Abstract
NAD-independent lactate dehydrogenases are commonly thought to be responsible for lactate utilization during the stationary phase of aerobic growth in Lactobacillus plantarum. To substantiate this view, we constructed single and double knockout mutants for the corresponding genes, loxD and loxL. Lactate-to-acetate conversion was not impaired in these strains, while it was completely blocked in mutants deficient in NAD-dependent lactate dehydrogenase activities, encoded by the ldhD and ldhL genes. We conclude that NAD-dependent but not NAD-independent lactate dehydrogenases are involved in this process.
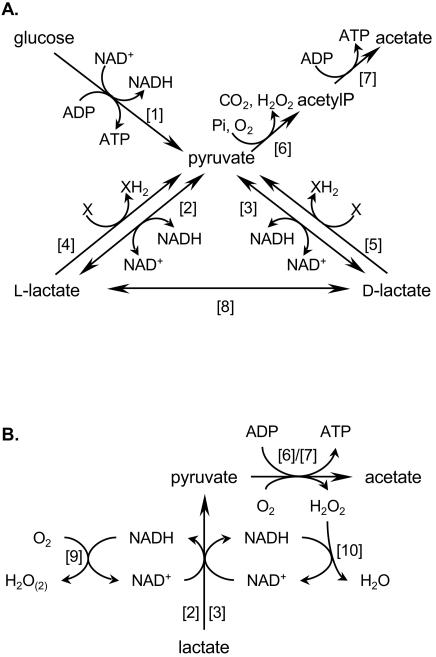
Under anaerobic conditions, Lactobacillus plantarum produces lactate as the major fermentation end product through the reduction of glycolytic pyruvate by NAD-dependent lactate dehydrogenases (nLDHs) (7, 8). When grown in the presence of oxygen, L. plantarum ferments glucose into lactate until glucose becomes limiting. Lactic acid produced during fermentation is then converted to acetate with concomitant hydrogen peroxide, carbon dioxide, and ATP production (Fig. 1A) (20). Acetate becomes the major fermentation end product, leading to homoacetic fermentation (18). This conversion is thought to play a role in pH homeostasis and survival during the stationary phase of growth under aerobic conditions (18, 20). The additional ATP generated through this pathway can also lead to increased biomass (20). The proposed pathway requires oxygen and involves three steps. Lactate is first converted to pyruvate, which is then decarboxylated by the pyruvate oxidase (Pox) to form acetylphosphate. In the third step, acetate is produced from acetylphosphate by an acetate kinase (Ack), generating ATP (20).
FIG. 1.
(A) Metabolic pathways of lactic acid production and utilization in L. plantarum in aerobic conditions as proposed in the literature. (B) Proposed pathway for d- and l-lactate utilization in the stationary phase of aerobic growth of L. plantarum. [1], glycolysis; [2], l-nLDH (LdhL); [3], d-nLDH (LdhD); [4], l-iLDH; [5], d-iLDH; [6], pyruvate oxidase; [7], acetate kinase; [8], lactate racemase; [9], NADH oxidase; [10], NADH peroxidase; X, unknown electron acceptor.
The key role played by Pox in this pathway has been demonstrated recently (18). Yet, the enzymes responsible for the conversion of lactate to pyruvate have not been identified. This oxidation could be catalyzed by nLDHs or NAD-independent lactate dehydrogenases (iLDHs) (Fig. 1A). In L. plantarum, two stereospecific nLDHs are present (LdhD and LdhL), whose role in lactate production has been established (7, 8). The LdhL enzyme is responsible for l-lactate production, while d-lactate can be produced in two ways: NADH-dependent reduction of pyruvate by the LdhD enzyme or racemization of l-lactate by an l-lactate-inducible lactate racemase (7, 8) (Fig. 1A). The possible involvement of nLDHs in lactate utilization after glucose exhaustion in the presence of oxygen has not been addressed in vivo, although these enzymes have been shown to catalyze the in vitro oxidation of lactate at high pH in the presence of high substrate concentrations (3, 10).
The iLDHs, also called respiratory lactate dehydrogenases, are stereospecific enzymes that catalyze the oxidation of lactate to pyruvate by a flavin-dependent mechanism (flavin mononucleotide and flavin adenine dinucleotide for l- and d-iLDH, respectively). These enzymes have been studied extensively in Escherichia coli and Saccharomyces cerevisiae, where they play an essential role in the aerobic utilization of lactate through the respiratory electron transport chain (2, 6, 9). In these species, the external electron acceptor is a membrane quinone (E. coli) or cytochrome c (S. cerevisiae). In E. coli, the iLDHs are peripheral membrane proteins (6, 9), while in S. cerevisiae, they are located in the mitochondrial intermembrane space (2). In most lactic acid bacteria, these enzymes appear to be soluble (10), although it has been recently shown that the lactate oxidase (l-iLDH) activity from Streptococcus pyogenes was associated with the membrane (21).
The iLDHs from lactobacilli have been divided in two groups (13). The enzymes from Lactobacillus curvatus, Lactobacillus sakei, Lactobacillus acidophilus, and Lactobacillus bulgaricus use molecular oxygen as the external electron acceptor and belong to the lactate oxidase group, which also includes the lactate oxidases from Streptococcus iniae (11) and S. pyogenes (21). The second group contains enzymes whose external electron acceptor has not been identified. This includes the iLDHs from Lactobacillus casei (19, 25) as well as the d- and l-iLDHs from L. plantarum, which are flavin-containing soluble proteins (22, 23). However, the amino acid sequences of proteins from the two groups are closely related, and the ability to use molecular oxygen as an electron acceptor cannot be inferred from their primary sequence (16).
Although the in vitro reduction of pyruvate to lactate was reported for some iLDHs (19), these are generally considered enzymes that catalyze only the oxidation of lactate to pyruvate in vivo (10). Based on in vitro experiments, some authors hypothesized that both iLDH and nLDH activities might be able to efficiently catalyze the oxidation of lactate (20).
The involvement of theses enzymes in lactate utilization in lactic acid bacteria has never been investigated by a genetic approach. The present study was thus aimed at determining which of the iLDH or nLDH enzymes are involved in the oxidation of lactate to pyruvate during the lactate consumption phase that takes place after glucose exhaustion in aerated cultures of L. plantarum.
Cloning and knockout of the loxD and loxL genes of L. plantarum.
We first assayed the d- and l-iLDH activities of L. plantarum NCIMB8826 as a function of growth phase to check whether their expression pattern was compatible with a role in the conversion of lactate to acetate during the stationary phase of aerobic growth. As previously reported for other strains of L. plantarum, both activities increase gradually during exponential phase until they reach their maximum in early stationary phase (20; data not shown). The expression pattern of the iLDH enzyme activities is thus similar to that of pyruvate oxidase activity (18, 20), supporting their postulated role in the lactate-to-acetate conversion pathway.
The corresponding genes were identified in the L. plantarum WCFS1 (single colony isolate of NCIMB8826) genome (14). The loxL gene (lp_3586) is annotated as a lactate oxidase, and its encoded protein displays a high level of identity (49%) with the l-lactate oxidase LctO of S. iniae (11). It contains the catalytic histidine residue at position 264 as well as the substrate-binding arginine residue (position 267, L. plantarum LoxL numbering), as described for the l-iLDH (CYB2) of S. cerevisiae (30). Of the four residues involved in flavin mononucleotide binding (K429, D459, R493, and R513 in CYB2 of S. cerevisiae), three are conserved, while the last one is replaced by an asparagine (N270, L. plantarum LoxL numbering). This residue, however, is also not conserved in the l-iLDH enzyme of Aerococcus viridans, which nevertheless has been shown to be fully functional (4). The loxL gene is located downstream of two pyruvate oxidase genes (lp_3589 [poxB] and lp_3587 [poxC]) (14, 18). Moreover, the two pox genes and loxL are part of a single transcription unit whose expression is negatively regulated by glucose and induced in early stationary phase (F. Lorquet, L. Muscariello, P. Goffin, M. Sacco, M. Kleerebezem, and P. Hols, unpublished data), corroborating the possible role of LoxL in the conversion of lactate to acetate after glucose exhaustion.
The L. plantarum gene coding for LoxD (lp_0291) is annotated as an oxidoreductase in the WCFS1 genome (14) and was identified by homology with the S. cerevisiae DLD1 gene encoding d-lactate ferricytochrome c oxidoreductase (d-iLDH [17]). The overall sequence identity of LoxD from L. plantarum with the S. cerevisiae d-iLDH enzyme is 29%. The loxD gene is probably monocistronic, and no gene recognizably involved in lactate or acetate metabolism is located in its direct vicinity.
Knockout mutants for the loxD and loxL genes were constructed by a two-step homologous recombination procedure (7, 8). The 5′- and 3′-recombination segments were amplified by PCR from NCIMB8826 chromosomal DNA, with use of the primers listed in Table 1. The amplicons (around 1.5 kb) were then digested with the appropriate enzymes and inserted between the corresponding sites of vector pUC18Ery (27). In the case of loxL, the loss of function was achieved by the in-frame deletion of the active-site-encoding region (261VSNHGGRQ→GT) of loxL (plasmid pGIZ950), in order to minimize polar effects such as mRNA instability on the upstream pyruvate oxidase gene. Since the loxD gene seems to be monocistronic, a disruption strategy was chosen to inactivate this gene. The chloramphenicol acetyltransferase (cat) gene from plasmid pGK12 (15) was amplified by PCR and transcriptionally fused to the strong P32 promoter amplified from plasmid pMG36e (26). The P32-cat fusion was inserted between the two loxD recombination segments, generating the loxD::cat disruption vector pGIZ850.
TABLE 1.
Bacterial strains, plasmids, and oligonucleotides used in this study
| Strain, plasmid, or oligonucleotide | Characteristic(s)a | Source or reference |
|---|---|---|
| Strains | ||
| L. plantarum | ||
| NCIMB8826 | Wild-type strain | NCIMBb |
| TF101 | NCIMB8826 ΔldhL | 7 |
| TF102 | NCIMB8826 ldhD::cat | 8 |
| TF103 | NCIMB8826 ΔldhL ldhD::cat | 8 |
| PG901 | NCIMB8826 ΔloxL | This study |
| PG801 | NCIMB8826 loxD::cat | This study |
| PG701 | NCIMB8826 ΔloxL loxD::cat | This study |
| E. coli | ||
| TG1 | supE hsdΔ5 thi Δ(lac-proAB) F′[traD36 proAB+lacIqlacZΔM15] | 12 |
| Plasmids | ||
| pUC18Ery | Emr Apr; pUC18 derivative with a 1.1-kb insert containing the erm gene | 27 |
| pGIZ950 | Emr Apr; pUC18Ery with a 3.063-kb insert containing a mutated copy of loxL with an in-frame deletion of the active site | This study |
| pGIZ850 | Emr Apr; Cmr; pUC18Ery with a 4.215-kb insert containing the loxD gene disrupted by a cat gene under control of the P32 promoter | This study |
| Oligonucleotides | ||
| LoxLKOA1 | 5′-CCGGAATTCAAGGAGGGTTGACGGCTGAAGG-3′c | This study |
| LoxLKOB12 | 5′-CGGGGTACCATAGATACCTTGAGCACCAGC-3′c | This study |
| LoxLKOA2 | 5′-CGGGGTACCTTAAATGGTGGTCCAGCTTCC-3′c | This study |
| LoxLKOB2 | 5′-GCTCTAGATTTGCCATATTACAACACCTCC-3′c | This study |
| LoxLKOA13 | 5′-AAGCTGCTCAACGTGATAAGC-3′ | This study |
| LoxLKOB14 | 5′-ATCCGGTGTCAGCGATTGTCC-3′ | This study |
| LoxDKOA1 | 5′-CCGGAATTCATGCGTCGCTAACCCAACAAGG-3′c | This study |
| LoxDKOB1 | 5′-CGGGGTACCTTACTTAACTTCGCGACCATCG-3′c | This study |
| LoxDKOA2 | 5′-CGCGGATCCTAAACGCAATTGATGATTGGTTCG-3′c | This study |
| LoxDKOB2 | 5′-ACGCGTCGACATTCTCGACTGTCTTGACATCG-3′c | This study |
| P32A1 | 5′-CGGGGTACCATGCAGTTTAAATTCGGTCCTCG-3′c | This study |
| P32B1 | 5′-CGCGGATCCGAATGCATTCTGCTGAAACGATTGCCATTTC-3′c | This study |
| CATA1 | 5′-AAAACTGCAGATGAAGAAAGCAGACAAGTAAGC-3′c | This study |
| CATB1 | 5′-CGCGGATCCAAGTACAGTCGGCATTATCTC-3′c | This study |
| LoxDKOA13 | 5′-TCTGATAGGAACTCGTATTGATCC-3′ | This study |
| LoxDKOB14 | 5′-ATGCAAACTCTGATAGCCATTAGC-3′ | This study |
Emr, Apr, and Cmr indicate resistance to erythromycin, ampicillin, and chloramphenicol, respectively.
NCIMB, The National Collections of Industrial and Marine Bacteria Ltd., Aberdeen, Scotland.
EcoRI, KpnI, XbaI, BamHI, SalI, NsiI, and PstI restriction sites introduced in the primers are underlined.
Both knockout vectors were then transformed separately in L. plantarum NCIMB8826, and the mutants bearing the desired mutations were obtained as previously described (7, 8). The loxL and loxD mutant strains were designated PG901 and PG801, respectively. A double loxD loxL knockout mutant was constructed by repeating the loxD disruption strategy in the loxL mutant PG901, resulting in strain PG701. The anticipated mutant genotypes were confirmed by PCR in these strains (data not shown).
d- and l-iLDH activities of the lox mutants.
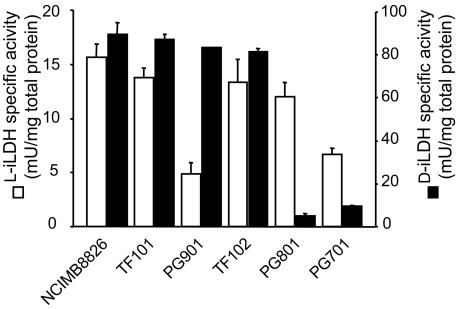
Cells of the wild-type NCIMB8826 and its loxD, loxL, and double loxD loxL derivatives were grown at 28°C with shaking in a modified DeMan-Rogosa-Sharpe (MRS) broth that lacks acetate and citrate (MRS-CA) with 0.2% (wt/vol) glucose, as previously described (18). The cultures were started at an optical density (OD; 600 nm) of 0.05 by inoculating fresh medium with a stationary-phase culture. Cells were harvested during the lactate-to-acetate conversion phase (9 h after inoculation; see Fig. 3), and d- and l-iLDH activities were measured in cell extracts with use of an assay with 2,6-dichlorophenolindophenol (DCPIP) as an electron acceptor in a Tris-maleate buffer, pH 6.0 (Fig. 2) (24). A threefold decrease in l-iLDH activity was observed in the loxL mutant compared to that in the wild-type strain, while d-iLDH activity remained unaffected in this mutant. In contrast, the loxD mutant displayed an 18-fold reduction of d-iLDH activity, while l-iLDH activity was maintained at the wild-type level. The same levels of residual d- and l-iLDH activities were found in the double loxD loxL mutant strain (Fig. 2). These phenotypes demonstrated that loxD and loxL encode d- and l-iLDHs, respectively.
FIG. 3.
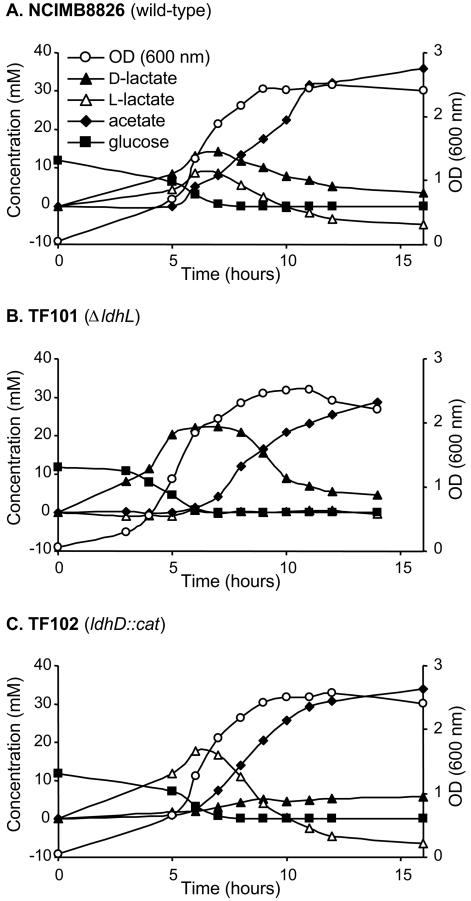
Growth curves of L. plantarum NCIMB8826 (A), TF101 (B), and TF102 (C) in MRS-CA with 0.2% (wt/vol) glucose and shaking at 28°C. The growth was measured by monitoring the OD (600 nm). Concentrations of d-lactate, l-lactate, acetate, and glucose were determined in the supernatant. Concentrations of lactate and acetate are given as the difference between the measured concentration and the initial concentration in the culture medium.
FIG. 2.
l-iLDH (white bars) and d-iLDH (black bars) activities in L. plantarum NCIMB8826 (wild type), TF101 (ΔldhL), TF102 (ldhD::cat), PG901 (ΔloxL), PG801 (loxD::cat), and PG701 (ΔloxL loxD::cat). One unit corresponds to 1 μmol of DCPIP reduced min−1 mg of total protein−1. Total protein concentration was measured using the Bradford method (1). These data represent average values (from at least three individual measurements) with their standard deviations.
We do not have a clear explanation for the residual d- and l-iLDH activities in these mutants. The assay was specific for iLDH activity since no DCPIP reduction was observed in the absence of lactate. In silico analysis of the L. plantarum genome sequence did not reveal any other recognizable candidate gene that could encode iLDH enzymes. Possibly, the residual iLDH activities could be due to other uncharacterized oxidoreductases that can oxidize lactate in the presence of DCPIP.
Conversion of lactate to acetate by cell suspensions of the lox and ldh mutants.
The ability of the lox mutants to produce acetate from d- or l-lactate was compared to that of the wild-type NCIMB8826 strain with use of cell suspensions. The strains were grown until the lactate-to-acetate conversion phase, as described in the previous section. Cells were harvested, washed twice with sodium phosphate buffer (0.1 M, pH 5.0), and resuspended in the same buffer containing 20 mM d- or l-lactate at an approximate OD (600 nm) of 10. Cell suspensions were incubated at 28°C with shaking, and samples were taken at time zero and after 2 h. Total lactate and acetate concentrations were determined by high-performance liquid chromatography analysis as previously described (18). The concentrations of d- and l-lactate were determined using an enzymatic kit (Boehringer, Mannheim, Germany; kit no. 1112821). In order to evaluate a possible role of nLDHs in the utilization of lactate, the ldhL mutant (TF101 [7]) and the ldhD mutant (TF102 [8]) were also included in the study. Notably, the d- and l-iLDH activities appeared unaffected in these two mutants (Fig. 2).
Both d- and l-lactate were effectively converted to acetate by the wild-type strain (Table 2). No difference was observed between the wild type and any of the single lox mutant strains with either d- or l-lactate as a substrate (Table 2). However, this could be due to the residual iLDH activities observed in the loxD and loxL mutants. In these three strains, the conversion of lactate to acetate was nearly complete after 2 h. As for the double loxD loxL mutant (PG701), neither lactate utilization nor acetate production was affected (Table 2). Remarkably, the ldhL mutant appeared to be unable to use l-lactate, while the ldhD mutant could not use d-lactate (Table 2). In the latter strain, lactate racemase activity had been suggested to be responsible for d-lactate production in commercial MRS broth containing 2% (wt/vol) glucose (8). No such activity could be detected after glucose exhaustion under the growth conditions used in the present study (data not shown), explaining why d-lactate could not be used by a combination of lactate racemase and LdhL activities. These results show that the enzymes responsible for the conversion of lactate to pyruvate in stationary phase in L. plantarum are not the LoxD and LoxL enzymes, but rather the nLDH enzymes, LdhD and LdhL. Moreover, it seems that the iLDH activities are not involved in this process, since the double ldhD ldhL mutant (TF103 [8]) was completely deficient in lactate utilization (Table 2).
TABLE 2.
Conversion of lactate to acetate by cell suspensions of wild-type and lox and ldh mutant strains of L. plantarum
| Strain | Product concn (mM) from:
|
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
d-Lactate (20 mM)
|
l-Lactate (20 mM)
|
|||||||||||
|
d-Lactate
|
l-Lactate
|
Acetate
|
d-Lactate
|
l-Lactate
|
Acetate
|
|||||||
| 0 h | 2 h | 0 h | 2 h | 0 h | 2 h | 0 h | 2 h | 0 h | 2 h | 0 h | 2 h | |
| NCIMB8826 (wild type) | 15.5 | 0.8 | 0.9 | 0.3 | 0.6 | 18.9 | 0.8 | 0.4 | 15.0 | 0.5 | NDa | 18.3 |
| TF101 (ΔldhL) | 16.3 | 1.2 | ND | ND | ND | 19.5 | ND | ND | 16.7 | 16.9 | ND | ND |
| PG901 (ΔloxL) | 15.7 | 0.9 | 0.4 | 0.3 | ND | 18.9 | 0.4 | 0.4 | 15.4 | 0.5 | ND | 19.0 |
| TF102 (ldhD::cat) | 16.7 | 17.3 | ND | 0.2 | ND | ND | ND | 0.2 | 15.6 | 0.5 | ND | 19.0 |
| PG801 (loxD::cat) | 16.0 | 0.9 | 0.3 | 0.3 | 0.4 | 18.5 | 0.5 | 0.4 | 15.4 | 0.4 | ND | 19.3 |
| PG701 (ΔloxL loxD::cat) | 17.3 | 1.8 | 0.4 | 0.8 | 0.4 | 21.2 | 0.8 | 0.6 | 16.8 | 0.7 | 0.6 | 22.2 |
| TF103 (ΔldhL ldhD::cat) | 18.0 | 18.2 | ND | ND | ND | ND | ND | ND | 17.2 | 17.9 | ND | ND |
ND, not detected.
In order to investigate a possible role of d- and l-iLDH activities in lactate utilization at higher pH values, cell suspensions were carried out at pHs ranging from 5.0 to 8.0. In the wild-type strain, lactate utilization was more efficient at low pH (5.0), while lactate consumption decreased concomitantly with pH increase until it became barely detectable at pH 8.0. However, increasing the pH did not allow iLDH activities to contribute to lactate utilization in any of the strains, as already observed at pH 5.0 (data not shown).
The role of LoxD and LoxL enzymes in L. plantarum remains unclear. These enzymes are able to oxidize lactate in vitro, but they do not seem to play a role in the stationary-phase utilization of lactate in vivo. Their electron acceptor is unknown. Studies have shown that Lactococcus lactis and other lactic acid bacteria are able to undergo respiration if heme is added to the culture medium (5, 29). In L. lactis, this process has been shown to require the cydA gene encoding cytochrome bd oxidase (5). This gene has been identified in the genome sequence of L. plantarum, together with the other cyd genes required for cytochrome bd oxidase synthesis (14). Although it has been shown that some strains of L. plantarum are able to synthesize cytochromes in the presence of heme (28), no experimental data are available about a possible quinone synthesis pathway, and more generally about the respiration capacity of this species. The role of LoxD and LoxL could be to serve as primary electron donors to the electron transport chain under respiration-permissive conditions. However, repeated attempts to induce aerobic respiration by addition of heme to various culture media were unsuccessful (data not shown).
Lactate utilization by growing cells of the lox and ldh mutants.
Since the same nLDH enzymes are responsible for lactate production and utilization, it was expected that no metabolic phenotype could be observed for the wild type and the lox mutants upon acetate and d- and l-lactate measurements in the culture supernatant of these strains (Fig. 3). In the wild-type strain, d- and l-lactate produced during glucose excess were converted to acetate during the glucose starvation phase. Notably, the final l-lactate concentration appeared to be even lower than the initial l-lactate concentration of the medium. The d-lactate concentration also decreased strongly but remained higher than the initial concentration in the culture medium (Fig. 3A). As expected, similar results were found for the loxD, loxL, and double loxD loxL mutant strains (data not shown), confirming that the LoxD and LoxL enzymes are not involved in lactate utilization under growing conditions.
In agreement with previously published results (7), the ldhL mutant produced the same amount of lactate as the wild-type strain did, but only d-lactate was formed. The d-lactate was subsequently used to form acetate. The production-utilization of d-lactate in this strain followed the same time course as that observed for d- and l-lactate in the wild type and the lox mutants. However, the l-lactate originally present in the culture medium was not consumed (Fig. 3B).
In the case of the ldhD mutant, l-lactate was initially produced, followed by the appearance of small amounts of d-lactate (Fig. 3C). This d-lactate production could be a consequence of l-lactate-mediated induction of the lactate racemase activity, responsible for the conversion of l-lactate to d-lactate, which was detected at a low level during the exponential growth phase (data not shown) (8). During glucose starvation, l-lactate was then converted to acetate by LdhL, while the d-lactate concentration remained stable.
During the sugar consumption phase, no significant difference in total lactate production could be observed between the different strains. Likewise, the final amount of acetate produced during the lactate consumption phase was similar in all strains. Similar to what has been reported previously (18), the calculated carbon balances for both phases were in excess, suggesting that some other compound(s) from the growth medium could be used for growth.
Concluding remarks.
The results presented here clearly establish that the LdhD and LdhL enzymes of L. plantarum are responsible for the conversion of lactate to acetate during glucose starvation in the presence of oxygen. Under the experimental conditions used, the NAD-independent lactate dehydrogenase activities do not seem to be involved in this process. Oxidation of lactate to pyruvate by LdhD and LdhL coincides with NAD+ consumption. This cofactor must be regenerated in order to allow further lactate utilization by the nLDH enzymes, which could potentially be achieved by the NADH peroxidase enzyme (Npr), as proposed by Kandler (13) (Fig. 1B). This reaction would also prevent deleterious effects of the hydrogen peroxide produced by the Pox enzyme(s). Alternatively, NADH oxidases (Nox, Fig. 1B) could recycle NADH in the presence of oxygen (13). Two genes potentially encoding Npr enzymes and six potential nox genes have been identified in the genome of L. plantarum WCFS1 (14).
In conclusion, our results clearly falsify the role of LoxD and LoxL in lactate utilization, which was predicted on the basis of the similarity of these proteins to demonstrated iLDH enzymes. It remains to be established whether such a predicted role of LoxD and LoxL is detectable under conditions where an alternative electron acceptor is available.
Acknowledgments
This research has been carried out with financial support from the Commission of the European Communities, specific RTD project NUTRACELLS (QLRT-1999-00053). P.G. and F.L. hold a doctoral fellowship from FRIA. P.H. is a scientific collaborator at FNRS.
We are grateful to K. Schanck and E. Viaene for their skillful help in high-performance liquid chromatography analyses. We warmly thank J. Delcour for critically reading the manuscript.
This paper does not necessarily reflect the views of the Commission of the European Communities and in no way anticipates the Commission's future policy in this area.
REFERENCES
- 1.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 2.Chapman, S. K., G. A. Reid, C. Bell, D. Short, and S. Daff. 1996. Flavocytochrome b2: an ideal model for studying protein-mediated electron transfer. Biochem. Soc. Trans. 24:73-77. [DOI] [PubMed] [Google Scholar]
- 3.Dennis, D., and N. O. Kaplan. 1960. d- and l-lactic acid dehydrogenases in Lactobacillus plantarum. J. Biol. Chem. 235:810-818. [PubMed] [Google Scholar]
- 4.Duncan, J. D., J. O. Wallis, and M. R. Azari. 1989. Purification and properties of Aerococcus viridans lactate oxidase. Biochem. Biophys. Res. Commun. 164:919-926. [DOI] [PubMed] [Google Scholar]
- 5.Duwat, P., S. Sourice, B. Cesselin, G. Lamberet, K. Vido, P. Gaudu, Y. Le Loir, F. Violet, P. Loubière, and A. Gruss. 2001. Respiration capacity of the fermenting bacterium Lactococcus lactis and its positive effects on growth and survival. J. Bacteriol. 183:4509-4516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dym, O., E. A. Pratt, C. Ho, and D. Eisenberg. 2000. The crystal structure of d-lactate dehydrogenase, a peripheral membrane respiratory enzyme. Proc. Natl. Acad. Sci. USA 97:9413-9418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ferain, T., D. Garmyn, N. Bernard, P. Hols, and J. Delcour. 1994. Lactobacillus plantarum ldhL gene: overexpression and deletion. J. Bacteriol. 176:596-601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ferain, T., J. N. Hobbs, Jr., J. Richardson, N. Bernard, D. Garmyn, P. Hols, N. E. Allen, and J. Delcour. 1996. Knockout of the two ldh genes has a major impact on peptidoglycan precursor synthesis in Lactobacillus plantarum. J. Bacteriol. 178:5431-5437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Futai, M., and H. Kimura. 1977. Inducible membrane-bound l-lactate dehydrogenase from Escherichia coli. Purification and properties. J. Biol. Chem. 252:5820-5827. [PubMed] [Google Scholar]
- 10.Garvie, E. I. 1980. Bacterial lactate dehydrogenases. Microbiol. Rev. 44:106-139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gibello, A., M. D. Collins, L. Dominguez, J. F. Fernandez-Garayzabal, and P. T. Richardson. 1999. Cloning and analysis of the l-lactate utilization genes from Streptococcus iniae. Appl. Environ. Microbiol. 65:4346-4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gibson, T. J. 1984. Ph.D. thesis. Cambridge University, Cambridge, United Kingdom
- 13.Kandler, O. 1983. Carbohydrate metabolism in lactic acid bacteria. Antonie Leeuwenhoek 49:209-224. [DOI] [PubMed] [Google Scholar]
- 14.Kleerebezem, M., J. Boekhorst, R. van Kranenburg, D. Molenaar, O. P. Kuipers, R. Leer, R. Tarchini, S. A. Peters, H. M. Sandbrink, M. W. E. J. Fiers, W. Stiekema, R. M. Klein Lankhorst, P. A. Bron, S. M. Hoffer, M. N. Nierop Groot, R. Kerkhoven, M. de Vries, B. Ursing, W. M. de Vos, and R. J. Siezen. 2003. Complete genome sequence of Lactobacillus plantarum WCFS1. Proc. Natl. Acad. Sci. USA 100:1990-1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kok, J., J. M. van der Vossen, and G. Venema. 1984. Construction of plasmid cloning vectors for lactic streptococci which also replicate in Bacillus subtilis and Escherichia coli. Appl. Environ. Microbiol. 48:726-731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lindqvist, Y., C.-Y. Bränden, F. S. Mathews, and F. Lederer. 1991. Spinach glycolate oxidase and yeast flavocytochrome b2 are structurally homologous and evolutionarily related enzymes with distinctly different function and flavin mononucleotide binding. J. Biol. Chem. 266:3198-3207. [PubMed] [Google Scholar]
- 17.Lodi, T., and I. Ferrero. 1993. Isolation of the DLD1 gene of Saccharomyces cerevisiae encoding the mitochondrial enzyme d-lactate ferricytochrome c oxidoreductase. Mol. Gen. Genet. 238:315-324. [DOI] [PubMed] [Google Scholar]
- 18.Lorquet, F., P. Goffin, L. Muscariello, J.-B. Baudry, V. Ladero, M. Sacco, M. Kleerebezem, and P. Hols. 2004. Characterization and functional analysis of the poxB gene, which encodes pyruvate oxidase in Lactobacillus plantarum. J. Bacteriol. 186:3749-3759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mizushima, S., and K. Kitahara. 1962. Purification and properties of lactic dehydrogenase of Lactobacillus casei. J. Gen. Appl. Microbiol. 8:130-141. [Google Scholar]
- 20.Murphy, M. G., L. O'Connor, D. Walsh, and S. Condon. 1985. Oxygen dependent lactate utilization by Lactobacillus plantarum. Arch. Microbiol. 141:75-79. [DOI] [PubMed] [Google Scholar]
- 21.Seki, M., K.-I. Iida, M. Saito, H. Nakayama, and S.-I. Yoshida. 2004. Hydrogen peroxide production in Streptococcus pyogenes: involvement of lactate oxidase and coupling with aerobic utilization of lactate. J. Bacteriol. 186:2046-2051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Snoswell, A. M. 1959. Flavins of Lactobacillus arabinosus 17.5. A lactic dehydrogenase containing a flavin prosthetic group. Aust. J. Exp. Biol. 37:49-64. [Google Scholar]
- 23.Snoswell, A. M. 1963. Oxidized nicotinamide-adenine dinucleotide-independent lactate dehydrogenases of Lactobacillus arabinosus 17.5. Biochim. Biophys. Acta 77:7-19. [DOI] [PubMed] [Google Scholar]
- 24.Snoswell, A. M. 1966. dl-Lactate dehydrogenases (NAD+-independent) from Lactobacillus arabinosus. Methods Enzymol. 9:321-327. [Google Scholar]
- 25.Strittmatter, C. F. 1959. Flavin-linked oxidative enzymes of Lactobacillus casei. J. Biol. Chem. 234:2794-2800. [PubMed] [Google Scholar]
- 26.van de Guchte, M., J. M. van der Vossen, J. Kok, and G. Venema. 1989. Construction of a lactococcal expression vector: expression of hen egg white lysozyme in Lactococcus lactis subsp. lactis. Appl. Environ. Microbiol. 55:224-228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.van Kranenburg, R., J. D. Marugg, I. I. van Swam, N. J. Willem, and W. M. de Vos. 1997. Molecular characterization of the plasmid-encoded eps gene cluster essential for exopolysaccharide biosynthesis in Lactococcus lactis. Mol. Microbiol. 24:387-397. [DOI] [PubMed] [Google Scholar]
- 28.Whittenbury, R. 1964. Hydrogen peroxide formation and catalase activity in the lactic acid bacteria. J. Gen. Microbiol. 35:13-26. [DOI] [PubMed] [Google Scholar]
- 29.Winstedt, L., L. Frankenberg, L. Hederstedt, and C. von Wachenfeldt. 2000. Enterococcus faecalis V583 contains a cytochrome bd-type respiratory oxidase. J. Bacteriol. 182:3863-3866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xia, Z.-X., and F. S. Mathews. 1990. Molecular structure of flavocytochrome b2 at 2.4 Å resolution. J. Mol. Biol. 212:837-863. [DOI] [PubMed] [Google Scholar]