GJ4652 glnE465::λplacMu55(Kan) |
Phages and plasmids.
The transposon phage λplacMu55(Kan) encoding kanamycin resistance and its helper phage λpMu507 have been described previously (29), as have the plasmid vectors (i) pCL1920 (pSC101 based, low copy number, spectinomycin and streptomycin resistant) (27), (ii) pACYC184 (p15A based, medium copy number, chloramphenicol and tetracycline resistant) (9), (iii) pBR329 (pMB9 based, high copy number, ampicillin, tetracycline, and chloramphenicol resistant) (10), and (iv) pBluescriptII-KS (pMB9 based, very high copy number, ampicillin resistant) (Stratagene, La Jolla, Calif.).
Plasmid pHYD909 was constructed by subcloning a 1.6-kb BamHI-HindIII fragment encoding an N-terminally truncated FNR protein from plasmid pGS198 (41) into the appropriate sites of pACYC184. Plasmids pHYD916, pHYD942, and pHYD943 are derivatives of pCL1920 carrying the following subcloned fragments from recombinant λ phage clones of the ordered genomic library of Kohara et al. (23): in pHYD916, 5.2-kb BamHI-KpnI fragment with glnE+ from λ508 in the corresponding sites of the vector; in pHYD942, 0.78-kb PstI-HpaI fragment with apaG+ from λ105 in the BamHI-SalI sites of the vector; and in pHYD943, 1.06-kb ClaI-EcoRI fragment with apaH+ from λ105 in the BamHI-SalI sites of the vector (the last two via an intermediate step of cloning into pBluescriptII-KS). Plasmids pHYD915, pHYD953, and pHYD954 have been described previously (34) and carry argP+ cloned in pCL1920, the dominant gain-of-function argP (argP-S94L) allele cloned in pBR329, and the argO cis regulatory region cloned in pBluescriptII-KS, respectively.
Isolation and molecular genetic characterization of NaCl-sensitive mutants.
By using the infection protocol described previously (29), populations of clones were obtained that carried random transpositions of the lac fusion phage λplacMu55(Kan) in the genome of strain GJ2530/pHYD909. Approximately 105 independent Kanr insertions were obtained in each of two different experiments. Each of the populations was inoculated at a concentration of 107 cells per ml into 10 ml of glucose-minimal A medium supplemented with 0.7 M NaCl and glycine betaine and incubated with shaking until the A600 was around 0.1; after this ampicillin was added to a concentration of 100 μg/ml. Surviving cells after incubation for an additional 4 h were harvested by filtration and grown overnight in glucose-minimal A medium, and the cycle used for ampicillin enrichment for cells unable to grow in NaCl-supplemented medium was repeated. The cultures were then plated on glucose-minimal A medium, and individual colonies were tested for NaCl sensitivity.
Molecular mapping of the mutation in each of the derivatives in which the NaCl sensitivity phenotype was 100% cotransducible with the Kanr marker of the transposon was performed by using an inverse PCR approach. Chromosomal DNA was digested to completion with HhaI, ligated at a high dilution, and then subjected to PCR amplification with a pair of divergently oriented primers (MuC1 [5′-TGCGTTTTTCTTCAGGTAATG-3′] and MuC2 [5′-TCCCGAATAATCCAATGTCCTCCCG-3′]) from the sequence at the c end of phage Mu. This procedure was expected to amplify the HhaI fragment at the junction of the Mu c end of the insertion and the chromosome. (If the terminal nucleotide pair at the c end of Mu is counted as bp 1, the MuC1 and MuC2 primers are designed to read inward from bp 438 and outward from bp 56, respectively, and the HhaI site is at bp 704.) Following agarose gel electrophoresis, the PCR products were eluted from the gel and sequenced with an automated DNA sequencer by using primer MuC2. The identity of the mutated gene was then established by a BLAST search analysis of the sequence determined against the E. coli genome sequence (2).
GDH assays.
For GDH assays, cultures initially grown in glucose-minimal A medium with glycine betaine to an A600 of around 0.2 were each split into two parts. To one part (high osmolarity grown) NaCl was added, from a 2.4 M stock solution prepared in glucose-minimal A medium with glycine betaine, to a final concentration of 0.6 M, and incubation was continued. The other part (low osmolarity grown) was incubated without further manipulation. All cultures were harvested at an A600 of around 0.5.
Cell extracts for determination of GDH activity were prepared as described previously (19), with the following modifications: (i) after harvested cells were washed, they were resuspended in 4 ml of ice-cold 50 mM Tris-Cl buffer (pH 7.6)-10 mM β-mercaptoethanol and (ii) cells were disrupted by sonication. The extracts were used for enzyme activity measurements immediately. The assay method was the method described by Meers et al. (32), except that the reactions were performed at room temperature in 0.5-ml (total volume) mixtures. Protein concentrations in cell extracts were determined by the method of Bradford (4). Enzyme specific activities were expressed in milliunits per milligram of protein in the cell extracts (after correction for endogenous NADPH oxidase activity); 1 U was defined as the amount of enzyme required to oxidize 1 μmol of NADPH (extinction coefficient at 340 nm, 6,220 M−1 cm−1) per min.
Estimation of intracellular glutamate and glutamine pools.
For estimation of intracellular glutamate and glutamine pools, cultures grown at low and high osmolarities were prepared in the way described above for the GDH assays. (When cultures were supplemented with Lys or Lys-Ala, they were supplemented at all stages of growth.) A 0.2-ml portion of each culture was added to 0.8 ml of ice-cold methanol as recommended in the no-harvest method described by Kustu and coworkers (14, 25, 43). Cell debris in the mixture was removed by low-speed centrifugation, and the supernatant was lyophilized, resuspended in 0.05 ml of water, and filtered through a 0.45-μm-pore-size filter. Proteins in the sample were precipitated by addition of 0.05 ml of 10% trichloroacetic acid, and the supernatant obtained after low-speed centrifugation was lyophilized. The content of glutamate and glutamine in the sample was then estimated, after precolumn derivatization with phenylisothiocyanate, by reverse-phase high-performance liquid chromatography with a Waters Pico · Tag column for free amino acids (Millipore Corp, Milford, Mass.) and detection of absorbance at 254 nm; the protocols for sample derivatization and column chromatography were similar to those described in the manufacturer's instructions. As standards for quantitation, 250 pmol of glutamate and 250 pmol of glutamine were injected after derivatization into the column, and, under the conditions employed, they eluted at 2.99 and 5.02 min, respectively.
For determination of the dry weight of bacteria in the culture, a 15- to 20-ml portion of a culture was filtered through a preweighed 0.45-μm-pore-size filter (diameter, 47 mm); the filter was washed with an equal volume of water and then dried at room temperature to a constant weight. By this method, we calculated the mean bacterial dry weights to be 0.82 and 0.46 mg ml−1 A600 unit−1 for cultures grown in glucose-minimal A medium with glycine betaine and in glucose-minimal A medium with glycine betaine and 0.6 M NaCl, respectively.
Other methods.
Mutations were transferred between strains by P1 transduction (17). Recombinant DNA procedures were performed as described previously (38).
RESULTS
Isolation of NaCl-sensitive mutants from gltBD strain.
As described above, an ampicillin enrichment strategy was employed with a population of transposon-mutagenized clones of a gltBD strain in order to identify insertions that conferred an NaCl-sensitive phenotype on glycine betaine-supplemented medium. Since glycine betaine is osmoprotective for E. coli (12, 13), its inclusion permitted ampicillin selection to be performed at a sufficiently high NaCl concentration, 0.7 M. (The starting strain also carried a Δfnr mutation, which is known to confer osmosensitivity in the gltBD background [39], along with a plasmid encoding an N-terminally truncated FNR protein that is proficient for complementing the fnr mutant for osmosensitivity; however, our subsequent studies [data not shown] indicated that neither the fnr mutation nor its complementing plasmid was relevant to the phenotypes described below.) The Kanr insertions were each transduced into ΔgltBD strain GJ4652 in order to establish that NaCl sensitivity is 100% linked to Kanr.
The sites of transposon insertions in the mutants were then determined by two complementary approaches, inverse PCR (as described above) and transductional mapping (Tables 2 and 3). One Kanr insertion in apaG, two Kanr insertions each in proU and argP, and three Kanr insertions in glnE were identified that conferred an NaCl-sensitive phenotype in the gltBD background.
TABLE 2.
Molecular characterization and NaCl tolerance of λplacMu55(Kan) insertion mutants
| Insertion allele | Genomic position (bp)a | Mu orientationb | lacZ orientationc | Distance from start of gene (total gene length) (bp)d | NaCl tolerancee
|
|
|---|---|---|---|---|---|---|
| gltBD (strain) | gltBD+ (strain) | |||||
| apaG11 | 51249 | CCW | − | 358 (375) | S (GJ4661) | NDf |
| argP202 | 3057872 | CCW | + | 104 (891) | S (GJ4654) | T (GJ4536) |
| argP203 | 3057754 | CCW | + | −19 (891) | S (GJ4655) | T (GJ4540) |
| glnE463 | —g | − | S (GJ4663) | T (GJ4545) | ||
| glnE464 | 3196273 | CW | + | 1,014 (2,838) | S (GJ4664) | T (GJ4547) |
| glnE465 | 3194695 | CCW | − | 2,588 (2,838) | S (GJ4929) | T (GJ4549) |
| proU610 | 2803134 | CCW | + | 303 (1,200) | S (GJ2560) | T (GJ4534) |
| proU611 | —g | + | S (GJ4928) | T (GJ4544) | ||
The nucleotide position in the E. coli genome at the junction with the Mu c end of each insertion is indicated. The genome sequence is from reference 2 (GenBank accession number U00096).
CW and CCW refer to the two alternative orientations, in which the Mu S end of the insertion is clockwise and counterclockwise, respectively, from the Mu c end in the E. coli chromosome.
+ and −, orientations in which the direction of the lacZ reporter gene within the transposon is the same as and opposite that of the host gene disrupted by the insertion, respectively.
Distance from the start codon to the Mu insertion site and length of the wild-type gene. The fact that Mu transposition is associated with a 5-bp target sequence duplication (33) was also taken into account when the distance was calculated.
Growth of the gltBD and gltBD+ derivatives was compared with that of the control strains GJ4652 (gltBD) and MC4100 (gltBD+) on glucose-minimal A agar plates supplemented with glycine betaine and several concentrations of NaCl. S, sensitive; T, tolerant.
ND, not determined.
—, not molecularly characterized (the Mu c end in glnE463 is 6 bp away from an HhaI site in glnE).
TABLE 3.
P1 transductional mapping of λplacMu55(Kan) insertionsa
| λplacMu55(Kan) allele(s) (min) | Donor marker (min) | % Linkage |
|---|---|---|
| apaG11 (1.1) | zab-3051::Tn10 (1.8) | 22 |
| argP202, −203 (65.9) | ΔdsbC::Cm (65.4) | 12-33 |
| galP::Tn10 (66.5) | 16-19 | |
| mutY::Tn10 (66.8) | 6-10 | |
| glnE463, −464, −465 (68.9) | metC162::Tn10 (67.9) | 11-16 |
| proU610, −611 (60.4) | zga-900::Tn10 (60.3)b | 67-100 |
In each cross, linkage was determined as the proportion of Kans derivatives among the transductants inheriting the donor marker (chloramphenicol resistance in the case of ΔdsbC::Cm and tetracycline resistance in the case of all other markers). For the loci with two or more λplacMu55(Kan) insertion alleles, the range of linkage values obtained is indicated.
The zga-900::Tn10 insertion was designated zfi-900::Tn10 in a previous study (17).
On the E. coli chromosome, the apaG gene is upstream of and is cotranscribed with apaH. The latter gene encodes an Ap4A hydrolase (28). We were able to demonstrate by using the following two criteria that the NaCl-sensitive phenotype conferred by the apaG insertion in the gltBD strain is the result of a polar effect on apaH expression (data not shown): (i) an apaH::Kan mutation described previously (28) conferred NaCl sensitivity in the gltBD strain, which was more pronounced than the NaCl sensitivity observed with apaG; and (ii) NaCl tolerance in both the apaG gltBD (GJ4661) and apaH gltBD (GJ4735) strains was restored upon introduction of the minimal apaH+ plasmid pHYD943 but not upon introduction of the minimal apaG+ plasmid pHYD942. However (in a finding which is different from the finding reported by Leveque et al. [28] in a different E. coli strain background), we observed that the apaG and apaH insertions (in both gltBD and gltBD+ derivatives of MC4100) were associated with slow growth even in glucose-minimal A and succinate-minimal A media that were not supplemented with NaCl (data not shown). The apaH mutant also exhibited a reduced growth rate in hypotonic medium, such as 0.2× minimal A medium with NH4+ added to a concentration of 15 mM and glucose. Therefore, we could not exclude the possibility that the NaCl sensitivity of the apaG or apaH mutants was just a manifestation of their nonspecific slow-growth phenotype, not unlike the behavior of other slowly growing control strains, such as recA or ruvABC strains (see below), which are not expected to be affected in NaCl tolerance. Accordingly, further studies with the apaGH mutants were not pursued.
argP gltBD and glnE gltBD mutants are osmosensitive.
By using plate growth tests, insertions in each of the other three loci (proU, argP, and glnE) were shown (i) to confer NaCl sensitivity only in the gltBD background and not when they were transduced into the isogenic gltBD+ strain MC4100 (Table 2) and (ii) not to affect growth on media not supplemented with NaCl in either strain background (data not shown). Since the role in osmoregulation of the glycine betaine/proline transporter encoded by the three constituent genes (proV, proW, and proX) of the proU operon is well established (12, 13), the two new proU insertion mutants identified in this study were not characterized further.
Of the two insertions in argP, the one located in the interval between the putative promoter and the start of the structural gene (argP203) conferred a less severe phenotype than the one in the coding region of the gene (argP202). All three glnE gltBD mutants were equally compromised for NaCl tolerance. The argP gltBD and glnE gltBD mutants were complemented for NaCl tolerance by pSC101-based plasmids carrying the argP+ (pHYD915) and glnE+ (pHYD916) genes, respectively. Neither of these plasmids (nor derivatives of pBR329 with the cloned argP+ or glnE+ genes) had any significant effect on NaCl tolerance of gltBD+ or gltBD strains that were argP+ glnE+ (data not shown).
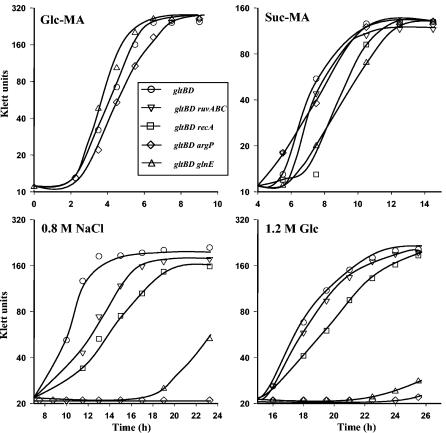
We compared the growth rates of argP202 gltBD and glnE463 gltBD mutants (test strains) on the one hand and the gltBD, recA gltBD, and ΔruvABC gltBD strains (control strains) on the other hand in (i) glucose minimal medium, (ii) succinate minimal medium, and (iii) glucose minimal medium rendered hyperosmolar with either 0.8 M NaCl or 1.2 M glucose (Fig. 1). It is known that the gltBD mutant is only marginally osmosensitive compared with the wild-type strain (14, 39, 43). The recA and ruvABC derivatives were chosen as mutants whose growth deficiency is not related to the osmolarity of the growth medium, and likewise succinate minimal medium was chosen to represent a growth stress not related to osmotic stress. It was reasoned that a true osmosensitive mutant was a mutant that was specifically defective for growth only in media with elevated osmolarity. By using these criteria, both test strains were classified as osmosensitive, since (i) in the succinate minimal medium, the argP gltBD strain's growth rate was similar to that of the gltBD single mutant, while the glnE gltBD strain grew at least as well as the recA gltBD derivative; and (ii) in the 0.8 M NaCl- or 1.2 M glucose-supplemented cultures, the test strains exhibited significantly reduced growth rates compared with all three control strains (Fig. 1).
FIG. 1.
Osmosensitivity of glnE gltBD and argP gltBD mutants. Semilogarithmic growth curves were plotted for strains GJ4652 (gltBD), GJ4654 (gltBD argP), GJ4663 (gltBD glnE), GJ4842 (gltBD ruvABC), and GJ4843 (gltBD recA) in different media after 1:100 inoculation from cultures grown to the stationary phase in glucose-minimal A medium. The media employed were 0.2% glucose-minimal A medium (Glc-MA), 0.2% succinate-minimal A medium (Suc-MA), glucose-minimal A medium supplemented with glycine betaine and 0.8 M NaCl (0.8 M NaCl), and glucose-minimal A medium supplemented with glycine betaine and 1.2 M glucose (1.2 M Glc).
The test strains were also significantly more sensitive than the wild-type or gltBD control strains to other ionic or nonionic impermeant solutes, including KCl, K2SO4, NH4Cl, (NH4)2SO4, and sucrose, but they were not more sensitive than the control strains to equiosmolar concentrations of freely permeable solutes, such as glycerol or ethylene glycol (data not shown). These observations indicated that the test strains are deficient for osmotic stress adaptation. The argP glnE gltBD triple mutant GJ4891 was even more osmosensitive than the argP gltBD and glnE gltBD double mutants (data not shown).
Exacerbation by argP of NH4+ assimilation deficiency in gltBD mutant.
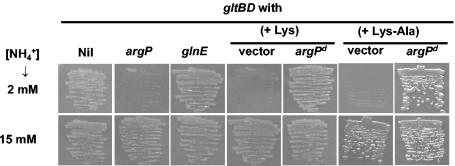
As mentioned above, a gltBD mutant (in which the GDH pathway of NH4+ assimilation alone is functional) is not significantly nitrogen limited in a medium containing NH4+ at a concentration of 1 mM or more (35-37). We observed that the argP gltBD mutant was unable to grow even on medium containing 2 mM NH4+, whereas a glnE gltBD strain and the gltBD single mutant were able to grow on this medium (Fig. 2). On the other hand, with 15 mM NH4+ (Fig. 2) or with aspartate as the nitrogen source (which readily donates its amino group for glutamate synthesis) (data not shown), the growth of the argP gltBD mutant was similar to that of the gltBD strain. Thus, osmosensitivity in the argP gltBD strain could be correlated with a deficiency in NH4+ assimilation into glutamate. The growth of the argP gltBD+ strain was not affected even in media containing <1 mM NH4+ (data not shown).
FIG. 2.
Effects of argP mutations and Lys or Lys-Ala supplementation on NH4+ assimilation in gltBD derivatives. Each strain was streaked on a pair of agar plates containing W salts medium supplemented with 2 mM NH4+ (top row) and 15 mM NH4+ (bottom row) (NH4+ was added as ammonium sulfate to concentrations of 1 and 7.5 mM, respectively). Additional genetic markers present in the gltBD derivatives are indicated above each pair of panels, and the strains employed were GJ4652 (Nil), GJ4654 (argP), GJ4663 (glnE), GJ4652/pBR329 (vector), and GJ4652/pHYD953 (argPd, that is, with the dominant argP-S94L mutation). The plasmid-bearing strains were streaked on ampicillin-containing medium with Lys or Lys-Ala, as indicated. The plates were incubated for 24 h.
Reduced GDH activity in argP gltBD strain.
GDH specific activities were then determined by the method described above for wild-type strain MC4100, as well as its gltBD, argP gltBD, and glnE gltBD derivatives, which were grown in NH4+-replete cultures at low or high osmolarity (Table 4). We observed that osmotic stress was associated with a twofold increase in GDH specific activity in GJ4652 (gltBD) but not in MC4100 (wild type), a difference that may perhaps reflect the existence in the latter strain of redundant pathways for increased glutamate synthesis for osmoregulation. The GDH activities in cultures of the glnE gltBD strain GJ4663 at both low and high osmolarities were not significantly different from those in cultures of GJ4652. On the other hand, for the argP gltBD strain GJ4654, the GDH activity in the low-osmolarity culture was only one-half that of GJ4652, and furthermore there was no increase in enzyme activity in the strain following imposition of NaCl stress. These results established that NH4+ assimilation deficiency and reduced GDH activity are correlated in the argP gltBD strain.
TABLE 4.
GDH activities in argP gltBD and glnE gltBD derivativesa
| Strain | Genotype | GDH sp act (mμ mg of protein−1)
|
|
|---|---|---|---|
| Low osmolarity | High osmolarity | ||
| MC4100 | Wild type | 90 | 105 |
| GJ4652 | gltBD | 111 | 224 |
| GJ4654 | argP gltBD | 54 | 46 |
| GJ4663 | glnE gltBD | 107 | 170 |
GDH specific activities were measured in cells that were grown as described in the text and were harvested from (i) glucose-minimal A medium with glycine betaine (low osmolarity) or (ii) glucose-minimal A medium with glycine betaine and 0.6 M NaCl (high osmolarity). Each value is the average of at least two independent determinations, and the variation between individual measurements was less than 20%.
Osmosensitivity is correlated with decreased glutamate accumulation in both argP gltBD and glnE gltBD mutants.
The fact that the argP and glnE mutants were affected for NH4+ assimilation and GS regulation, respectively, combined with the fact that each of the mutations acted synergistically with gltBD for osmosensitivity, made it likely that insufficient glutamate accumulation was responsible for the osmosensitive phenotypes of the mutants. Accordingly, we measured the intracellular glutamate and glutamine pools in the cultures of these mutants (along with those of the wild-type, gltBD, and proU gltBD strains as controls) grown in NH4+-replete medium at low and elevated osmolarities (Table 5).
TABLE 5.
Glutamate and glutamine pools in gltBD derivativesa
| Strain genotypeb | Supplement | Concn (nmol mg [dry wt] of cells−1) of:
|
|||
|---|---|---|---|---|---|
| Glutamate
|
Glutamine
|
||||
| Low osmolarity | High osmolarity | Low osmolarity | High osmolarity | ||
| Wild type | None | 36 | 212 | 5 | 26 |
| gltBD | None | 26 | 117 | 7 | 43 |
| proU gltBD | None | 30 | 118 | 5 | 36 |
| argP gltBD | None | 18 | 50 | 7 | 40 |
| glnE gltBD | None | 21 | 39 | 19 | 236 |
| gltBD | Lys | NDc | 46 | ND | 21 |
| gltBD | Lys-Ala | ND | 40 | ND | 18 |
| gltBD argP-S94L | Lys | ND | 107 | ND | 44 |
| gltBD argP-S94L | Lys-Ala | ND | 111 | ND | 52 |
Intracellular levels of glutamate and glutamine were measured in cells that were grown as described in the text and were harvested from (i) glucose-minimal A medium with glycine betaine (low osmolarity) or (ii) glucose-minimal A medium with glycine betaine and 0.6 M NaCl (high osmolarity) without any additional supplement (None) or with Lys or Lys-Ala. Each value is the average of at least two independent determinations, and the variation between individual measurements was less than 20%.
The strains employed were wild-type strain MC4100, gltBD strain GJ4652, proU gltBD strain GJ2560, argP gltBD strain GJ4654, glnE gltBD strain GJ4663, and strain GJ4652/pHYD953 with the dominant argP mutation (gltBD argP-S94L).
ND, not determined:
Consistent with previous reports (14, 30, 31, 43), there was substantial accumulation of glutamate in cells of wild-type strain MC4100 grown at the elevated osmolarity, which was accompanied by a small increase in the size of the intracellular glutamine pool. Compared to the level in MC4100, the intracellular glutamate levels in NaCl-grown cultures of the gltBD and proU gltBD mutants were about 40% lower, whereas the reduction was even more pronounced (around 80%) for the argP gltBD and glnE gltBD strains. At the same time, the size of the glutamine pool in high-osmolarity-grown cells of the glnE gltBD derivative was markedly increased (nearly 10-fold) compared with that in MC4100. Our results therefore indicate that osmosensitivity in the argP gltBD and glnE gltBD mutants (but not in the proU gltBD strain) is correlated with a failure to accumulate sufficient glutamate at an elevated osmolarity.
Exogenous Lys (or Lys-Ala) inhibits NH4+ assimilation and confers osmosensitivity in gltBD mutant.
A recent study in our laboratory (34) showed (i) that ArgP is a transcriptional activator of the argO gene (previously called yggA) that encodes a putative arginine exporter in E. coli; (ii) that intracellular Lys accumulation apparently correlates with abolition of the activator function of ArgP, as evidenced by the shutting down of argO transcription in argP+ strains grown in medium supplemented with either Lys or the dipeptide Lys-Ala (which releases Lys by hydrolysis within the cells after uptake from the medium and hence serves to distinguish the [indirect] extracellular effects of Lys from the [direct] intracellular effects); and (iii) that a dominant gain-of-function argP mutation (argP-S94L), predicted to cause a Ser-to-Leu substitution at residue 94 of the encoded protein, renders argO expression high and constitutive with respect to Lys or Lys-Ala supplementation.
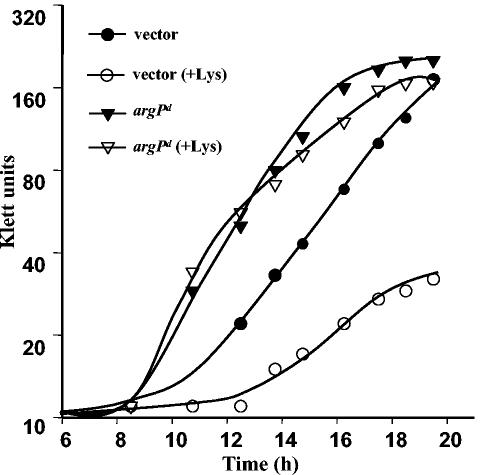
In light of the findings in the present study that an argP null mutation confers both osmosensitivity and an NH4+ assimilation deficiency in the gltBD background, the effects of exogenous Lys or Lys-Ala supplementation on the growth of the argP+ gltBD strain were examined. We found that Lys addition inhibited growth of the gltBD mutant, both in medium with 2 mM NH4+ as a nitrogen source (as shown in Fig. 2 for a gltBD derivative carrying the plasmid vector pBR329) and in a high-osmolarity medium (Fig. 3), just as an argP null mutation did. Inhibition by Lys of both NH4+ assimilation and osmotolerance was also observed in a gltBD derivative carrying argP+ on a multicopy plasmid (data not shown). Lys supplementation did not further exacerbate the same phenotypes in an argP gltBD double mutant (data not shown).
FIG. 3.
Dominant argP-S94L (argPd) mutation and Lys effects on osmosensitivity in derivatives of gltBD strain GJ4652. Semilogarithmic growth curves were plotted for pBR329 (vector) (• and ○) and pHYD953 (argPd) (▾ and ▿) derivatives of GJ4652 after 1:100 inoculation (from cultures grown to the stationary phase in glucose-minimal A medium) into glucose-minimal A medium containing 0.75 M NaCl, glycine betaine, and ampicillin without (• and ▾) or with (○ and ▿) Lys.
The effects of exogenous Lys on the gltBD mutant were also mimicked by the effects of the dipeptide Lys-Ala for both inhibition of NH4+ assimilation (Fig. 2) and osmosensitivity (data not shown), whereas a control dipeptide histidylalanine had no effect (data not shown). Supplementation with Lys or Lys-Ala was also associated with a reduction in intracellular glutamate accumulation in high-osmolarity-grown cultures of the gltBD single mutant, to much the same extent that was observed upon introduction of the argP null mutation (Table 5).
Suppression by the dominant gain-of-function argP-S94L mutation of Lys and Lys-Ala effects in a gltBD strain.
The inhibitory effects of Lys and Lys-Ala on argO transcription are abolished in strains with a dominant constitutive argP-S94L mutation (34). Accordingly, we tested whether the inhibition by these two compounds of NH4+ assimilation and osmotolerance in gltBD strains was affected by the dominant argP-S94L mutation. Growth comparisons between a pair of isogenic gltBD derivatives, one carrying plasmid pHYD953 with the dominant argP-S94L mutation (test strain) and the other carrying plasmid vector pBR329 (control strain), demonstrated that the dominant argP-S94L mutant allele conferred insensitivity to Lys or Lys-Ala supplementation on low-NH4+ medium (Fig. 2). The test strain was also insensitive to the inhibitory effect of Lys in high-osmolarity growth medium and indeed was somewhat more osmotolerant even in presence of Lys than the control strain was in its absence (Fig. 3); likewise, osmosensitivity in the gltBD strain associated with Lys-Ala supplementation was suppressed by the dominant argP-S94L mutation (data not shown). Concomitantly, the accumulation of cytoplasmic glutamate at high osmolarity was not affected by Lys or Lys-Ala supplementation in the gltBD argP derivative with the dominant argP mutation, unlike the accumulation of cytoplasmic glutamate in the gltBD argP+ strain (Table 5). These results suggest that the inhibitory effect of intracellular Lys on growth of the gltBD strain in either low-NH4+ or high-osmolarity medium, like the effect on argO expression, is mediated by the product of the argP+ gene.
argO is not involved in osmoregulation.
Given the parallels between the effects of argP (and the dominant argP mutation) on transcriptional regulation of the argO-encoded arginine exporter on the one hand (34) and on NH4+ assimilation and osmotolerance of gltBD strains on the other hand, we performed experiments to test whether the latter effect is in some way mediated by the former. Null mutants with mutations in argO, unlike mutants with mutations in argP, exhibited neither osmosensitivity nor NH4+ assimilation deficiency in the gltBD background (data not shown). Suppression by the dominant argP-S94L mutation of the inhibitory effect of Lys supplementation on osmotolerance and NH4+ assimilation in gltBD strains was not affected by introduction of an argO insertion mutation (data not shown). These results are consistent with the hypothesis (see below) that the gltBD-related phenotypes reflect ArgP-mediated regulation of a gene different from argO that is involved in NH4+ assimilation through a GOGAT-independent pathway.
Additional evidence for the putative dual role of argP, for regulation of argO on the one hand and for regulation of a gene(s) involved in osmoregulation on the other, was obtained from an activator titration experiment. When the multicopy test plasmid pHYD954 with the cloned cis regulatory region of argO or the control plasmid vector pBluescriptII-KS was introduced into the argP+ gltBD strain GJ4652, derivatives with the former plasmid but not derivatives with the latter plasmid exhibited both osmosensitivity and a deficiency in NH4+ assimilation comparable to the results obtained with the argP gltBD derivatives described above (data not shown). Our interpretation is that the argO regulatory region in multiple copies titrates the ArgP protein so that this protein is not available for activation of the gene involved in NH4+ assimilation.
DISCUSSION
GOGAT-deficient (gltBD) mutants have previously been shown to be osmosensitive in medium with limiting NH4+ (14, 43). In this study, starting from a population of transposon-mutagenized cells of a gltBD strain, we identified E. coli derivatives with null mutations in proU, argP, or glnE that were osmosensitive in NH4+-replete medium. In all these mutants, the absence of GOGAT was necessary for osmosensitivity.
The proU and proP loci encode active uptake systems that mediate the osmoprotectant effects of glycine betaine and proline in the enterobacteria (12, 13). The two transporters have overlapping functions, so that loss of either one is not associated with significant osmosensitivity (11, 17, 18, 24). Therefore, the isolation in the present study of osmosensitive proU insertion mutants in the gltBD background is a validation of the hypothesis that a GOGAT-deficient strain is sensitive to mutations affecting osmoregulation; Cayley et al. (5) have shown that glycine betaine accumulation in high-osmolarity-grown cells of a proU+ proP+ strain is associated with a reduction in intracellular glutamate levels.
Insertions at the other two loci, glnE and argP, also conferred osmosensitivity only in the GOGAT-deficient strain. We propose below that each of these genes is synergistic (for different reasons) with gltBD in reducing intracellular glutamate accumulation under osmotic stress conditions. Additional experimental support for the notion of synergism with the gltBD mutation has come from our findings (data not shown) that (i) in selection in transduction for osmotolerant derivatives of the glnE gltBD and argP gltBD strains, a large proportion of the transductants in both crosses had become gltBD+; and (ii) osmotolerance in both the argP gltBD and glnE gltBD mutants was fully restored in medium supplemented with aspartate, which serves a bypass route for glutamate synthesis. Therefore, it may be concluded that the strains are osmosensitive because they are limited for glutamate as a compatible solute and counterion for K+. (The alternative possibility, that glutamate is limiting for protein synthesis in these strains, is unlikely given the data shown in Table 5 for substantial intracellular pools of the amino acid, unless one assumes that the Km for charging of glutamyl-tRNA is increased at an elevated osmolarity.) It may also be noted that although glycine betaine is known to mediate a K+- and glutamate-sparing effect in osmoregulation (5, 26), the NaCl concentrations used in this study were high enough to override this effect.
Osmosensitivity of glnE gltBD mutants.
Whereas in a wild-type strain, increased activity of GS permits (and indeed is required for) growth on low levels of NH4+ or alternative nitrogen sources (35-37), excessive GS activity (particularly in a GOGAT-deficient strain) traps the assimilated nitrogen in glutamine. Adenylylation by the glnE-encoded adenylyltransferase is one mechanism by which the activity of GS is down-regulated, but glnE mutants are nevertheless phenotypically almost normal because of the presence of additional mechanisms for glnA transcriptional regulation. Accordingly, glnE mutants have previously been shown to exhibit a growth deficiency only (i) when GS is constitutively expressed because of a glnA promoter mutation (25) or (ii) transiently upon shift-up from nitrogen-poor to high-NH4+ conditions (25, 43).
The synergism that was observed in this study between glnE and gltBD in conferring osmosensitivity may be explained by the following model: (i) during steady-state growth of the wild-type strain in NH4+-replete media, the adenylylation of GS (which is synthesized only at a basal level) is dispensable at low osmolarity but is required at high osmolarity in order to prevent the channeling of the accumulated glutamate into glutamine; and (ii) in the glnE gltBD+ strain subjected to osmotic stress, GOGAT is able to catalyze the conversion of glutamine to glutamate. Goss et al. (16) similarly explained the inability of gltBD mutants to utilize as nitrogen sources even compounds, such as proline, that can be catabolized to yield glutamate (37), on the grounds that GOGAT is needed to enable reconversion of glutamine to glutamate. Our model is consistent with the data in Table 5 which show that, compared with the wild-type strain or even the gltBD single mutant, the glnE gltBD double mutant had a reduced glutamate pool and a greatly increased glutamine pool after growth in the high-osmolarity medium.
Another implication of our model is that intracellular glutamine cannot substitute for glutamate in osmoregulation. This conclusion is consistent with the findings obtained in a previous study (14), as well as with the hypothesis that glutamate's primary role in osmoregulation is to serve as the counterion to K+ (12, 13).
Osmosensitivity of argP gltBD mutants.
Null mutations in argP, unlike null mutaions in glnE, were associated with a reduction in the ability of the gltBD mutant to grow in medium with a low NH4+ concentration. Compared with the gltBD single mutant, the argP gltBD strain exhibited reduced GDH activity in NH4+-replete medium, particularly at elevated osmolarity (Table 4). Thus, we concluded that the GDH-catalyzed pathway of NH4+ assimilation is compromised in the argP mutants and that the reduced capacity for glutamate synthesis in the argP gltBD strains may be sufficient for growth in low-osmolarity media but not for growth in high-osmolarity media. A defect in glutamate accumulation in the double mutant was directly demonstrated in this study (Table 5). Whether an NH4+ uptake defect also might contribute to the argP mutant phenotypes remains to be determined.
A previous study (39) showed that fnr mutations also reduce NH4+ assimilation and confer osmosensitivity in the gltBD background and that multiple copies of spoT+ reverse both phenotypes in an fnr gltBD strain. We found in this work that multiple copies of spoT+ did not affect either phenotype in an argP gltBD strain (data not shown).
The mechanism by which a null mutation in argP reduces GDH activity, and consequently NH4+ assimilation and osmotolerance, in the gltBD strain is not known. Evidence from a study recently completed in our laboratory (34) suggests that ArgP is ordinarily a transcriptional activator protein for a target gene, argO (involved in arginine efflux), and that in the presence of intracellular Lys as a coeffector it represses argO expression. Thus, the results described here for the effects of argP mutations and of Lys or Lys-Ala supplementation in gltBD strains are most simply explained by the hypothesis that there is another gene (which perhaps is gdhA itself) whose regulation mirrors that of argO, which is involved in determining or modulating GDH activity in the gltBD strain. The hypothesis that intracellular Lys likely phenocopies the argP null mutation for decreased glutamate accumulation and osmosensitivity is strongly supported by our observation that the effects of exogenous Lys or Lys-Ala are suppressed in strains carrying a Lys-insensitive dominant gain-of-function argP allele.
Interestingly, Bender and coworkers have shown that gdhA transcription in Klebsiella aerogenes is repressed about threefold upon Lys supplementation; to explain this, these authors postulated that an unidentified regulator protein which is Lys sensitive activates gdhA in the strain (15, 22). Based on our results, it is likely that this regulator protein is the ortholog of E. coli ArgP.
Acknowledgments
We thank S. Blanquet, E. Bremer, K. Isono, and C. G. Lerner for providing strains, phages, and plasmids. We acknowledge V. Vamsee Krishna and T. Giri Babu for technical assistance, Mehar Sultana for primer synthesis, N. Nagesh for DNA sequencing, S. Jamal Mohamed Nurul Jain and S. M. Naushad for help with reverse-phase high-performance liquid chromatography analysis, and members of the laboratory of J.G. for advice and discussions.
This work was supported by research fellowships to M.R.N. and R.S.L. from the Council of Scientific and Industrial Research and by a grant-in-aid from the Department of Biotechnology, Government of India (project BT/PR2430). J.G. is an honorary faculty member of the Jawaharlal Nehru Centre for Advanced Scientific Research.
REFERENCES
- 1.Berlyn, M. K. B. 1998. Linkage map of Escherichia coli K-12, edition 10: the traditional map. Microbiol. Mol. Biol. Rev. 62:814-984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Blattner, F. R., G. Plunkett III, C. A. Bloch, N. T. Perna, V. Burland, M. Riley, J. Collado-Vides, J. D. Glasner, C. K. Rode, G. F. Mayhew, J. Gregor, N. W. Davis, H. A. Kirkpatrick, M. A. Goeden, D. J. Rose, B. Mau, and Y. Shao. 1997. The complete genome sequence of Escherichia coli K-12. Science 277:1453-1462. [DOI] [PubMed] [Google Scholar]
- 3.Botsford, J. L., M. Alvarez, R. Hernandez, and R. Nicholas. 1994. Accumulation of glutamate by Salmonella typhimurium in response to osmotic stress. Appl. Environ. Microbiol. 60:2568-2574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 5.Cayley, S., B. A. Lewis, and M. T. Record, Jr. 1992. Origins of the osmoprotective properties of betaine and proline in Escherichia coli K-12. J. Bacteriol. 174:1586-1595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Celis, R. T. F. 1990. Mutant of Escherichia coli K-12 with defective phosphorylation of two periplasmic transport proteins. J. Biol. Chem. 265:1787-1793. [PubMed] [Google Scholar]
- 7.Celis, R. T. F. 1999. Repression and activation of arginine transport genes in Escherichia coli K-12 by the ArgP protein. J. Mol. Biol. 294:1087-1095. [DOI] [PubMed] [Google Scholar]
- 8.Celis, R. T. F., H. J. Rosenfeld, and W. K. Maas. 1973. Mutant of Escherichia coli K-12 defective in the transport of basic amino acids. J. Bacteriol. 116:619-626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chang, A. C., and S. N. Cohen. 1978. Construction and characterization of amplifiable multicopy DNA cloning vehicles derived from the P15A cryptic mini-plasmid. J. Bacteriol. 134:1141-1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Covarrubias, L., and F. Bolivar. 1982. Construction and characterization of new cloning vehicles. VI. Plasmid pBR329, a new derivative of pBR328 lacking the 482-base-pair inverted duplication. Gene 17:79-89. [DOI] [PubMed] [Google Scholar]
- 11.Csonka, L. N. 1982. A third l-proline permease in Salmonella typhimurium which functions in media of elevated osmotic strength. J. Bacteriol. 151:1433-1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Csonka, L. N. 1989. Physiological and genetic responses of bacteria to osmotic stress. Microbiol. Rev. 53:121-147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Csonka, L. N., and W. Epstein. 1996. Osmoregulation, p. 1210-1223. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed. American Society for Microbiology, Washington, D.C.
- 14.Csonka, L. N., T. P. Ikeda, S. A. Fletcher, and S. Kustu. 1994. The accumulation of glutamate is necessary for optimal growth of Salmonella typhimurium in media of high osmolality but not induction of the proU operon. J. Bacteriol. 176:6324-6333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Goss, T. J., B. K. Janes, and R. A. Bender. 2002. Repression of glumatate dehydrogenase formation in Klebsiella aerogenes requires two binding sites for the nitrogen assimilation control protein, NAC. J. Bacteriol. 184:6966-6975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Goss, T. J., A. Perez-Matos, and R. A. Bender. 2001. Roles of glutamate synthase, gltBD, and gltF in nitrogen metabolism of Escherichia coli and Klebsiella aerogenes. J. Bacteriol. 183:6607-6619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gowrishankar, J. 1985. Identification of osmoresponsive genes in Escherichia coli: evidence for participation of potassium and proline transport systems in osmoregulation. J. Bacteriol. 164:434-445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gowrishankar, J. 1986. proP-mediated proline transport also plays a role in Escherichia coli osmoregulation. J. Bacteriol. 166:331-333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gowrishankar, J., and J. Pittard. 1982. Construction from Mu d1 (lac Apr) lysogens of lambda bacteriophage bearing promoter-lac fusions: isolation of λ ppheA-lac. J. Bacteriol. 150:1122-1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hwang, D. S., and A. Kornberg. 1990. A novel protein binds a key origin sequence to block replication of an E. coli minichromosome. Cell 63:325-331. [DOI] [PubMed] [Google Scholar]
- 21.Hwang, D. S., B. Thony, and A. Kornberg. 1992. IciA protein, a specific inhibitor of initiation of Escherichia coli chromosomal replication. J. Biol. Chem. 267:2209-2213. [PubMed] [Google Scholar]
- 22.Janes, B. K., P. J. Pomposiello, A. Perez-Matos, D. J. Najarian, T. J. Goss, and R. A. Bender. 2001. Growth inhibition caused by overexpression of the structural gene for glutamate dehydrogenase (gdhA) from Klebsiella aerogenes. J. Bacteriol. 183:2709-2714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kohara, Y., K. Akiyama, and K. Isono. 1987. The physical map of the whole E. coli chromosome: application of a new strategy for rapid analysis and sorting of a large genomic library. Cell 50:495-508. [DOI] [PubMed] [Google Scholar]
- 24.Koo, S. P., and I. R. Booth. 1994. Quantitative analysis of growth stimulation by glycine betaine in Salmonella typhimurium. Microbiology 140:617-621. [DOI] [PubMed] [Google Scholar]
- 25.Kustu, S., J. Hirschman, D. Burton, J. Jelesko, and J. C. Meeks. 1984. Covalent modification of bacterial glutamine synthetase: physiological significance. Mol. Gen. Genet. 197:309-317. [DOI] [PubMed] [Google Scholar]
- 26.Larsen, P. I., L. K. Sydnes, B. Landfald, and A. R. Strom. 1987. Osmoregulation in Escherichia coli by accumulation of organic osmolytes: betaines, glutamic acid, and trehalose. Arch. Microbiol. 147:1-7. [DOI] [PubMed] [Google Scholar]
- 27.Lerner, C. G., and M. Inouye. 1990. Low copy number plasmids for regulated low-level expression of cloned genes in Escherichia coli with blue/white insert screening capability. Nucleic Acids Res. 18:4631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Leveque, F., S. Blanchin-Roland, G. Fayat, P. Plateau, and S. Blanquet. 1990. Design and characterization of Escherichia coli mutants devoid of Ap4N-hydrolase activity. J. Mol. Biol. 212:319-329. [DOI] [PubMed] [Google Scholar]
- 29.May, G., E. Faatz, M. Villarejo, and E. Bremer. 1986. Binding protein dependent transport of glycine betaine and its osmotic regulation in Escherichia coli K12. Mol. Gen. Genet. 205:225-233. [DOI] [PubMed] [Google Scholar]
- 30.McLaggan, D., T. M. Logan, D. G. Lynn, and W. Epstein. 1990. Involvement of γ-glutamyl peptides in osmoadaptation of Escherichia coli. J. Bacteriol. 172:3631-3636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McLaggan, D., J. Naprstek, E. T. Buurman, and W. Epstein. 1994. Interdependence of K+ and glutamate accumulation during osmotic adaptation of Escherichia coli. J. Biol. Chem. 269:1911-1917. [PubMed] [Google Scholar]
- 32.Meers, J. L., D. W. Tempest, and C. M. Brown. 1970. ‘Glutamine (amide):2-oxoglutarate amino transferase oxidoreductase (NADP)', an enzyme involved in the synthesis of glutamate by some bacteria. J. Gen. Microbiol. 64:187-194. [DOI] [PubMed] [Google Scholar]
- 33.Miller, J. H. 1992. A short course in bacterial genetics: a laboratory manual and handbook for Escherichia coli and related bacteria. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 34.Nandineni, M. R., and J. Gowrishankar. 2004. Evidence for an arginine exporter encoded by yggA (argO) that is regulated by the LysR-type transcriptional regulator ArgP in Escherichia coli. J. Bacteriol. 186:3539-3546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Reitzer, L. 2003. Nitrogen assimilation and global regulation in Escherichia coli. Annu. Rev. Microbiol. 57:155-176. [DOI] [PubMed] [Google Scholar]
- 36.Reitzer, L., and B. L. Schneider. 2001. Metabolic context and possible physiological themes of σ54-dependent genes in Escherichia coli. Microbiol. Mol. Biol. Rev. 65:422-444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Reitzer, L. J. 1996. Ammonia assimilation and the biosynthesis of glutamine, glutamate, aspartate, asparagine, l-alanine, and d-alanine, p. 391-407. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed. American Society for Microbiology, Washington, D.C.
- 38.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 39.Saroja, G. N., and J. Gowrishankar. 1996. Roles of SpoT and FNR in NH4+ assimilation and osmoregulation in GOGAT (glutamate synthase)-deficient mutants of Escherichia coli. J. Bacteriol. 178:4105-4114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Smith, G. R., Y. S. Halpern, and B. Magasanik. 1971. Genetic and metabolic control of enzymes responsible for histidine degradation in Salmonella typhimurium. J. Biol. Chem. 246:3320-3329. [PubMed] [Google Scholar]
- 41.Spiro, S., and J. R. Guest. 1988. Inactivation of the FNR protein of Escherichia coli by targeted mutagenesis in the N-terminal region. Mol. Microbiol. 2:701-707. [DOI] [PubMed] [Google Scholar]
- 42.Thony, B., D. S. Hwang, L. Fradkin, and A. Kornberg. 1991. iciA, an Escherichia coli gene encoding a specific inhibitor of chromosomal initiation of replication in vitro. Proc. Natl. Acad. Sci. USA 88:4066-4070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yan, D., T. P. Ikeda, A. E. Shauger, and S. Kustu. 1996. Glutamate is required to maintain the steady-state potassium pool in Salmonella typhimurium. Proc. Natl. Acad. Sci. USA 93:6527-6531. [DOI] [PMC free article] [PubMed] [Google Scholar]