Abstract
Genomic scans of clones isolated from long-term stab cultures of Escherichia coli K-12 showed the loss of two large segments of the genome, with each lost segment being approximately 20 kb long. A detailed analysis of one of the deletions, located between 5.4 and 5.9 min, revealed that similar deletions had arisen in several other stab cultures. All deletions of this type exhibited a right terminus ending precisely at an IS5A element and a left terminus that varied over an ∼5-kb range but was bordered in all but two cases by sequences belonging to the preferred consensus target sequence for IS5, YTAR. The ubiquity of such deletions in independent stab cultures and the increase in their frequency over time argue that they have a selective advantage. It is speculated that the loss of the crl locus is responsible for the selective advantage of the deletions.
The gram-negative bacterium Escherichia coli is one of the most intensively studied organisms in biology. Our wealth of knowledge concerning this organism has come about in no small part from the ease of growing clones in laboratory cultures with defined media under easily replicated environmental conditions. Yet this ease of experimental manipulation has not encouraged the study of this organism outside of these simple environments, and as a result our understanding of the biology of this organism is largely restricted to the biology of mid-log-phase laboratory cultures, for which all components of the medium are nonlimiting. However, such conditions are rarely encountered by E. coli in natural environments. Savageau (36) estimated that the average generation time of E. coli in nature is ∼40 h. Since the generation time of E. coli in a rich medium under ideal conditions is ∼40 min, we must conclude that this organism spends the vast majority of its life cycle under conditions of starvation.
It is now known that E. coli cultures entering stationary phase undergo many complex physiological and biochemical changes (17, 28). Evidence is now accumulating that cultures maintained in stationary phase continue to undergo genetic changes. Kolter and colleagues (41, 42) have shown that mutants carrying loss-of-function alleles of both lrp and rpoS predominate in stationary-phase cultures, while Naas et al. (26, 27) have described extensive genetic variations arising from insertion sequence (IS) element transposition in stab cultures of E. coli maintained for up to 30 years. In a series of communications, Eisenstark and colleagues (13, 20, 30, 37) showed that archival cultures of Salmonella have accumulated variations in genome size and gene order as well as variations at the gene level. For this communication, we used comparative gene hybridization to systematically scan for changes in the genome of E. coli K-12 maintained in long-term stab cultures.
MATERIALS AND METHODS
Strains.
The strains used for this study are listed in Table 1. All clones isolated from stabs were derivatives of E. coli K-12 strain W3110. All clones were stored at −80°C in a 20% glycerol solution.
TABLE 1.
Bacterial strains used for this study
| E. coli K-12 derivative(s) | Relevant characteristics | Reference or originb |
|---|---|---|
| W3110a | rph-1 IN(rrnD-rrnE)1 | PEGM |
| A59 | Isolated from a 30-year-old stab | 26 |
| E26, E41 | proB-IS30, isolated from a 42-day-old stab (stab E) | This work |
| F10 | proB-IS30, isolated from a 42-day-old stab (stab F) | This work |
| G92, G95, G113 | proB-IS30, isolated from a 183-day-old stab (stab G) | This work |
| H122, H148 | proB-IS30, isolated from a 183-day-old stab (stab H) | This work |
| B52 | proB-IS30, isolated from a 183-day-old stab (stab B) | PEGM |
| D22 | proB-IS30, isolated from a 183-day-old stab (stab D) | PEGM |
| Y4, Y5, Y15, Y24, Y26, Y34, Y35, Y39, Y42, Y45, Y46, Y49, Y50, Y51, Y52, Y53, Y54, Y58, Y59, Y60 | proB-IS30, isolated from a 5-year-old stab (stab Y) | PEGM |
| Z14, Z20, Z22, Z24, Z27, Z28, Z29, Z57 | proB-IS30, isolated from a 4.5-year-old stab (stab Z) | PEGM |
| W2914a | Δ(gpt-proA) (also called proA2) | CGSC |
| W2915 | proA2 | CGSG |
| HB101 | proA2 | PEGM |
Strains W2914 and W3110 were derived from E. coli K-12 by two distinct lineages (5).
PEGM, Plasticité et Expression des Génomes Microbiens, Université Joseph Fourier, Grenoble, France, CGSC, Coli Genetic Stock Center (Yale University).
Media and culture conditions.
Cultures were maintained at 37°C in Luria-Bertani (LB) or M9 minimal medium supplemented with 0.004% proline when necessary (35). LB and M9 solid media contained 12 g of agar/liter. Stabs consisted of capped glass tubes (12-mm diameter) containing 2 ml of LB medium with agar (10 g/liter). The stabs were inoculated from overnight cultures by use of a loop. The inoculated stabs were incubated for 24 h at 37°C and then stored at room temperature. Twelve stabs were inoculated with the W3110 original strain, and at different stages (2, 42, and 183 days), four tubes were opened and a small plug (ca. 20 mm3) was removed from the surface of each. Serial dilutions were spread onto LB plates to isolate 30 independent clones from each of the stabs.
Molecular methods.
Standard molecular biology procedures were used (35). The digested total DNAs of the bacteria were hybridized with different digoxigenin-labeled probes. The IS5 and IS30 probes were prepared as previously described (26). IS911 DNA was obtained by PCR amplification with the pOF139 plasmid DNA as a template (32) and with primers IS911-1 (5′-GAAGTGGCACACTGAATTTGG) and IS911-2 (5′-TGAAGTGGTCAACAAAAACTGC). The other DNA probes were obtained by PCR amplification with pairs of internal primers for the following genes: for lcpA, primers LcpA1 (5′-AACGAACTGAACGAAGCGGCG) and LcpA2 (5′-AATCTCCTGAATGCGGTCGGC); for dinB, primers DinB1 (5′-AAATTATGCCCACATCTCACC) and DinB2 (5′-CGCGGTGGCGATTAGATCAGC); for yafO, primers YafO1 (5′-CAACCGGTTGCGGTTCTTTC) and YafO2 (5′-GGTTATGCTTGCGTCCACCG); for pepD, primers PepD1 (5′-CGTTCAGGGAGGTTTCAACC) and PepD2 (5′-CACGAAAGATCCTATCCAGC); for crl, primers Crl1 (5′-GCAACCCGAAGAGCAGATTG) and Crl2 (5′-CCGTTAACTTCACCGGCTCG); for phoE, primers PhoE1 (5′-TGCTTCTGCACGCTTGCCTG) and PhoE2 (5′-CGCCAGTAAAGATGGCGACC); for proB, primers ProB1 (5′-GTTGAACTTGTTCGCGAGTG) and ProB2 (5′-CCGAAAATCCAGCGTTTACGG); and for ykfC, primers YkfC1 (5′-AGCTTGCCACATGGGCAGCC) and YkfC2 (5′-GTACGCCACCGCGGGTTTCC). The nucleotide sequence of the region flanking the proB-IS30 deletion was determined with the purified PCR product obtained with an internal IS5 primer (5′-GACTGAGTCAGCCGAGAAG) in combination with primer PepD1, PepD3 (5′-GCTGGATAGGATCTTTCGTG), or YafO1.
Comparative genomic hybridization.
Genomic DNAs were isolated by standard procedures from overnight cultures grown in LB medium (35). After digestion with AluI, the DNAs were labeled by random priming with 33P-labeled nucleotides as described previously. The genomic DNAs were then hybridized to E. coli DNA array high-density nylon filters (Sigma-Genosys) by the procedures recommended by the manufacturer. The filters were then exposed overnight on a phosphorimaging screen. The filters were stripped by standard procedures as recommended by the manufacturer. Each high-density array was probed a maximum of six times. The arrays were analyzed with Array Gauge, v. 1.21, software (Fuji Medical Systems, Stamford, Conn.). Intensity measurements at each spot for each exposure were then obtained and were normalized by use of the following equation: normalized spot intensity = (spot intensity reading) × (10,000/average intensity of the reference spots). For each comparison, a total of four measured spot intensities were used. Modifying the work of Riehle et al. (34), we used a regression model, W1 = mW2 + b, to fit the strain W3110 data to obtain a total blot intensity-corrected replicate-averaged value for each gene by the equation W = (W1 + mW2 + b)/2, where W1 and W2 represent different exposures of one strain. In the same way, intensity-corrected replicate-averaged values were calculated for strains A59 and Y34. A residual was then calculated by fitting the data to a second regression model, y = mW + b, and determining the difference between the predicted and measured spot intensities. The residuals were sorted according to their gene order in E. coli K-12, and moving averages were calculated based on a window size of 20. Confidence intervals were calculated as ±3 standard deviations, determined as previously described (34).
RESULTS
We selected two clones isolated from long-term stab cultures to screen for duplications and deletions by comparative genomic hybridization. A59 was isolated from a stab culture of W3110 maintained for approximately 30 years in the laboratory of W. Arber (26). The second clone analyzed, Y34, was isolated from a stab culture (stab Y) (Table 1) that was maintained for 5 years and was initially inoculated with 22 independent clones (including A59) isolated from the same stab culture at the same time at which A59 was isolated (Table 1). Y34 was derived from clone A83, one of the 22 clones that were used to initiate the mixed stab culture Y. Stab culture Y (as well as stab culture Z [see below]), comprised of a series of isolates from a 30-year-old stab culture, was originally constructed to analyze fitness relationships between the clones. Independent mixed stab culture experiments indicated that A59 and A83 were the most abundant clones isolated from mixed stab cultures after several years.
Both A59 and Y34 were compared to a culture of the ancestral strain of W3110, which had been stored at −80°C. The comparative genomic hybridization intensities for W3110 and Y34 indicated that two large deletions occurred in strain Y34. Deletion I spans ∼19 kb and is located between 5.4 and 5.9 min on the E. coli K-12 map, while deletion II spans ∼23 kb and is located between 72.1 and 72.5 min on the map (Fig. 1). The genes located within these two deletions are listed in Table 2. Comparative genomic hybridization intensities for W3110 and the intermediate culture A59 indicated that A59 does not possess any deletions (data not shown).
FIG. 1.
Comparative genomic hybridization of strain W3110 and strain Y34. Genes are plotted by their coordinates according to the E. coli K-12 genome map (6). Confidence intervals were calculated as described in the text.
TABLE 2.
Deletions detected in strain Y34
| Deletion | Deleted genes | Size (kb) | Location on E. coli K-12 map (min) |
|---|---|---|---|
| I | gpt, yafA, crl, phoE, proB, proA, b0245, yafW, b0247, yafX, b0249, ykfB, yafY, yafZ, ykfA, perR, b0255, tra8_1, b0257, ykfc, yi52_10 | 19.1 | 5.4-5.9 |
| II | ptsN, b3205, ptsO, b3207, yrbM, yhbL, arcB, yhcC, gltB, gltD, gltF, yhcA, yhcD, yhcE, yi52_10 | 23.4 | 72.1-72.5 |
Deletion I, found in Y34, includes the proBA operon, which is required for proline biosynthesis. We confirmed that this clone was auxotrophic for proline. Since proline auxotrophy therefore provided a convenient screen for strains that had suffered a deletion of this section of the chromosome, we elected to screen isolates from other stab cultures for similar deletions.
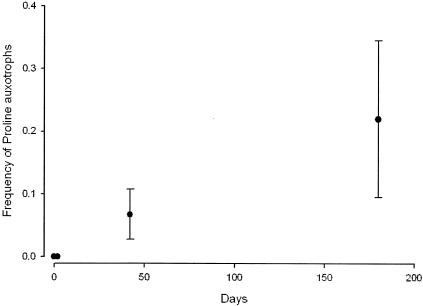
An analysis of the 22 strains used to inoculate stab Y (including A59 and A83) revealed that none were proline auxotrophs. We therefore concluded that deletion I arose during the 5 years of maintenance of this stab culture. To determine the dynamics of the proline auxotroph phenotype in stab cultures, we tested a total of 360 clones isolated from a series of 12 independent stab cultures maintained for 2, 42, or 183 days. Figure 2 shows that the frequency of proline auxotrophs increased steadily, to an average frequency of 0.22 after 183 days. In addition, we tested a total of 201 clones isolated from two additional mixed stab cultures maintained for 5 years (stab Y) (Table 1) and 4.5 years (stab Z) (Table 1). Among these 201 clones, the frequency of proline auxotrophs reached 0.21 and 0.40 after 4.5 and 5 years of starvation, respectively.
FIG. 2.
Increase in average frequency of proline auxotrophs with age of stab culture. Error bars indicate standard errors.
Since the occurrence of type I deletions occurred repeatedly in independent stab cultures and since their frequency increased steadily over time, we analyzed a subset of 38 clones isolated from these stab cultures, all of which were proline auxotrophs. To determine the exact end points of the deletions, we digested the genomic DNAs with EcoRV and hybridized them with probes for IS5, IS30, IS911, lcpA, dinB, yafO, pepD, crl, phoE, proB, and ykfC. For each clone, the restriction map of the region was refined by PCR amplification with the IS5 primer in combination with the PepD1, PepD3, or YafO1 primer (see Materials and Methods). The results confirmed the existence of the type I deletion (Table 2) in Y34 and allowed us to more precisely determine its right and left boundaries. Based on restriction analysis of the lcpA-IS1A region and the lengths of the PCR products, the 38 clones identified as proline auxotrophs fell into 12 groups (Fig. 3). All mutants shared a deleted region of approximately 16 kb between the crl and ykfC loci. At the left extremity, all deletions terminated upstream of crl and downstream of dinB. With the 16-kb deletion (the shortest one), the following characterized loci were lost: crl, phoE, proBA, thrW, and IS30A. With the largest deletions, which were approximately 21 kb long, the gpt and pepD genes were also lost. Some stab cultures were observed to be polymorphic, containing up to three different type I deletions (e.g., E26 and E41 in stab E; Y39, Y52, and Y54 in stab Y; and Z20, Z22, and Z29 in stab Z). The same deletion was found in two different stab cultures, B52 and D22, but otherwise the deletions found in each culture were unique to that culture.
FIG. 3.
The type I deletions can be grouped into the following 12 categories: B52 (D22), H122 (H148), F10, G113 (G92 and G95), Y54 (Y34, Y42, Y50, and Y59), E26 (HB101, W2914, and W2915), Z22 (Z27, Z28, and Z57), Z14 (Z29), Y39 (Y4, Y5, Y15, Y24, Y26, Y35, Y45, Y46, Y49, Y58, and Y60), Y52 (Y51 and Y53), Z20 (Z24), and E41. The deletions are displayed as thick lines. Comparisons of the deletion lengths distinguish variable and common deleted regions. At the opposite end from the IS element, the last open reading frame in the common deleted region is crl. The IS30 element is inserted inside an inactivated single copy of IS911. The distances are given (in kilobases) according to the published E. coli K-12 genome (6).
In order to precisely determine the boundaries of the 12 type I deletions, we sequenced the regions encompassing the 12 different junctions. The results are shown in Fig. 4. The right boundary was invariant in all of the deletion types sequenced and was located exactly at the left terminus of the IS50A sequence. The locations of the left boundaries of the deletions varied over 5 kb, confirming the Southern analysis results presented above. Interestingly, although the left boundaries of the 12 deletions were variable, all but two terminated in a four-nucleotide consensus TTAR sequence, a subset of the YTAR target consensus sequence seen with IS5 transposition.
FIG. 4.
Sequences of the flanking regions of type 1 deletions. The sequences at the left boundary for H122, B52, F10, G113, Y54, E26, HB101, Y39, Y52, Z20, and E41 matched the consensus preferred target sequence, YTAR, for IS5. The sequences for Y14 and Z52 differed from this consensus sequence by one base. The end of IS5A is underlined and shown in lowercase, and the target sequences of the IS5 insertion are shown in bold. The nucleotide positions on the chromosome are given according the E. coli K-12 genomic sequence (6).
The commonly used E. coli K-12 strain HB101 is known to be auxotrophic for proline and to harbor a deletion termed either Δ(gpt-proA)62 or proA2 (7). In addition, this strain has been used to identify the function of the crl gene by complementation (4, 29). We therefore characterized this deletion by sequencing the region encompassing the junction. The results (Fig. 4) revealed that the deletion present in HB101 is identical to the E26 deletion. In the HB101 lineage (5), strain W2915 was the first ancestral clone to be described as a proline auxotroph, but it has been suggested (M. Berlyn, personal communication) that this phenotype was already present in W2914, the direct ancestor of W2915. We verified that both of these strains indeed carry the same deletion.
DISCUSSION
Genomic rearrangements arising in long-term culture.
We have presented evidence here that long-term cultures in stationary phase accumulate large-scale deletions spanning many kilobases of the E. coli K-12 chromosome. These results thus extend earlier observations that cultures in stationary phase continue to undergo genetic changes. However, all of the previous work has reported on the accumulation of single gene mutations (41, 42), transpositions of IS elements (26, 27), or genomic rearrangements (43) rather than the loss of genetic material.
Porwillok et al. (30) have described archival strains of Salmonella enterica that have apparently suffered deletion as well as the amplification of genetic material. Although not common, there have been earlier reports of selection for deletions arising in long-term culture. Riehle et al. (34) used comparative genomic hybridization and observed two deletions in six lines of E. coli B evolving for ∼2,000 generations under high-temperature stress. Modi et al. (24) described the selection of a plasmid deletion arising in a long-term glucose-limited chemostat culture under conditions in which the so-called hunger response and the starvation response overlapped (14). Similarly, Adams et al. (1) and Dunham et al. (12) observed gross chromosomal changes and rearrangements in evolving yeast populations, some of which were large deletions. The evolutionary significance of deletions and gene loss has been revealed by comparative evolutionary studies, in which the inactivation of nonfunctional or inessential genes often precedes gene loss. This pattern is seen most dramatically in the evolution of parasitic or symbiotic prokaryotic genomes, which show a significant reduction in size (23, 25).
Involvement of IS5 in the generation of type I deletions.
The location of an IS5A sequence at the right-hand boundary of all of the type I deletions seen points to the involvement of this element in the generation of the deletions. First described in the case of IS1 (33), several other IS elements have been shown to generate deletions that terminate precisely at one end of the IS element. The mechanism of the formation of such deletions and the transposition of the element appear to share a common pathway (15). Deletions are thought to be generated by a two-step process in which the insertion of a new IS copy near a preexisting IS is followed by homologous recombination between the two copies. IS5 transposes nonrandomly, and a preferred consensus target sequence, YTAR, has been described (21). All but two of the deletions possessed the sequence TTAR, belonging to the preferred consensus sequence YTAR, at the left boundary. The two deletions that did not possess the TTAR sequence at the left boundary had a four-base sequence at the left boundary that differed by only one base from the YTAR consensus. It is therefore hard to escape the conclusion that the IS5 transposition machinery is involved in the generation of these deletions.
Many reports have shown that repeated elements in the genome can often be recombinogenic, facilitating the formation of both duplications and deletions (3, 10, 11, 16, 18, 19). In a recent report, Dunham et al. (12) showed that deletions and major chromosomal rearrangements arising in Saccharomyces cerevisiae were all bordered by sigma, tau, or delta repeated elements. Gene duplications, by providing regions of adjacent homology, are intrinsically unstable and can result in frequent amplifications and deletions of duplicated regions via unequal recombination events (2).
Selective advantage of type I deletions.
Four lines of evidence point to the selective advantage of type I deletions in stab cultures. (i) A type I deletion was observed both in Y34, a clone isolated from a stab culture that was maintained for 5 years, and in several other stab cultures. (ii) The frequency of type I deletions increased steadily over time in short-term stab cultures (Fig. 2). (iii) The variety of different type I deletions seen, which were often characteristic of the particular stab culture, indicates that such deletions arose repeatedly during the evolution of the cultures. Thus, their ubiquity cannot be explained by the occurrence of a unique event occurring in a clone containing a selectively favored mutation (“hitchhiking”). (iv) A type I deletion is present in the widely used laboratory strain of E. coli K-12, HB101, as well as in other strains of the HB101 lineage, such as W2914 and W2915. In addition, a deletion in the same region occurs in strain χ711 (8).
Deletion of the crl locus.
It is noteworthy that the right boundaries of type I deletions vary over a 5-kb range but that in all 12 categories, the crl gene was deleted and appeared to define the limit of the smallest deletion. We therefore speculate that the selective advantage of type I deletions resides in the loss of the crl gene. In all but two cases, the left boundary of the deletion was characterized by the existence of the YTAR consensus preferred target sequence for IS5 transposition. YTAR sequences are observed frequently in the E. coli genome, occurring every 50 to 100 bases, and can be found adjacent to the left boundary of the deletions. Thus, the absence of sequences of the YTAR family cannot be explained to invoke the lower size limit of type I deletions. Although we cannot rule out mechanical constraints for the lower size limit of type I deletions, the available information for the crl gene implicates it as a likely candidate. First identified as a transcriptional activator of csgAB, the operon encoding the surface protein curli (4, 29), the cytoplasmic Crl factor also modulates the expression of rpoS during starvation (31). Mutations in rpoS that confer a selective advantage during starvation in liquid cultures have already been described (41), suggesting that other mutations modulating the rpoS regulon may be advantageous under starvation conditions.
The dinB locus is maintained by selection.
While the lower size limit of type I deletions can be speculated to be defined by the crl locus, the upper size limit of these deletions appears to be defined by the presence of the dinB locus, which encodes DNA polymerase IV. As with the lower limit defined by the crl locus, the paucity of YTAR sequences in the genome cannot explain this upper limit, as they occur frequently in the E. coli genome, on an average of once every 50 to 100 bases. Similarly, it is unlikely that mechanical constraints define this upper limit, as Brooke and Valvano (8) have described a large IS5-mediated deletion of the crl-dinB-lpcA region in the E. coli K-12 derivative χ711. However, it is noteworthy that DNA polymerase IV is involved in SOS-induced untargeted mutagenesis (38, 39) and in the appearance of mutations in cells that are not growing (22). Furthermore, the loss of dinB activity results in a marked reduction in fitness compared with dinB+ strains (40). It is therefore tempting to suggest that the upper size limit of type I deletions is determined by dinB activity and the generation of adaptive genetic variation in stab cultures undergoing low rates of cell turnover. The selective advantage of mutator strains in continuous cultures has likewise been shown to be due to an increase in the rate of generation of genetic variations among mutator strains (9).
In conclusion, the use of comparative genomic hybridization has allowed us to identify large-scale deletions occurring in cultures evolving under long-term conditions of extreme starvation. The analysis of one category of deletion has further allowed us to identify loci that we speculate are both selectively lost and selectively maintained in such cultures.
Acknowledgments
We thank H. Geiselmann, V. Horn, and J. P. Alcaraz for technical assistance and O. Fayet and M. Chandler for the gifts of IS911-specific primers and the pOF139 plasmid. We give special thanks to M. Berlyn for her help in reconstituting the genetic events arising in HB101 lineage and to M. Chapman for critically reading the manuscript.
This work was supported in part by the Centre National de la Recherche Scientifique, the Université Joseph Fourier—Grenoble 1, a national program of the Bureau des Ressources Génétiques (1999-2000), and NIH grant AI55756.
Footnotes
This paper is dedicated to the memory of Michel Blot, whose contributions to the microbial evolution community will long be remembered.
REFERENCES
- 1.Adams, J., S. Puskas-Rozsa, J. Simlar, and C. M. Wilke. 1992. Adaptation and major chromosomal changes in populations of Saccharomyces cerevisiae. Curr. Genet. 22:13-19. [DOI] [PubMed] [Google Scholar]
- 2.Anderson, R. P., and J. R. Roth. 1977. Tandem genetic duplication in bacteria and phage. Annu. Rev. Microbiol. 31:473-505. [DOI] [PubMed] [Google Scholar]
- 3.Arber, W. 1983. Bacterial “inserted sequence” elements and their influence on genetic stability and evolution. Prog. Nucleic Acids Res. Mol. Biol. 29:27-33. [DOI] [PubMed] [Google Scholar]
- 4.Arnqvist, A., A. Olsen, J. Preifer, D. G. Russell, and S. Normark. 1992. The Crl protein activates cryptic genes for curli formation and fibronectin binding in Escherichia coli HB101. Mol. Microbiol. 6:2443-2452. [DOI] [PubMed] [Google Scholar]
- 5.Bachmann, B. J. 1996. Derivations and genotypes of some mutant derivatives of Escherichia coli K-12, p. 2460-2488. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed. American Society for Microbiology, Washington, D.C.
- 6.Blattner, F. R., G. R. Plunkett, C. A. Bloch, N. T. Perna, V. Burland, M. Riley, et al. 1997. The complete genome sequence of Escherichia coli K-12. Science 277:1453-1474. [DOI] [PubMed] [Google Scholar]
- 7.Boyer, H. W., and D. Roulland-Dussoix. 1969. A complementation analysis of the restriction and modification of DNA in Escherichia coli. J. Mol. Biol. 41:459-472. [DOI] [PubMed] [Google Scholar]
- 8.Brooke, J. S., and M. A. Valvano. 1996. Biosynthesis of inner core polysaccharide in enteric bacteria: identification and characterization of a conserved phosphoheptose isomerase. J. Biol. Chem. 271:3608-3614. [DOI] [PubMed] [Google Scholar]
- 9.Chao, L., and E. C. Cox. 1983. Competition between high and low mutating strains of Escherichia coli. Evolution 37:125-134. [DOI] [PubMed] [Google Scholar]
- 10.Charlier, D., Y. Severne, M. Zafarullah, and N. Glansdorff. 1983. Turn-on of inactive genes by promoter recruitment in Escherichia coli: inverted repeats resulting in artificial divergent operons. Genetics 105:469-488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Clugston, C. K., and A. P. Jessop. 1991. A bacterial position effect: when the F factor in E. coli K12 is integrated in cis to a chromosomal gene that is flanked by IS1 repeats, the elements are activated so that amplification and other regulatory changes that affect the gene can occur. Mutat. Res. 248:1-15. [DOI] [PubMed] [Google Scholar]
- 12.Dunham, M. J., H. Badrane, T. Ferea, J. Adams, P. O. Brown, R. F. Rosenzweig, and D. Botstein. 2002. Characteristic genome rearrangements accompany experimental evolution of S. cerevisiae Proc. Natl. Acad. Sci. USA 99:16144-16149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Edwards, K., I. Linetsky, C. Hueser, and A. Eisenstark. 2001. Genetic variability among archival cultures of Salmonella typhimurium. FEMS Microbiol. Lett. 199:215-219. [DOI] [PubMed] [Google Scholar]
- 14.Ferenci, T. 1999. Regulation by nutrient limitation. Curr. Opin. Microbiol. 2:208-213. [DOI] [PubMed] [Google Scholar]
- 15.Galas, D. J., and M. Chandler. 1989. Bacterial insertion sequences, p. 109-162. In D. E. Berg and M. M. Howe (ed.), Mobile DNA. American Society for Microbiology, Washington, D.C.
- 16.Hill, C. W., R. H. Graftstrom, B. W. Harnish, and B. S. Hillman. 1977. Tandem duplications resulting from recombination between ribosomal RNA genes in Escherichia coli. J. Mol. Biol. 116:407-428. [DOI] [PubMed] [Google Scholar]
- 17.Huisman, G. W., D. A. Siegele, M. M. Zambrano, and R. Kolter. 1996. Morphological and physiological changes during stationary phase, p. 1672-1682. In F. C. Neidhardt, R. Curtiss III, J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, W. S. Reznikoff, M. Riley, M. Schaechter, and H. E. Umbarger (ed.), Escherichia coli and Salmonella: cellular and molecular biology, 2nd ed. American Society for Microbiology, Washington, D.C.
- 18.Jessop, A. P., and C. Clugston. 1985. Amplification of the argF region in strain HfrP4X of E. coli K-12. Mol. Gen. Genet. 201:347-350. [DOI] [PubMed] [Google Scholar]
- 19.Lin, R., M. Capage, and C. W. Hill. 1984. A repetitive DNA sequence, rhs, responsible for duplications within the Escherichia coli chromosome. J. Mol. Biol. 177:1-18. [DOI] [PubMed] [Google Scholar]
- 20.Liu, G.-R., K. Edwards, A. Eisenstark, Y.-M. Fu, W.-Q. Liu, K. E. Sanderson, R. N. Johnston, and S.-U. Liu. 2003. Genomic diversification among archival strains of Salmonella enterica serovar Typhimurium LT7. J. Bacteriol. 185:2131-2142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mahillon, J., and M. Chandler. 1998. Insertion sequences. Microbiol. Mol. Biol. Rev. 62:725-774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McKenzie, G. J., P. L. Lee, M. J. Lombardo, P. J. Hastings, and S. M. Rosenberg. 2001. SOS mutator DNA polymerase IV functions in adaptive mutation and not adaptive amplification. Mol. Cell 7:571-579. [DOI] [PubMed] [Google Scholar]
- 23.Mira, A., H. Ochman, and N. A. Moran. 2001. Deletional bias and the evolution of bacterial genomes. Trends Genet. 17:589-596. [DOI] [PubMed] [Google Scholar]
- 24.Modi, R. I., C. M. Wilke, R. F. Rosenzweig, and J. Adams. 1991. Plasmid macro-evolution: selection of spontaneous deletions in a pBR322 derivative. Genetica 84:195-202. [DOI] [PubMed] [Google Scholar]
- 25.Moran, N. A. 2003. Tracing the evolution of gene loss in obligate bacterial symbionts. Curr. Opin. Microbiol. 6:512-518. [DOI] [PubMed] [Google Scholar]
- 26.Naas, T., M. Blot, W. M. Fitch, and W. Arber. 1994. Insertion sequence-related genetic variation in resting Escherichia coli K-12. Genetics 136:721-730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Naas, T., M. Blot, W. M. Fitch, and W. Arber. 1995. Dynamics of IS-related genetic rearrangements in resting Escherichia coli K-12. Mol. Biol. Evol. 12:198-207. [DOI] [PubMed] [Google Scholar]
- 28.Nystrom, T. 2003. Conditional senescence in bacteria: death of the immortals. Mol. Microbiol. 48:17-23. [DOI] [PubMed] [Google Scholar]
- 29.Olsen, A., A. Jonsson, and S. Normak. 1989. Fibronectin binding mediated by a novel class of surface organelles on Escherichia coli. Nature 338:652-655. [DOI] [PubMed] [Google Scholar]
- 30.Porwillok, S., R. M.-Y. Wong, R. A. Helm, K. K. Edwards, M. Calcutt, A. Eisenstark, and M. McClelland. 2004. DNA amplification and rearrangements in archival Salmonella enterica serovar Typhimurium LT2 cultures. J. Bacteriol. 186:1678-1682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pratt, L. A., and T. J. Silhavy. 1998. Crl stimulates RpoS activity during stationary phase. Mol. Microbiol. 29:1225-1236. [DOI] [PubMed] [Google Scholar]
- 32.Prère, M. F., M. Chandler, and O. Fayet. 1990. Transposition in Shigella dysenteriae: isolation and analysis of IS911, a new member of the IS3 group of insertion sequences. J. Bacteriol. 172:4090-4099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Reif, H. J., and H. Saedler. 1975. IS1 is involved in deletion formation in the gal region of E. coli K-12. Mol. Gen. Genet. 137:17-28. [DOI] [PubMed] [Google Scholar]
- 34.Riehle, M. M., A. F. Bennett, and A. D. Long. 2001. Genetic architecture of thermal adaptations in Escherichia coli. Proc. Natl. Acad. Sci. USA 98:525-530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 36.Savageau, M. 1983. Escherichia coli habitats, cell types and molecular mechanisms of gene control. Am. Nat. 122:732-744. [Google Scholar]
- 37.Sutton, A., B. Buencamino, and A. Eisenstark. 2000. rpoS mutants in archival culture of Salmonella enterica serovar Typhimurium. J. Bacteriol. 182:4375-4379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tang, M., P. Pham, X. Shen, J. S. Taylor, M. O'Donnell, R. Woodgate, and M. F. Goodman. 2000. Roles of E. coli DNA polymerase IV and V in lesion-targeted and untargeted SOS mutagenesis. Nature 404:1014-1018. [DOI] [PubMed] [Google Scholar]
- 39.Wagner, J., P. Gruz, S. R. Kim, M. Yamada, K. Matsui, R. P. P. Fuchs, and T. Nohmi. 1999. The dinB gene encodes a novel E. coli DNA polymerase, DNA Pol IV, involved in mutagenesis. Mol. Cell 4:281-286. [DOI] [PubMed] [Google Scholar]
- 40.Yeiser, B., E. D. Pepper, M. F. Goodman, and S. E. Finkel. 2002. SOS-induced DNA polymerases enhance long-term survival and evolutionary fitness. Proc. Natl. Acad. Sci. USA 99:8737-8741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zambrano, M. M., D. A. Siegele, M. Almiron, A. Tormo, and R. Kolter. 1993. Microbial competition: Escherichia coli mutants that take over stationary phase cultures. Science 259:1757-1760. [DOI] [PubMed] [Google Scholar]
- 42.Zinser, E. R., and R. Kolter. 2000. Prolonged stationary-phase incubation selects for lrp mutation in Escherichia coli K-12. J. Bacteriol. 182:4361-4365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zinser, E. R., D. Schneider, M. Blot, and R. Kolter. 2003. Bacterial evolution through the selective loss of beneficial genes: trade-offs in expression involving two loci. Genetics 164:1271-1277. [DOI] [PMC free article] [PubMed] [Google Scholar]