Abstract
Mycobacterium tuberculosis, an obligate mammalian pathogen, adapts to its host during the course of infection via the regulation of gene expression. Of the regulators of transcription that play a role in this response, several alternative sigma factors of M. tuberculosis have been shown to control gene expression in response to stresses, and some of these are required for virulence or persistence in vivo. For this study, we examined the role of the alternative sigma factor SigD in M. tuberculosis gene expression and virulence. Using microarray analysis, we identified several genes whose expression was altered in a strain with a sigD deletion. A small number of these genes, including sigD itself, the gene encoding the autocrine growth factor RpfC, and a gene of unknown function, Rv1815, appear to be directly regulated by this sigma factor. By identifying the in vivo promoters of these genes, we have determined a consensus promoter sequence that is putatively recognized by SigD. The expression of several genes encoding PE-PGRS proteins, part of a large family of related genes of unknown function, was significantly increased in the sigD mutant. We found that the expression of sigD is stable throughout log phase and stationary phase but that it declines rapidly with oxygen depletion. In a mouse infection model, the sigD mutant strain was attenuated, with differences in survival and the inflammatory response in the lung between mice infected with the mutant and those infected with the wild type.
Mycobacterium tuberculosis is an obligate mammalian pathogen that is believed to infect roughly one-third of the world's population (33). While capable of causing disease in a substantial proportion of those infected, resulting in approximately eight million cases of active tuberculosis in the world each year, this bacillus causes an asymptomatic infection in most individuals. After an initial period of rapid replication, the infection is typically contained by the host immune system, resulting in the apparent eradication of the infection in some individuals but in the persistence of small numbers of bacteria in others, resulting in asymptomatic chronic infections. These latent infections may subsequently become active, often in the setting of decreased host immunity, with increased bacterial replication and extensive tissue damage.
During these several stages of infection, M. tuberculosis encounters a changing host environment, in response to which the bacillus must activate defense and repair mechanisms and reprogram its physiology to ensure survival. The large number of putative transcription regulators identified in the M. tuberculosis genome sequence indicate that much of the regulation required for these adaptations by M. tuberculosis occurs at the level of transcription (6). Among the transcription regulators that have been implicated in these processes are the alternative sigma factors of this organism, 12 of which are encoded in the M. tuberculosis genome. In previous work, our laboratory and others have implicated several alternative sigma factors in the mycobacterial response to a variety of stresses, most notably oxidative stress (13, 17-19, 24, 34). A role for alternative sigma factor-regulated gene expression in stationary-phase adaptation and in vivo replication in late-stage infections in mice has also been demonstrated (5, 11).
In addition to oxidative and nitrosative stresses, in vitro models of M. tuberculosis infection and latency have focused on two environmental conditions that are thought to be encountered by M. tuberculosis during infection, i.e, nutrient limitation (starvation) and hypoxia. A shift in carbon source utilization requiring the enzyme isocitrate lyase has been associated with the ability of M. tuberculosis to persist in vivo (20). Similarly, an intact relA gene, encoding ppGpp synthase, which is required for the induction of the stringent response, has been shown to be essential for the in vivo persistence of M. tuberculosis in mice (8). The expression of sigD appears to be linked to the stringent response, with an increased expression in response to starvation that is at least partly Rel dependent (3, 8). Microarray data have demonstrated substantial, though incomplete, similarities between the M. tuberculosis transcription profiles produced in response to hypoxia, sublethal concentrations of nitric oxide, and macrophage infection (26, 27, 31). In contrast, these transcription responses show little overlap with starvation- or stringent response-induced alterations in transcription. These data suggest that both hypoxia and starvation are important for M. tuberculosis survival in vivo but that they provoke distinct physiologic adaptations that may be relevant at different stages of the dynamic process of infection by M. tuberculosis.
In this work, we present an initial characterization of the M. tuberculosis alternative sigma factor SigD. After constructing a sigD deletion (ΔsigD) strain of M. tuberculosis, we examined the role of this sigma factor in growth and viability in vitro and its effects on global and specific gene transcription. Using in vivo promoter analysis, we have identified a consensus SigD recognition sequence. Using a mouse model of infection, we assessed the role of this sigma factor in M. tuberculosis virulence. Based on the identity of genes that it regulates, its effects on global transcription, and the response of its gene to starvation and hypoxia, our data indicate that this sigma factor plays a role in optimal growth both under nutrient replete conditions and, paradoxically, in response to starvation. These data suggest that SigD, while nonessential for viability in vitro, may play a role at several stages during infection to optimize bacterial replication and survival.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
M. tuberculosis H37Rv was used as the parental strain for generating an isogenic ΔsigD strain and as the wild type (wt) for all experiments. Escherichia coli DH5α (Life Technologies) and XL1 Blue (Stratagene) were used as host strains for cloning experiments. Mycobacterium smegmatis strain mc2-155 or its derivatives were used for all experiments involving this mycobacterial species (29). M. tuberculosis and M. smegmatis were grown in flasks with shaking or as standing cultures in Middlebrook 7H9 broth supplemented with albumin-dextrose complex (7H9-ADC) plus 0.05% Tween 80 and 0.2% glycerol. For hypoxia studies, the uniform gradual oxygen depletion model of Wayne and Hayes was used, as described below (32). Colony counts from mouse organs were performed by plating bacteria on Middlebrook 7H11 selective agar medium (Hardy Diagnostics). E. coli was grown on L agar or in L broth. Ampicillin (100 μg/ml), apramycin (30 μg/ml), kanamycin (50 μg/ml), and zeocin (50 μg/ml) were added to culture media as indicated.
Construction of M. tuberculosis sigD mutant strain and complementation of the mutation.
The M. tuberculosis sigD mutant was constructed by targeted mutagenesis with the temperature-sensitive sacB counterselection system. Briefly, the sigD gene, from annotated codon 22 through codon 201, was removed by overlap PCR and replaced with a kanamycin resistance gene. The resulting disrupted gene and flanking DNA were sequenced and blunt cloned into the XbaI site of pRH1351, a derivative of pPR23 harboring the sacB and xylE genes (7, 22, 24). The resulting vector, pRH1450, was electroporated into M. tuberculosis H37Rv. The recombinants were selected at 30°C on kanamycin plates. Individual colonies were picked, grown at the permissive temperature in broth culture, and then plated on plates containing 10% sucrose at 39°C. The resulting colonies were screened for XylE activity as described previously (7). Genomic DNAs from xylE-negative colonies were analyzed by PCR, and the presence of the sigD deletion and the absence of the wt allele were confirmed by Southern analysis.
To complement the sigD mutation, we placed an intact copy of sigD in an apramycin-resistant derivative of the integrating vector pMH94 (15, 24). Because of previous results indicating poor expression from this type of construct (24), the constitutive hsp70 promoter was placed 5′ of the sigD coding sequence. This construct was electroporated into the sigD mutant, followed by plating and selection for apramycin-resistant colonies. The presence of the intact sigD allele was confirmed by PCR.
RNA isolation and primer extension.
Total RNAs were isolated from wt M. tuberculosis and the ΔsigD mutant strains grown to mid-log phase (optical density at 600 nm [OD600], 0.4). The bacterial cells from 50 ml of culture were centrifuged, and the pellet was resuspended in 1 ml of Trizol (Invitrogen) and transferred to a 2 ml-tube containing ceramic and silica beads. The tubes were agitated in an FP120 instrument (QBiogene) or in a bead beater (Biospec), with cooling of the tubes on ice between each agitation. The lysate was removed and place into a fresh tube, and RNA was prepared according to the supplier's recommendations (Invitrogen). The final RNA pellet was dissolved in water and treated with DNase I. The DNase-treated RNA was further purified by use of an RNeasy clean-up kit according to the manufacturer's protocol (Qiagen).
For primer extension analysis, 0.5 pmol of a γ-32P-labeled primer (sigD [5′-CACGGCCTCCGCAACCACAGCGTCGAGA-3′], rpfC [5′-CGTGGGCAACGGCGGTGGAGAGCGACAT-3′], or Rv1815 [5′-GGATTTCCATGCCGGGGAACACCAAGAC-3′]) was mixed with 5 μg of RNA in a 6-μl volume, and reverse transcription (RT) was performed as previously described (24). The reaction products were separated by polyacrylamide gel electrophoresis in a sequencing gel. Sequencing reactions, performed with the same primer used for primer extension, were run in adjacent lanes to determine the sizes of the transcripts generated. Transcripts were quantified with a phosphorimager and ImageQuant software (Molecular Dynamics).
Microarray experiments and analysis.
M. tuberculosis PCR arrays and oligoarrays were used to identify SigD-regulated genes. PCR arrays, provided by Eric Rubin, were produced with gene-specific PCR products generated by the use of amino-modified primers coupled to aldehyde-coated 3D-Link slides (Motorola) (25). The M. tuberculosis oligoarrays were obtained from The Institute for Genomic Research (TIGR) through the National Institute of Allergy and Infectious Diseases-sponsored Pathogen Functional Genomics Resource Center. These microarrays consisted of 4,750 70-mer oligonucleotides representing 4,127 open reading frames from M. tuberculosis strain H37Rv and 623 unique open reading frames from strain CDC1551 that are not present in the H37Rv strain's annotated gene complement. RNAs were prepared as described above, and cDNAs were synthesized by RT with Superscript II (Invitrogen) and amino-allyl dUTP incorporation, followed by coupling to Cy-3 or Cy-5 fluorescent dyes (Amersham). TIGR protocols were followed for probe synthesis, hybridization, and washing procedures (http://www.tigr.org/tdb/microarray/protocolsTIGR.shtml). The hybridized slides were scanned with an Axon 4000B scanner, and spot intensities were defined and quantified with Genepix software (Axon, Union City, Calif.).
Four hybridization values from oligoarray experiments (two replicates from each of two independent RNA preparations) were used for statistical analysis. The raw data median values were normalized as described by Churchill and colleagues (14). The data were reformatted to use the signals from duplicate spots. Statistical analysis was performed with the National Institute of Aging array analysis tool (http://lgsun.grc.nia.nih.gov/ANOVA/). We used the most conservative error model, the maximum of averaged and actual error variance, in order to reduce the number of false-positive results. Up-regulated and down-regulated genes were selected based on false discovery rates (FDRs) of <0.05 (FDR = P × N/rank, where P is the P value, N is the total number of genes, and rank is the rank of the gene, as ordered by increasing P values).
Real-time PCR and mRNA quantitation.
RNAs extracted from M. tuberculosis H37Rv grown under several conditions were prepared as described above, except that an additional DNase treatment was added. Primers were designed with Primer Express software (Applied Biosystems), and RT-PCR was performed by use of an iScript cDNA synthesis kit, iTaq SYBR green reagents (Bio-Rad), and an ABI7000 sequence detection instrument. Duplicate reactions containing 100 ng of template were performed for sigD and other genes of interest. The expression of sigA was quantified for each sample to allow for comparisons with the relatively stable expression of this gene (17). An RNA sample that had not been reverse transcribed was included in all experiments to exclude significant DNA contamination. Standard curves were obtained by performing PCRs with SYBR green detection on serial dilutions of spectrophotometrically quantified genomic DNA or plasmid DNA. hspX was used as a positive control for oxygen depletion experiments.
Oxygen depletion experiments.
For RT-PCR experiments and viability studies, the stirred model of gradual oxygen depletion was used (32). Early-log-phase cultures of bacteria were inoculated into 25-ml sealed bottles with 17 ml of medium and a stir bar (headspace ratio = 0.5). The bottles were stirred by use of a magnetic stir plate at low speed to keep the cells in suspension and to keep the dissolved oxygen concentration uniform in the medium. One set of cultures was harvested at each time point for RNA extraction, and a second set of cultures was used for plating serial dilutions.
Computer database searching.
Searches of the M. tuberculosis H37Rv genome sequence for consensus promoter elements were performed by utilizing the “search pattern” program available at the TubercuList web site of the Pasteur Institute (http://genolist.pasteur.fr/TubercuList) and the motif-finding function of the Bioprospector program (http://robotics.stanford.edu/∼xsliu/BioProspector/).
Oxidative stress and isoniazid exposure.
Susceptibilities to diamide, plumbagin, cumene hydroperoxide, and isoniazid (INH) were determined by a disk diffusion assay as previously described (24). Plating efficiency experiments were performed with Middlebrook 7H10 agar supplemented with ADC enrichment and 0.05 μg of INH/ml. To determine the effect of INH on gene expression, we grew broth cultures to mid-exponential phase (OD600 = 0.4 to 0.5), split the cultures into two aliquots, added INH to one aliquot at a concentration of 1 μg/ml, and incubated the cultures for 4 h prior to RNA isolation as described above.
Mouse infection and virulence assay.
Six-week-old BALB/c mice were purchased from Jackson Laboratory (Bar Harbor, Maine). All mice were housed in microisolator cages in a biosafety level 3 animal facility at the Dana Farber Cancer Institute (Boston, Mass.) under specific-pathogen-free conditions. A vial of frozen bacteria was thawed to room temperature and then dispersed by sonication in a cup horn sonicator twice for 10 s. The bacterial suspension was then triturated through a 27-gauge insulin syringe three times to disrupt clumps. The bacteria were then diluted to 107 CFU/ml in 0.9% saline with 0.02% Tween 80. Twelve mice from each group were injected with 106 CFU in the lateral tail vein. The weights and morbidity of the mice were monitored for the survival experiment. Mice that became moribund were euthanized, with the time of survival counted until the day of euthanasia.
For bacterial replication and histopathology experiments, six mice from each group were sacrificed after 9, 24, and 59 days. The right lungs were removed and homogenized in saline-Tween by use of a bead beater. Serial dilutions of the homogenates were plated for CFU, and colonies were counted after 3 to 4 weeks at 37°C. For histological studies, the left lungs were removed and fixed for at least 48 h in 10% buffered formalin. The tissues were processed and embedded in paraffin. Tissues for slides were cut into 5-μm-thick sections and then stained with hematoxylin and eosin or acid fast stained for bacteria by standard techniques. All mouse experiments were performed according to protocols approved by the Animal Care and Use Committees of Children's Hospital Boston and the Dana Farber Cancer Institute.
For all data presented, representative results of one experiment are shown, with all experiments repeated at least once to confirm the results.
RESULTS
Construction of ΔsigD mutant and complemented strains of M. tuberculosis and identification of SigD-regulated genes.
A deletion of the sigD coding sequence from annotated codon 22 through codon 201 was created, and a kanamycin resistance gene was inserted. This deletion allele was introduced into the chromosome of H37Rv by homologous recombination, with counterselection to achieve replacement of the wt allele, as described in Materials and Methods. Gene replacement was confirmed by Southern blot analysis of the genomic DNA. To create a strain in which the ΔsigD mutation was complemented, we introduced a single copy of the intact sigD coding region into the chromosome at the phage L5 attB site (15), as described in Materials and Methods. Expression of sigD in H37Rv and the complemented strain (ΔsigD attB::sigD) was nearly identical, as determined by quantitative RT-PCR using RNAs isolated from broth cultures at two different time points (OD600 = 0.4 and 0.6) (not shown).
To identify genes regulated by SigD in M. tuberculosis, we performed a whole-genome transcription analysis by using microarrays to compare the transcription profiles of the ΔsigD mutant and the parental strain H37Rv. A comparison of transcription in these two strains, using a conservative error model with an FDR of <0.05, revealed a total of 471 genes that differed between the wt and the sigD mutant. Of these, 265 showed significantly decreased expression in the sigD mutant and 206 showed increased expression in the mutant. This number of affected genes represents about 10% of the total M. tuberculosis genome. Table 1 shows the 47 genes whose expression was ≥1.7 times higher in the wt than in the sigD mutant. Among these, hypothetical genes of unknown function were the most common. sigD and its adjacent 3′ gene (Rv3413c), Rv1815, Rv1816, rpfC and its adjacent 3′ gene (Rv1883c), and iniB and its 3′ genes showed the highest expression ratios. Also notable among the genes showing high expression ratios were several major chaperone/heat shock genes. Among the genes whose expression was significantly increased in the ΔsigD strain relative to H37Rv, PE-PGRS (proline glutamate [PE] motif and polymorphic GC-rich sequence) family genes, hypothetical genes of unknown function, and genes involved in cell wall processes and lipid metabolism were the most common (Table 2). See Table S1 in the supplemental material for a list of the 471 significantly regulated genes.
TABLE 1.
Functional categories of genes with 1.7-fold more expression in M. tuberculosis H37Rv than in the sigD mutant strain
| Gene family and name | Fold change | FDR | Product, its function, and additional comments |
|---|---|---|---|
| Regulatory | |||
| Rv3414c | 4.4 | <0.001 | SigD, ECF sigma factor |
| Rv3413c | 2.8 | <0.001 | RsdA, probable anti-sigma factor |
| MT1932 | 8.0 | <0.001 | RpfC, resuscitation promoting factor |
| MT1931.1 | 2.6 | <0.001 | Downstream gene of rpfC, probable part of rpfC operon |
| Rv1883c | 2.7 | <0.001 | Same as MT1931.1, except for a five-amino-acid gap |
| Rv1815 | 3.0 | <0.001 | Probable transcriptional regulation associated with Rv1816 |
| Rv1816 | 1.7 | 0.008 | Transcriptional regulatory protein, similar to tetR family |
| Heat shock response | |||
| Rv0350 | 2.6 | <0.001 | DnaK, heat shock protein Hsp70 |
| Rv0351 | 2.5 | 0.001 | GrpE, Hsp70 cofactor, plays a role in adaptation |
| MT0366 | 2.3 | 0.008 | Same as Rv0351 |
| Rv0440 | 4.7 | <0.001 | GroEL2, chaperonin 2, prevents misfolding and promotes refolding |
| Rv3417c | 2.1 | <0.001 | GroEL1, chaperonin 1, prevents misfolding and promotes refolding |
| Rv3418c | 3.7 | <0.001 | GroES, chaperonin |
| Lipid metabolism | |||
| MT0137 | 2.0 | 0.013 | FbpC, fibronectin binding protein, required for biogenesis of cord factor, maintains cell wall integrity |
| MT0870 | 1.7 | <0.001 | LpqS, probable lipoprotein |
| MT1041 | 1.9 | 0.001 | Putative polyketide synthase |
| Rv1182 | 1.7 | <0.001 | PapA3, probable polyketide synthase-associated protein |
| Rv1270 | 2.0 | 0.005 | LprA, lipoprotein |
| Rv1750 | 1.9 | <0.001 | FadD1, fatty acid-CoA synthase |
| Rv2187 | 1.9 | <0.001 | FadD15, fatty acid CoA synthase, involved in fatty acid degradation |
| Cell wall process | |||
| Rv0341 | 7.2 | 0.001 | IniB, INH-inducible protein |
| Rv0342 | 2.4 | <0.001 | IniA, INH-inducible protein |
| Rv1860 | 1.8 | 0.001 | ModD, fibronectin attachment protein |
| Rv2301 | 1.8 | 0.004 | Probable cutinase, hydrolysis cutin |
| Rv3890c | 1.8 | 0.002 | EsxC, ESAT-6-like protein 11 |
| Rv3891c | 2.4 | 0.004 | EsxD, ESAT-6-like protein |
| Intermediary metabolism and respiration | |||
| MT0455 | 1.8 | <0.001 | Probable dehydrogenase |
| Rv2455c | 1.7 | 0.048 | Possible oxidoreductase |
| Rv3914 | 2.2 | 0.035 | TrxC, thioredoxin, participates in various redox reactions |
| Adaptation | |||
| Rv3846 | 2.5 | 0.002 | SodA, superoxide dismutase A, involved in detoxification and adaptation |
| Unknown function | |||
| MT0196 | 1.9 | <0.001 | Hypothetical protein |
| MT1020 | 1.8 | <0.001 | Hypothetical protein |
| MT1628 | 1.9 | 0.025 | Hypothetical protein |
| MT2422 | 1.8 | 0.004 | PPE family proteins |
| MT2890 | 1.9 | <0.001 | CRISPR-associated protein, TM1811 family |
| MT3258 | 2.2 | <0.001 | Hypothetical protein |
| Rv0257c | 2.0 | 0.041 | Hypothetical protein |
| Rv0276 | 2.0 | <0.001 | Hypothetical protein |
| Rv0309 | 1.7 | 0.004 | Conserved exported protein |
| Rv1687c | 1.9 | <0.001 | Hypothetical protein |
| Rv1779c | 1.7 | 0.050 | Hypothetical integral membrane protein |
| Rv2159c | 1.7 | 0.007 | Hypothetical protein |
| Rv2348c | 1.7 | 0.008 | Hypothetical protein |
| Rv2633c | 2.1 | <0.001 | Hypothetical protein |
| Rv2699c | 1.9 | 0.001 | Hypothetical protein |
| Rv2822c | 1.9 | <0.001 | Hypothetical protein |
| Rv3169 | 1.7 | <0.001 | Hypothetical protein |
| Rv3354 | 2.3 | <0.001 | Hypothetical protein |
| Rv3614c | 2.0 | 0.001 | Hypothetical protein |
| Rv3615c | 2.2 | <0.001 | Hypothetical protein |
| Rv3675 | 1.8 | 0.002 | Membrane protein |
TABLE 2.
Functional categories of genes with 1.7-fold more expression in the M. tuberculosis sigD mutant strain than in strain H37Rv
| Gene family and name | Fold change | FDR | Product, its function, and additional comments |
|---|---|---|---|
| Adaptation | |||
| Rv3811 | 1.7 | 0.001 | Cold shock protein |
| Rv3648 | 1.8 | 0.004 | Cold shock protein A |
| Rv3268 | 1.9 | <0.001 | Seems to be coregulated with CSPA-related protein Rv3267 (ratio = 1.5, FDR = 0.002) |
| Rv3269 | 1.9 | <0.001 | Heat shock protein, chaperonin, seems to be coregulated with Rv3267 and RV3268 |
| Lipid metabolism | |||
| Rv1094 | 2.4 | <0.001 | DesA2, Acyl-[ACP]-desaturase |
| Rv3229 | 4.4 | <0.001 | DesA3, linolenoil CoA desaturase |
| Cell wall process | |||
| MT1416 | 2.0 | 0.001 | Desaturase-related protein |
| Rv1614 | 2.2 | <0.001 | Lgt, prolipoprotein diacylglyceryl transferase |
| Rv1312 | 1.7 | 0.001 | Secreted protein |
| Rv3920c | 2.3 | <0.001 | Jag-like protein, involved in cell division |
| Regulatory | |||
| Rv0823c | 2.5 | <0.001 | Transcriptional regulatory protein |
| Rv1297 | 2.2 | <0.001 | Rho, transcription termination factor, Rho homologue |
| Rv0704 | 1.8 | 0.002 | RplB, 50S ribosomal protein, peptidyltransferase activity |
| Respiration | |||
| Rv3230c | 1.9 | <0.001 | Oxidoreductase |
| Rv1644 | 2.1 | <0.001 | TsnR, 23S rRNA methyltransferase |
| Unknown | |||
| Rv0516c | 1.8 | 0.036 | Hypothetical protein |
| Rv2393 | 1.8 | 0.001 | Hypothetical protein |
| Rv3212 | 2.1 | <0.001 | Hypothetical protein |
| Rv3376 | 1.9 | <0.001 | Hypothetical protein |
| Rv3647c | 1.7 | 0.001 | Hypothetical protein |
| PE-PGRS family | |||
| Rv0124 | 2.4 | 0.004 | PE-PGRS 2 |
| MT0132 | 2.3 | 0.02 | PE-PGRS 2 |
| Rv0834c | 2.2 | 0.001 | PE-PGRS 14 |
| MT1123 | 2.1 | 0.001 | PE-PGRS 22 |
| Rv1396c | 2.1 | 0.008 | PE-PGRS 25 |
| Rv1450c | 1.9 | <0.001 | PE-PGRS 27 |
| MT1499 | 1.8 | 0.002 | PE-PGRS 28 |
| Rv2490c | 2.0 | 0.001 | PE-PGRS 43 |
| Rv3344c | 2.1 | <0.001 | PE-PGRS 49 |
| Rv3345c | 2.2 | <0.001 | PE-PGRS 50 |
| MT3612 | 1.9 | <0.001 | PE-PGRS 53 |
| MT3615.3 | 2.7 | 0.021 | PE-PGRS 57 |
| MT3615.1 | 2.3 | 0.023 | PE-PGRS |
| MT3448 | 2.1 | 0.001 | PE-PGRS |
| MT3582 | 1.7 | <0.001 | PPE 19 |
To verify that these results were effects of the mutation in sigD, we performed a microarray experiment comparing global transcription in the complemented strain with expression in the sigD mutant. Although some variation was observed in the rank order of the expression ratios, all of the genes that were most highly expressed in H37Rv relative to the ΔsigD strain were also highly expressed in the complemented strain relative to the ΔsigD strain (not shown). Rv3413c was among the genes that were highly expressed in the complemented strain, confirming the absence of a polar effect of the sigD mutation on this gene, which is immediately 3′ of sigD and expressed as part of an operon with sigD.
Identification of SigD-regulated promoters.
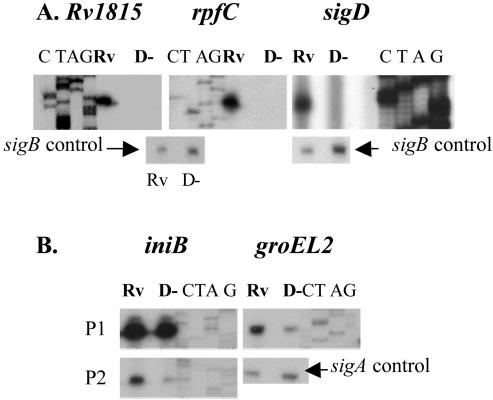
To identify promoters that were likely to be directly regulated by SigD, we focused on those with the highest expression ratios (H37Rv/ΔsigD ratios). Primer extension experiments were performed with more than 20 candidate SigD-regulated genes. Three classes of promoters were identified. One group, represented by sigD, rpfC, and Rv1815, showed strong transcript signals in experiments using RNAs from H37Rv but no signal, even with prolonged exposure, in experiments using RNAs from the sigD mutant (Fig. 1A). In vivo transcription from these promoters is thus completely dependent on SigD. For each of these genes, no other strong promoter was identified within approximately 300 bases 5′ of the annotated initiation codon. For both rpfC and sigD, the sigD-dependent promoter is within the predicted coding sequence of these genes, indicating that the translation of these transcripts is initiated at a site 3′ of the annotated start codon (6). There are potential start codons 51 bases downstream of the experimentally determined 5′ end of the sigD transcript and 36 bases downstream of the 5′ end of the rpfC transcript. As for M. tuberculosis, an analysis of the M. smegmatis sigD locus identified a promoter that was active in the wt but absent from a sigD mutant strain (not shown).
FIG. 1.
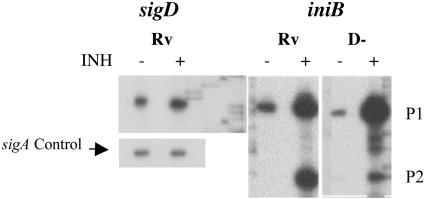
Primer extension analysis was performed with genes whose expression was increased in H37Rv relative to the ΔsigD strain to identify in vivo transcription start sites. RNAs were isolated from exponential-phase cultures of H37Rv (Rv) or the ΔsigD strain (D−) of M. tuberculosis. Sequencing ladders shown adjacent to each transcript were generated with the same primers as those used for RT. The results for promoters that were completely (A) or partially (B) SigD dependent are shown. Control experiments demonstrating sigA or sigB transcripts were performed with each RNA tested.
A second class of SigD-regulated promoters was represented by iniB and groEL2 (Fig. 1B). In these two cases, transcripts were identified that were present in experiments using wt RNAs and showed substantially decreased signals in experiments using RNAs from the ΔsigD strain. Thus, for these genes, expression from these promoters is partly regulated by SigD. In the case of iniB, a second transcript was present from a promoter whose activity did not differ between the wt and the sigD mutant, while for groEL2 no other transcript was seen. It is not known whether this class of SigD-regulated promoters is directly recognized by SigD and also by a second sigma factor or whether the regulation by SigD is indirect. For the third set of genes examined by primer extension, we did not identify any transcripts that were consistently different between H37Rv and the sigD mutant (not shown).
By examining the regions 5′ of the beginning of the SigD-regulated transcripts, we sought to identify consensus promoter elements that might be recognized by SigD. A clear consensus −35 element was identified that was present in all of the promoters whose expression was completely SigD dependent (Fig. 2). A second region, separated by 16 bases from the 3′ end of the consensus, did not show a specific consensus sequence but had a higher than expected A+T content. Alignment of the partially SigD-regulated promoter of iniB showed moderate similarity to the consensus −35 sequence of the other SigD-dependent promoters. We did not identify any clear similarity to this consensus in the groEL2 promoter region. This strong consensus identified in the promoters that were completely SigD dependent suggests that these promoters are directly recognized by the RNA polymerase holoenzyme incorporating SigD (EσD).
FIG. 2.
Alignment of promoter regions from the completely SigD-dependent in vivo promoters shown in Fig. 1 plus the experimentally defined SigD-dependent promoter of M. smegmatis (Ms-sigD). A strong consensus sequence in the −35 region is evident (bold letters), and an AT-rich −10 element is also present (underlined). Below the consensus line, the iniB P2 promoter, which is partially SigD dependent, is shown.
To attempt to identify additional promoters that might be directly regulated by SigD, we performed pattern searching with the −35 consensus sequence by using the search algorithm on the TubercuList web server (http://genolist.pasteur.fr/TubercuList). Although several partial matches were identified, none of the genes 3′ of these sequences were present among the genes whose expression was significantly decreased in the sigD mutant relative to H37Rv. Similarly, motif searching with the Bioprospector program (http://bioprospector.stanford.edu) of the 200 bases upstream and downstream of the start codons of the other genes with significantly decreased expression in the sigD mutant did not reveal additional sequences that matched the −35 consensus. Taken together, these data suggest that SigD directly regulates a relatively small number of genes and that much of the effect on transcription seen in the microarray experiments is likely to be indirect.
Growth and viability of ΔsigD strain and expression of sigD during growth and oxygen depletion.
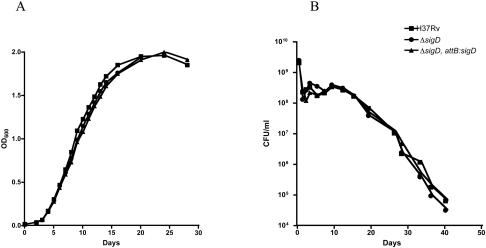
Our observation that rpfC is regulated by SigD led us to examine the growth and viability of M. tuberculosis in broth culture and in response to oxygen depletion. On several occasions, when we inoculated cultures from frozen stocks or from colonies, the sigD mutant strains of M. smegmatis or M. tuberculosis appeared to take longer to become visibly turbid, suggesting an effect of SigD on viability or the lag phase. We were unable, however, to document a consistent difference in lag phase between the mutant and the wt in growth experiments in liquid medium, and no difference in exponential growth was observed (Fig. 3A). A comparison of the viabilities of long-term frozen cultures, measured by plating on solid medium, also showed no differences between H37Rv and the sigD mutant strain. During the course of gradual uniform oxygen depletion in the Wayne model of sealed stirred cultures, the sigD mutant strain showed a decrease in viability that was similar to those of H3Rv and the complemented strain (Fig. 3B). A strain that overexpressed sigD from a replicating plasmid in H37Rv also showed exponential growth and hypoxic survival that were not significantly different from those of the parental H37Rv and sigD mutant strains (not shown).
FIG. 3.
(A) Growth kinetics of M. tuberculosis H37Rv, the ΔsigD mutant, and the ΔsigD complemented strain, as determined in Middlebrook 7H9 broth supplemented with ADC and 0.05% Tween 80 incubated at 37°C. OD600 values were measured at serial time points for 4 weeks. (B) Survival of M. tuberculosis H37Rv, the ΔsigD mutant, and the ΔsigD complemented strain in an oxygen depletion system. Seventeen-milliliter samples of early-log-phase cultures (OD600 = 0.3) were placed in 25-ml sealed bottles to obtain a headspace ratio of 0.5. Aliquots were removed for CFU determinations and were plated in duplicate on Middlebrook 7H11 medium at serial time points for 40 days.
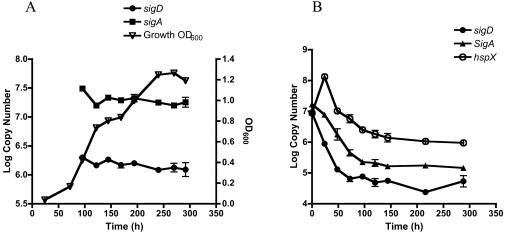
Two research groups have previously shown that sigD expression increases in response to starvation (3, 8). To further examine the possible role of SigD on growth and viability, we measured the expression of sigD during exponential growth and stationary phase and in response to oxygen depletion. As shown in Fig. 4A, sigD was expressed at a relatively high level throughout exponential growth and during stationary phase. This pattern closely followed the pattern of expression of sigA, the gene encoding the primary sigma factor of M. tuberculosis. These results are consistent with dot blot hybridization experiments showing the stable expression of sigD in exponential- and stationary-phase cultures (12). In cultures that were gradually depleted of oxygen, sigD expression decreased rapidly and to a greater extent than did sigA expression (Fig. 4B). The onset of this shift coincided with an increase in the expression of hspX, indicating that this change in expression occurred at a time when sufficient oxygen depletion had occurred to initiate the hypoxic response in these cultures.
FIG. 4.
Expression of sigD mRNA in M. tuberculosis H37Rv, as measured by quantitative real-time RT-PCR during exponential growth and stationary phase (A) and during gradual oxygen depletion (B). Aliquots were removed at each time point, RNAs were isolated, and RT-PCRs were performed with 100 ng of RNA as described in Materials and Methods. Quantification of sigA mRNA was performed with an aliquot of the same RNA preparation at each time point. For the oxygen depletion experiments, hspX mRNA was quantified as a control for hypoxia. For each gene, the relative copy number was determined by use of a standard curve of H37Rv genomic DNA. Errors bars indicate standard errors of the means.
Expression of resuscitation promoting factor genes in H37Rv and the ΔsigD strain.
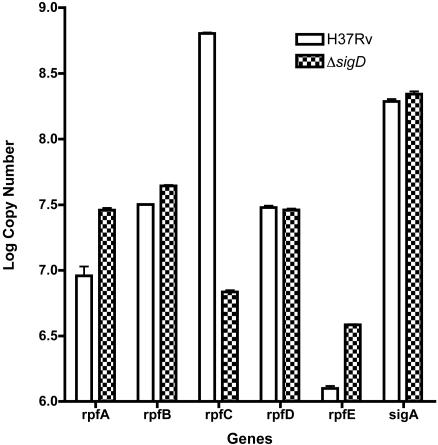
We identified rpfC as one of the genes that is likely to be directly regulated by SigD. M. tuberculosis has five rpf genes, and a recent study showed that the inactivation of rpfC, or any of the other individual rpf genes, did not have a measurable effect on growth, suggesting a possible redundancy of function of these genes (21, 30). We therefore measured the relative expression of all of the M. tuberculosis rpf genes in the wt and the sigD mutant strain, using quantitative real-time PCR, to see if there was a compensatory effect on the expression of any of the other rpf genes. As shown in Fig. 5, rpfC was by far the most abundant resuscitation promoting factor gene in M. tuberculosis; its mRNA level was approximately 50- to 500-fold higher than those of the other rpf genes. In this assay, the wt expressed about 100 times more rpfC than the ΔsigD mutant, consistent with the results from microarray experiments. Although no significant difference in the expression of rpfB or rpfD was seen between the ΔsigD mutant and H37Rv, small (four- to fivefold) but reproducible increases in the levels of rpfA and rpfE mRNAs were observed in the ΔsigD mutant.
FIG. 5.
Expression of each of the five M. tuberculosis rpf gene mRNAs during exponential growth, as measured in H37Rv and the ΔsigD strain by quantitative real-time RT-PCR. RNAs were isolated, and RT-PCRs were performed with 100 ng of RNA as described in Materials and Methods. Quantification of sigA mRNA was performed on aliquots of the same RNA preparations. Errors bars indicate standard errors of the means.
Response to oxidative stresses and INH.
Several alternative sigma factors of M. tuberculosis have been shown to play a role in the mycobacterial stress response, through alterations in survival following stress and through changes in gene expression in response to stress (9, 10, 13, 17-19, 24, 34). Although our microarray data did not suggest an obvious link to stress, we examined the role of sigD under a limited number of stresses by using a previously described disk diffusion assay. No consistent differences were observed between H37Rv and the ΔsigD mutant in experiments testing susceptibilities to the superoxide-generating agent plumbagin, the organic peroxide cumene hydroperoxide, or the disulfide stress agent diamide. Based on our observation that SigD regulates the iniBAC operon, we examined the susceptibility of these strains to INH by using disk diffusion experiments and by examining their plating efficiencies on medium containing 0.05 μg of INH/ml. Although the disk diffusion assays did not show a consistent difference, a small (two- to fourfold) but reproducible decrease in the plating efficiency of the sigD mutant was observed in the presence of INH (not shown).
To examine the effect of INH on iniB expression and the role of SigD in the transcription of this gene, we performed primer extensions with RNAs from H37Rv and the ΔsigD mutant, with and without INH induction (Fig. 6). By phosphorimager analysis of the primer extension products (confirmed by quantitative RT-PCR), we observed a twofold induction of sigD expression following exposure to 1 μg of INH/ml. For iniB, in agreement with previous results (1), we observed a significant induction of transcription from the previously identified iniB promoter (P1 in our experiments) in response to INH (1). In addition, however, we observed substantial basal transcription from this promoter. A strong INH-induced induction of this transcript was observed in both H37Rv and the ΔsigD strain. Transcription from the partially SigD-dependent promoter that we identified (P2) was also strongly induced in response to INH, and this induction was blunted in the ΔsigD mutant. These data indicate that INH induction of the iniB operon occurs from two promoters, one of which is partially SigD dependent, suggesting that distinct mechanisms of induction may affect the activities of these two promoters.
FIG. 6.
Primer extension analysis of sigD and iniB promoters in response to INH exposure. Cultures of H37Rv (Rv) and ΔsigD (D−) were grown to mid-log phase and split into two aliquots, and INH was added to one aliquot to a final concentration of 1 μg/ml. The cells were harvested after 4 hours for RNA isolation, and primer extension experiments were performed with primers specific for sigD, iniB, or sigA (control).
Virulence analysis of M. tuberculosis sigD mutant.
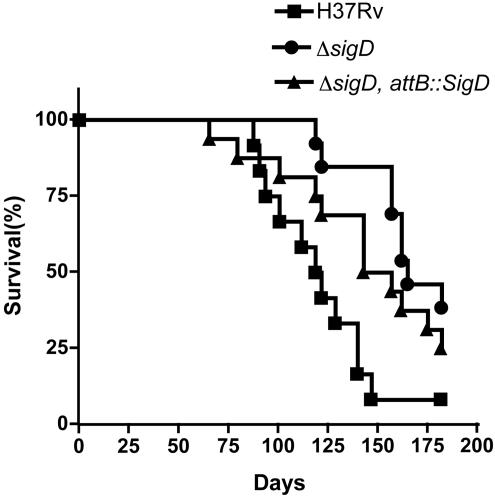
To assess the role of SigD in M. tuberculosis virulence, we used a mouse model of infection. BALB/c mice infected intravenously with the ΔsigD mutant displayed significantly prolonged survival relative to those infected with the parental H37Rv strain (P < 0.001 for the mutant versus the wt) (Fig. 7). Median survival times of 119, 164, and 149 days were obtained for the groups of mice infected with H37Rv, the ΔsigD mutant, and the complemented strain, respectively. In a second experiment, the median survival times of mice infected with H37Rv and the ΔsigD mutant were 117 and 151 days, respectively (P < 0.005).
FIG. 7.
Survival of mice infected with M. tuberculosis H37Rv, the ΔsigD mutant, and the ΔsigD complemented strain. BALB/c mice were infected by lateral tail vein injections with 106 CFU. Morbidity and weights were monitored throughout the experiment, and mice that became moribund were sacrificed. Twelve mice were infected in each group.
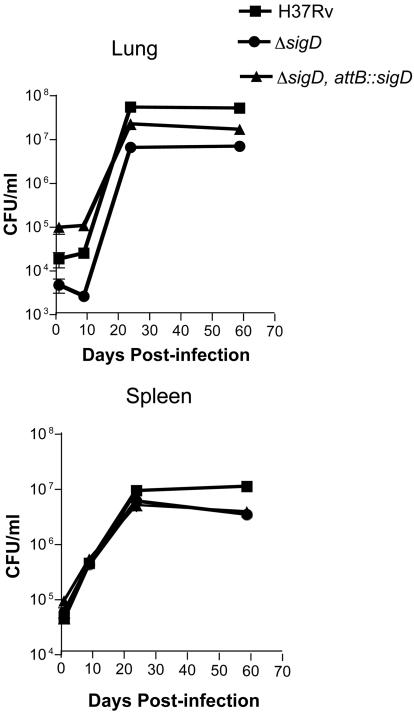
We examined the ability of these three M. tuberculosis strains to proliferate and persist in the tissues of BALB/c mice after intravenous infection with 106 CFU. The organ bacterial burdens in lungs and spleens were assessed by plating dilutions of the organ homogenates on days 1, 9, 24, and 59 after infection (Fig. 8). Although the inoculum sizes of the three strains varied less than twofold, as determined by plating on the day of infection, the bacterial burdens in the lungs measured on day 1 postinfection varied substantially, whereas very similar numbers of organisms were present in the spleens. The initial growth kinetics of the three strains were similar, although H37Rv appeared to replicate at a slightly higher rate between days 9 and 24, resulting in higher numbers of bacteria in both lungs and spleens on day 24. This difference persisted at 8 weeks in the lungs, with a slight decrease in the CFU in spleens of the ΔsigD and complemented strains during this plateau phase.
FIG. 8.
Replication and persistence of M. tuberculosis H37Rv, the ΔsigD mutant, and the ΔsigD complemented strain in mice. BALB/c mice were infected by lateral tail vein injections with 106 CFU, mice were sacrificed on days 1, 9, 24, and 59, and plating for bacterial burdens in the lungs and spleens was performed as described in Materials and Methods.
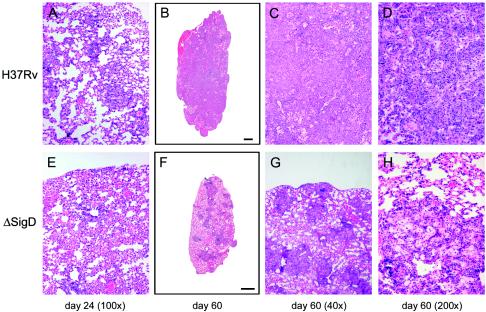
Hematoxylin and eosin-stained lung tissues from infected BALB/c mice were compared for their extents of tissue pathology. On day 24, the wt- and ΔsigD mutant-infected lung tissues showed similar degrees of lung pathology characterized by lymphocytic infiltration and a diffuse inflammation of the alveoli (Fig. 9A and E). On day 59, whole-lung cross sections showed extensive consolidation of the lung tissues from wt-infected mice (Fig. 9B). In the ΔsigD mutant-infected lungs, less diseased tissue and individual granulomatous lesions could be appreciated, and there appeared to be some resolution of the inflammatory response compared to day 24 (Fig. 9F). At a higher magnification, the wt-infected lung tissue showed extensive consolidation and lymphocytic infiltration, with almost no healthy lung tissue (Fig. 9C). In contrast, the ΔsigD mutant-infected lung tissue had significant areas of healthy lung space interspersed with smaller rounded granulomatous lesions (Fig. 9G). The wt-infected tissue had numerous monocytoid cellular infiltrates, consistent with the presence of macrophages, in addition to extensive lymphocyte infiltration (Fig. 9D). The ΔsigD mutant-infected lesions had a less intense lymphocyte infiltration and exhibited inflamed alveolar tissue with maintenance of some of the structural integrity of the alveoli (Fig. 9H).
FIG. 9.
Histopathology of lungs of BALB/c mice infected with M. tuberculosis H37Rv or the ΔsigD strain. The images show hematoxylin and eosin staining of lung sections from mice infected with H37Rv (A to D) or the ΔsigD mutant (E to H). The time points at which the mice were sacrificed are indicated below the columns of panels.
DISCUSSION
In this paper, we have shown that the M. tuberculosis alternative sigma factor SigD regulates a large number of genes, based on global transcription analysis, but that it appears to regulate relatively few genes directly. The breadth of the effect of SigD on transcription is likely mediated through its control of other regulators of transcription and through molecules that alter other cell processes, which then affect transcription indirectly. Among the SigD-controlled proteins, RpfC, Rv1816, and Rv3413c are likely to affect transcription. Though the function of Rv1816 has not been determined, sequence-based annotation indicates that it is a tetR family transcription factor. This gene is located immediately 3′ of Rv1815, a gene of unknown function whose promoter we have shown to be completely SigD dependent, and it appears to be part of an operon with Rv1815 based on similarities in their expression patterns. Similarly, Rv3413c is located immediately 3′ of sigD and is expressed as part of an operon from the SigD-dependent sigD promoter. Although it is also not functionally characterized, the protein encoded by this gene may function as a regulator of SigD activity, e.g., an anti-sigma factor. With predicted intra- and extracellular domains linked by a single transmembrane region, this protein may function to control SigD activity in response to extracellular or membrane-localized signals.
The resuscitation promoting factor RpfC may play a major role in the broad transcriptional effects observed in the sigD mutant. Our data and previous reports indicate that rpfC is the most highly expressed of the rpf genes (21, 30); here we have shown that its expression is highly dependent on SigD. Previous data indicated that RpfC is the second most potent of the M. tuberculosis Rpf proteins (21). Despite this observation, we were unable to identify strong effects of sigD inactivation on growth or survival under a variety of conditions. These results are similar to those of Tufariello et al., who were unable to identify growth phenotypes in any of the individual rpf mutant strains that they constructed (30). These data suggest a redundancy of function of the M. tuberculosis Rpf proteins. This interpretation is supported by our finding that rpfA and rpfE expression is increased in the sigD mutant strain, a result that suggests a compensatory mechanism regulating rpf expression in this strain. RpfA appears to be the most potent of the Rpf proteins, achieving a maximal reduction of lag phase in a Micrococcus resuscitation assay at concentrations that were 100-fold lower than those of RpfC, the second most potent M. tuberculosis Rpf in this assay (21). The relatively small increase in the expression of rpfA, together with this large difference in potency, could explain the lack of a consistent effect on lag phase of the sigD mutation in our experiments. Our data indicate that neither rpfA nor any of the rpf genes are other than rpfC is regulated by SigD.
Based on in vivo promoters that we identified among the genes that were most highly regulated by SigD, we identified a likely consensus recognition sequence for promoters transcribed by EσD. Like many other extracytoplasmic-function sigma factors (16), the main specificity for promoter recognition by SigD lies in the −35 region, where an eight-base consensus sequence was identified. In addition, an A+T-rich region is present at the optimal distance from the −35 element for a −10 promoter element and is therefore likely to contribute to the activities of these promoters. The SigD-regulated iniB promoter that we identified contains a region with some similarity to the SigD −35 consensus, but it lacks the −10 A+T-rich element. Whether this promoter is directly recognized by SigD in vivo is not certain. If it is, this would suggest that a broader range of promoters may be transcribed by EσD. Our inability to identify additional SigD-regulated in vivo promoters in more than 20 additional genes examined by primer extension, however, suggests that this is not the case. Interestingly, the SigD-regulated promoter of iniB is distinct from the iniB promoter identified by Alland et al. (1). In addition to confirming the induction by INH of the expression of this operon from the previously identified SigD-independent promoter, we determined that iniB expression from the partially SigD-dependent promoter is also induced by INH and that this induction is largely SigD dependent.
Our analysis of sigD expression showed that there is a moderately high level of expression throughout the exponential phase and into stationary phase, a result that is consistent with the dot blot data of Hu and Coates (12). Under hypoxic conditions resulting from gradual oxygen depletion, we found that the expression of sigD declined rapidly and to a larger degree than the expression of sigA, coincident with the onset of the hypoxic response, as indicated by the increased expression of hspX. In contrast, the expression of sigD was not found to be strongly affected in previously reported microarray experiments investigating gene expression after a rapid shift to hypoxic conditions (27). The expression of sigD was, however, significantly decreased following infection of both quiescent and activated macrophages (26).
The expression of sigD has been found to be significantly regulated, based on microarray analyses, in two other settings. Betts et al. found that sigD expression was increased two- to threefold 24 and 96 h after a shift to starvation conditions (3). Dahl et al. identified sigD and rpfC as having significantly increased expression in the wt relative to a relA mutant of M. tuberculosis in log-phase cultures, and they also showed that sigD and rpfC were up-regulated in response to starvation (8). Our data showing that the promoters of sigD and rpfC are SigD dependent indicate that the Rel-dependent starvation-induced expression of these genes is likely mediated via SigD. Consistent with this interpretation, the starvation-induced expression of sigD was blunted in a relA mutant strain, suggesting that sigD is part of the relA regulon. These results do not exclude an additional independent regulation of sigD expression under other conditions. These data, together with the stable, relatively high-level expression of sigD during growth and early stationary phase in our studies and those of Hu and Coates, suggest roles for SigD both in nutrient-replete growth conditions and in the adaptation to starvation or stringent response physiology, an apparent paradox that remains to be further defined.
In addition to sigD, Rv3413c, rpfC, Rv1815, and Rv1816, other genes that were up-regulated in the wt relative to the sigD mutant included several chaperone genes and genes involved in lipid metabolism and cell wall processes. SigD regulates gene expression for optimal growth in the absence of oxygen limitation, which might occur early during infection in vivo. Other sigma factors, most notably SigH, have been shown to regulate chaperone gene expression in response to oxidative and heat stress (24). In the setting of rapid growth in the absence of stress, the induction of chaperone expression regulated by SigD may be more important for nascent protein folding than for disaggregation and degradation or refolding of proteins that have been damaged by stress.
The genes whose expression was increased in the sigD mutant may also provide insight into SigD function. Most striking was the large number of genes encoding PE-PGRS family proteins. These genes represented >40% of the genes that were most highly expressed in the sigD mutant relative to H37Rv. It was interesting that several PE-PGRS family genes were also up-regulated in a relA mutant (8). The function of this large family of genes remains unknown, but some of them have been shown to be expressed in granulomas and to play a role in mycobacterial virulence (23). It has been hypothesized that this family of proteins may be important for immune modulation or surface interactions, and evidence indicates that members of this family of proteins are expressed at the cell surface and recognized by the immune system (2, 4, 28). The fact that the expression of many of these genes is repressed by SigD indicates that they are expressed in settings in which sigD expression is decreased. One such setting is during infection of both quiescent and activated macrophages (26), raising the possibility that these PE-PGRS genes may be important for bacterial survival during the early stages of infection.
We observed a moderate but significant decrease in the virulence of the sigD inactivated strain, with associated small decreases in replication in vivo. The most notable differences in histopathology occurred after the onset of immunity, as 8-week specimens showed more extensive inflammation in the lungs of mice infected with the wt than in those infected with the mutant. Differences in survival, replication, and histopathology are likely linked to the decreased expression of sigD in the mutant, although the partial complementation of survival and the lack of clear complementation of in vivo replication make this conclusion less certain. Whether the specific expression of one or a few key SigD-dependent genes causes this phenotype or whether it is the result of a more global disruption of growth regulation is not known. The less severe virulence phenotype of the sigD mutant relative to the relA mutant (8) indicates that SigD-dependent gene expression may account for part, but not all, of the virulence defect of the relA mutant.
The stable expression of sigD during exponential growth and stationary phase, the RelA-dependent increased expression of sigD in response to starvation, the down-regulation of sigD during macrophage infection and hypoxia, and the regulation by SigD of rpfC, which is thought to play a role in the renewal of growth after starvation or stationary phase, suggest a complex interaction of signals mediated by SigD and other regulators that allows M. tuberculosis to adapt its growth physiology during infection. The opposite effects on sigD expression of starvation versus hypoxia are particularly striking. These data suggest that these conditions are encountered at distinct stages or sites during infection and that activation or repression of the sigD regulons is important for M. tuberculosis adaptation in these settings. Since M. tuberculosis encounters a broad range of environmental conditions during infection, fine-tuning of its physiology by a large number of regulatory signals, including those mediated by SigD and other alternative sigma factors, is undoubtedly required to allow growth and survival in vivo. Further insight into the interplay of these regulatory networks should ultimately lead to a more integrated understanding of the pathogenesis of tuberculosis.
Supplementary Material
Acknowledgments
This work was supported by Public Health Service grant AI 37901 from the National Institute of Allergy and Infectious Diseases to R.N.H. and by a fellowship from the Warren Whitman Richardson fund to R.H. Oligonucleotide microarrays were provided by the Institute for Genomic Research through the NIAID-sponsored Pathogen Functional Genomics Resource.
We thank Xiaoling Puyang for excellent technical assistance, Eric Rubin for the generous gift of DNA microarrays, Eric Rubin, Chris Sassetti, and Seby L. Edassery for assistance with microarray methods and analysis, and Michael Brenner for the use of biosafety level 3 facilities.
Footnotes
Supplemental material for this article may be found at http://jb.asm.org.
REFERENCES
- 1.Alland, D., A. J. Steyn, T. Weisbrod, K. Aldrich, and W. R. Jacobs, Jr. 2000. Characterization of the Mycobacterium tuberculosis iniBAC promoter, a promoter that responds to cell wall biosynthesis inhibition. J. Bacteriol. 182:1802-1811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Banu, S., N. Honore, B. Saint-Joanis, D. Philpott, M. C. Prevost, and S. T. Cole. 2002. Are the PE-PGRS proteins of Mycobacterium tuberculosis variable surface antigens? Mol. Microbiol. 44:9-19. [DOI] [PubMed] [Google Scholar]
- 3.Betts, J. C., P. T. Lukey, L. C. Robb, R. A. McAdam, and K. Duncan. 2002. Evaluation of a nutrient starvation model of Mycobacterium tuberculosis persistence by gene and protein expression profiling. Mol. Microbiol. 43:717-731. [DOI] [PubMed] [Google Scholar]
- 4.Brennan, M. J., G. Delogu, Y. Chen, S. Bardarov, J. Kriakov, M. Alavi, and W. R. Jacobs, Jr. 2001. Evidence that mycobacterial PE_PGRS proteins are cell surface constituents that influence interactions with other cells. Infect. Immun. 69:7326-7333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen, P., R. E. Ruiz, Q. Li, R. F. Silver, and W. R. Bishai. 2000. Construction and characterization of a Mycobacterium tuberculosis mutant lacking the alternate sigma factor gene sigF. Infect. Immun. 68:5575-5580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cole, S. T., R. Brosch, J. Parkhill, T. Garnier, C. Churcher, D. Harris, S. V. Gordon, K. Eiglmeier, S. Gas, C. E. Barry III, F. Tekaia, K. Badcock, D. Basham, D. Brown, T. Chillingworth, R. Connor, R. Davies, K. Devlin, T. Feltwell, S. Gentles, N. Hamlin, S. Holroyd, T. Hornsby, K. Jagels, and B. G. Barrell. 1998. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature 393:537-544. [DOI] [PubMed] [Google Scholar]
- 7.Curcic, R., S. Dhandayuthapani, and V. Deretic. 1994. Gene expression in mycobacteria: transcriptional fusions based on xylE and analysis of the promoter region of the response regulator mtrA from Mycobacterium tuberculosis. Mol. Microbiol. 13:1057-1064. [DOI] [PubMed] [Google Scholar]
- 8.Dahl, J. L., C. N. Kraus, H. I. Boshoff, B. Doan, K. Foley, D. Avarbock, G. Kaplan, V. Mizrahi, H. Rubin, and C. E. Barry III. 2003. The role of RelMtb-mediated adaptation to stationary phase in long-term persistence of Mycobacterium tuberculosis in mice. Proc. Natl. Acad. Sci. USA 100:10026-10031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.DeMaio, J., Y. Zhang, C. Ko, D. Young, and W. Bishai. 1996. A stationary-phase stress-response sigma factor from Mycobacterium tuberculosis. Proc. Natl. Acad. Sci. USA 93:2790-2794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fernandes, N., Q.-L. Wu, D. Kong, X. Puyang, S. Garg, and R. Husson. 1999. A mycobacterial extracytoplasmic function sigma factor involved in survival following heat shock and oxidative stress. J. Bacteriol. 181:4266-4274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Geiman, D. E., D. Kaushal, C. Ko, S. Tyagi, Y. C. Manabe, B. G. Schroeder, R. D. Fleischmann, N. E. Morrison, P. J. Converse, P. Chen, and W. R. Bishai. 2004. Attenuation of late-stage disease in mice infected by the Mycobacterium tuberculosis mutant lacking the SigF alternate sigma factor and identification of SigF-dependent genes by microarray analysis. Infect. Immun. 72:1733-1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hu, Y., and A. R. Coates. 2001. Increased levels of sigJ mRNA in late stationary phase cultures of Mycobacterium tuberculosis detected by DNA array hybridisation. FEMS Microbiol. Lett. 202:59-65. [DOI] [PubMed] [Google Scholar]
- 13.Kaushal, D., B. G. Schroeder, S. Tyagi, T. Yoshimatsu, C. Scott, C. Ko, L. Carpenter, J. Mehrotra, Y. C. Manabe, R. D. Fleischmann, and W. R. Bishai. 2002. Reduced immunopathology and mortality despite tissue persistence in a Mycobacterium tuberculosis mutant lacking alternative sigma factor, SigH. Proc. Natl. Acad. Sci. USA 99:8330-8335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kerr, M. K., M. Martin, and G. A. Churchill. 2000. Analysis of variance for gene expression microarray data. J. Comput. Biol. 7:819-837. [DOI] [PubMed] [Google Scholar]
- 15.Lee, M., L. Pascopella, W. Jacobs, Jr., and G. Hatfull. 1991. Site-specific integration of mycobacteriophage L5: integration-proficient vectors for Mycobacterium smegmatis, Mycobacterium tuberculosis and bacille Calmette-Guerin. Proc. Natl. Acad. Sci. USA 88:3111-3115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lonetto, M., K. Brown, K. Rudd, and M. Buttner. 1994. Analysis of the Streptomyces coelicolor sigE gene reveals the existence of a subfamily of eubacterial RNA polymerase σ factors involved in the regulation of extracytoplasmic functions. Proc. Natl. Acad. Sci. USA 91:7573-7577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Manganelli, R., E. Dubnau, S. Tyagi, F. R. Kramer, and I. Smith. 1999. Differential expression of 10 sigma factor genes in Mycobacterium tuberculosis. Mol. Microbiol. 31:715-724. [DOI] [PubMed] [Google Scholar]
- 18.Manganelli, R., M. I. Voskuil, G. K. Schoolnik, E. Dubnau, M. Gomez, and I. Smith. 2002. Role of the extracytoplasmic-function sigma factor sigmaH in Mycobacterium tuberculosis global gene expression. Mol. Microbiol. 45:365-374. [DOI] [PubMed] [Google Scholar]
- 19.Manganelli, R., M. I. Voskuil, G. K. Schoolnik, and I. Smith. 2001. The Mycobacterium tuberculosis ECF sigma factor sigmaE: role in global gene expression and survival in macrophages. Mol. Microbiol. 41:423-437. [DOI] [PubMed] [Google Scholar]
- 20.McKinney, J. D., K. Honer zu Bentrup, E. J. Munoz-Elias, A. Miczak, B. Chen, W. T. Chan, D. Swenson, J. C. Sacchettini, W. R. Jacobs, Jr., and D. G. Russell. 2000. Persistence of Mycobacterium tuberculosis in macrophages and mice requires the glyoxylate shunt enzyme isocitrate lyase. Nature 406:735-738. [DOI] [PubMed] [Google Scholar]
- 21.Mukamolova, G. V., O. A. Turapov, D. I. Young, A. S. Kaprelyants, D. B. Kell, and M. Young. 2002. A family of autocrine growth factors in Mycobacterium tuberculosis. Mol. Microbiol. 46:623-635. [DOI] [PubMed] [Google Scholar]
- 22.Pelicic, V., M. Jackson, J.-M. Reyrat, W. J. Jacobs, B. Gicquel, and C. Guilhot. 1997. Efficient allelic exchange and transposon mutagenesis in Mycobacterium tuberculosis. Proc. Natl. Acad. Sci. USA 94:10955-10960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ramakrishnan, L., N. A. Federspiel, and S. Falkow. 2000. Granuloma-specific expression of Mycobacterium virulence proteins from the glycine-rich PE-PGRS family. Science 288:1436-1439. [DOI] [PubMed] [Google Scholar]
- 24.Raman, S., T. Song, X. Puyang, S. Bardarov, W. Jacobs, Jr., and R. Husson. 2001. The alternative sigma factor SigH regulates major components of the oxidative and heat stress responses in Mycobacterium tuberculosis. J. Bacteriol. 183:6119-6125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sassetti, C. M., D. H. Boyd, and E. J. Rubin. 2003. Genes required for mycobacterial growth defined by high density mutagenesis. Mol. Microbiol. 48:77-84. [DOI] [PubMed] [Google Scholar]
- 26.Schnappinger, D., S. Ehrt, M. I. Voskuil, Y. Liu, J. A. Mangan, I. M. Monahan, G. Dolganov, B. Efron, P. D. Butcher, C. Nathan, and G. K. Schoolnik. 2003. Transcriptional adaptation of Mycobacterium tuberculosis within macrophages: insights into the phagosomal environment. J. Exp. Med. 198:693-704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sherman, D. R., M. Voskuil, D. Schnappinger, R. Liao, M. I. Harrell, and G. K. Schoolnik. 2001. Regulation of the Mycobacterium tuberculosis hypoxic response gene encoding alpha-crystallin. Proc. Natl. Acad. Sci. USA 98:7534-7539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Singh, K. K., X. Zhang, A. S. Patibandla, P. Chien, Jr., and S. Laal. 2001. Antigens of Mycobacterium tuberculosis expressed during preclinical tuberculosis: serological immunodominance of proteins with repetitive amino acid sequences. Infect. Immun. 69:4185-4191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Snapper, S., R. Melton, S. Mustafa, T. Kieser, and W. Jacobs, Jr. 1990. Isolation and characterization of efficient plasmid transformation mutants of Mycobacterium smegmatis. Mol. Microbiol. 4:1911-1919. [DOI] [PubMed] [Google Scholar]
- 30.Tufariello, J. M., W. R. Jacobs, Jr., and J. Chan. 2004. Individual Mycobacterium tuberculosis resuscitation-promoting factor homologues are dispensable for growth in vitro and in vivo. Infect. Immun. 72:515-526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Voskuil, M. I., D. Schnappinger, K. C. Visconti, M. I. Harrell, G. M. Dolganov, D. R. Sherman, and G. K. Schoolnik. 2003. Inhibition of respiration by nitric oxide induces a Mycobacterium tuberculosis dormancy program. J. Exp. Med. 198:705-713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wayne, L. G., and L. G. Hayes. 1996. An in vitro model for sequential study of shiftdown of Mycobacterium tuberculosis through two stages of nonreplicating persistence. Infect. Immun. 64:2062-2069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.World Health Organization. 2002. Tuberculosis fact sheet. World Health Organization, Geneva, Switzerland.
- 34.Wu, Q.-L., D. Kong, K. Lam, and R. Husson. 1997. A mycobacterial extracytoplasmic function sigma factor involved in survival following stress. J. Bacteriol. 179:2922-2929. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.