Abstract
The par stability determinant, encoded by the Enterococcus faecalis plasmid pAD1, is the only antisense RNA regulated postsegregational killing system identified in gram-positive bacteria. Because of the unique organization of the par locus, the par antisense RNA, RNA II, binds to its target, RNA I, at relatively small, interspersed regions of complementarity. The results of this study suggest that, rather than targeting the antisense bound message for rapid degradation, as occurs in most other antisense RNA regulated systems, RNA I and RNA II form a relatively stable, presumably translationally inactive complex. The stability of the RNA I-RNA II complex would allow RNA I to persist in an untranslated state unless or until the encoding plasmid was lost. After plasmid loss, RNA II would be removed from the complex, allowing translational activation of RNA I. The mechanism of RNA I activation in vivo is unknown, but in vitro dissociation experiments suggest that active removal of RNA II, for example by a cellular RNase, may be required.
Regulation by small complementary RNAs now appears to be widespread in both prokaryotic and eukaryotic organisms (8, 14). The first such regulatory RNAs were identified on bacterial mobile genetic elements and were dubbed antisense RNAs (39, 42; for reviews, see references 7 and 45). In these systems, the antisense RNA is generally transcribed from the opposite strand overlapping the 5′ end of a target RNA. The antisense RNA binds to its target via extensive complementarity and negatively regulates gene expression by directly interfering with ribosome binding (33, 43), altering mRNA conformation to interfere with ribosome binding (2), facilitating premature transcriptional termination (9, 36), or interfering with appropriate processing (41). These antisense RNAs are referred to as cis encoded, since the genes for the antisense RNA and its target are overlapping. In general, regulation is irreversible because the antisense RNA-target RNA complex is rapidly degraded by the double-strand-specific endoribonuclease RNase III (5, 10, 18, 30, 46). trans-encoded antisense RNAs have been more recently defined as those which are not genetically linked to their targets (12). trans-encoded antisense RNAs were originally believed to be rare, but recent bioinformatic searches have suggested that they may be more widespread than originally thought (27, 44). Although complementarity is more limited in these systems, targeted complex degradation still appears to be a primary mechanism of regulation (1, 31, 34). However, RNase E rather than RNase III appears to degrade the RyhB/SodB complex, and the mechanism of target destabilization is unknown in the other systems. In a few cases, trans-encoded antisense RNAs have been found to positively regulate expression from the targeted message (31, 35). In these cases, the mRNA may be stabilized, but it is unclear whether this stabilization is directly due to RNA complex formation or to translational activation. Targeted RNA degradation is also the primary method of regulation by RNA interference in eukaryotic systems (4).
Two plasmid addiction systems have been shown to be regulated by an antisense RNA mechanism. Plasmid addiction systems function to stabilize plasmids within bacterial populations by programming for death any cell that loses a copy of the plasmid (for a recent review, see reference 25). This is accomplished by plasmid-encoded production of a stable toxin and an unstable antitoxin. As long as the plasmid is retained, the antitoxin inhibits the activity and/or synthesis of the toxin, but if the plasmid is lost the antitoxin is degraded and the toxin kills the cell. In most addiction systems, both toxin and antitoxin are proteins, but in the hok/sok system of Escherichia coli plasmid R1 (20) and related systems on other E. coli plasmids (19) and in the par system of Enterococcus faecalis plasmid pAD1 (50), the antitoxin is an antisense RNA that inhibits translation of the toxin message. Addiction systems present special problems for antisense RNA regulation, since rapid degradation of the RNA-RNA complex would leave no toxin message to be translated once the plasmid was lost. In the hok/sok system, this problem is solved by the accumulation of a pool of a conformation of the hok mRNA that neither binds the sok antisense regulator nor allows ribosome binding (15, 40). This pre-mRNA is then slowly degraded from the 3′ end, triggering a conformational switch to a sok- and ribosome-binding form (16). If the plasmid is still present, sok binds rapidly via a U-turn motif located within one of the loops of the hok target (17), and the complex is rapidly degraded by RNase III (18). If the plasmid is lost, the Hok toxin is translated because of the absence of the unstable sok antisense RNA, and the cell is killed.
The par addiction system is encoded on the pAD1 plasmid native to the gram-positive bacterium E. faecalis. pAD1 is the prototype of a family of conjugative plasmids whose transfer genes are induced by the production of peptide sex pheromones from potential recipient cells (13). The par locus blends features of cis- and trans-regulated antisense systems, since the genes for the antisense RNA, RNA II, and its target, RNA I, overlap only at the extreme 3′ end (Fig. 1) (50). The complementarity responsible for the inhibition of translation is provided by direct repeats located in the 5′ ends of the RNA I and RNA II genes that are transcribed in opposite directions (21). Therefore, rather than interacting at a single contiguous stretch of complementarity, the par RNAs interact at multiple dispersed regions (23). Despite multiple attempts, no evidence of a processed intermediate of RNA I has been found and no alternative conformations of RNA I has been indicated by either computer or experimental analysis (22). While RNA I does contain a structure that partially inhibits translation, this stem-loop does not appreciably affect binding by RNA II. No alternative conformations that would alter this structure are apparent (21, 23), suggesting that the mode of regulation of par may be quite different from that of hok/sok. Evidence is provided in this report indicating that, rather than targeting the RNA I toxin message for degradation, RNA II binding results in formation of a relatively stable complex that accumulates as a pool in the cell. RNA II was stabilized fourfold in the presence of RNA I; RNA I was unusually stable even in the absence of RNA II and was slightly stabilized in its presence. RNA I must be released from this complex in plasmid-free cells to allow toxin translation. While the mechanism of release remains uncertain, in vitro dissociation experiments suggested that active removal of RNA II may be required.
FIG. 1.
Genetic organization of the par locus. The par locus is approximately 400 bp in size, delimited by the promoters for the RNA I and RNA II genes located on the right and left ends, respectively, and is shown by a heavy black line. The relative lengths of the RNA I and RNA II transcripts are depicted as arrowed lines below and above the genetic map, respectively. The fst toxin-encoding gene is depicted on the RNA I gene and transcript with a hatched box. The direct repeats DRa and DRb and the inverted repeat of the bidirectional rho-independent transcription terminator are shown by white arrows. These sequences provide the complementary regions required for interaction of RNA I and RNA II, since transcription occurs in opposite directions across the repeats. Note also that the DRa and DRb sequences overlap the translation initiation region of fst in RNA I. The sequence of the toxic Fst peptide is shown below the map.
MATERIALS AND METHODS
Bacterial strains, media, and culture conditions.
E. coli strain DH5-α (Invitrogen) was used for constructing the plasmids used in this study. E. faecalis strain OG1X was used for the in vivo RNA stability analysis. OG1X is a streptomycin-resistant, gelatinase-negative derivative of OG1 (29). E. coli was routinely cultured in Luria broth (37), and E. faecalis was cultivated with Todd-Hewitt broth (Difco Laboratories). Antibiotics (Sigma Chemical) were used at the following concentrations: chloramphenicol, 10 to 25 μg ml−1; ampicillin, 100 μg ml−1; tetracycline, 10 μg ml−1; erythromycin, 10 μg ml−1; rifampin, 250 to 350 μg ml−1; and spectinomycin, 100 μg ml−1.
Plasmids, primers, DNA manipulation, and construction of mutants.
Plasmids used and constructed in this study are shown in Table 1. Oligonucleotide primers and probes are listed in Table 2. Small-scale plasmid purifications of E. coli were performed by the boiling prep method (28). Small-scale plasmid purifications of E. faecalis were performed by the modified alkaline lysis procedure (49). Large-scale plasmid purifications of E. coli used the QIAGEN midi-prep protocol according to the manufacturer's instructions. Restriction enzymes and T4 DNA ligase were obtained from Gibco/BRL or New England BioLabs and used according to the manufacturer's instructions. Plasmid constructs were introduced into E. faecalis by electroporation as described previously (23). Transformation to E. coli was accomplished with Subcloning Efficiency DH5-α chemically competent cells (Invitrogen) according to the manufacturer's instructions.
TABLE 1.
Plasmids
| Plasmid | Relevant phenotype | Marker | Reference or source |
|---|---|---|---|
| Vectors | |||
| pAM401 | E. coli-E. faecalis shuttle vector with pACYC184 and pIP501 replicons | Cmr, Tcr | 53 |
| pDL278 | E. coli-E. faecalis shuttle vector with pVA380-1 replicon | Spr | 32 |
| pGEM-T | E. coli TA cloning vector | Ampr | Promega |
| pGEM-T Easy | E. coli TA cloning vector | Ampr | Promega |
| pSPORT1/2 | E. coli cloning vector | Ampr | Gibco/BRL |
| par clones | |||
| pDAK606 | pDL278 carrying the wild-type par locus | Spr | This work |
| pDAK607 | pAM401 carrying the wild-type par locus | Cmr | 50 |
| pDAK608 | pSPORT1 containing the RNA II gene | Ampr | 50 |
| pDAK611 | pDL278 carrying the RNA II gene | Spr | 50 |
| pDAK616 | pSPORT2 containing the RNA II gene antisense to the T7 promoter | Ampr | This work |
| pDAK704 | pAM401 carrying the RNA I gene | Cmr | 21 |
| pDAK734 | pAM401 carrying the 19-stop mutation of the RNA I gene | Cmr | This work |
| pDAK752 | pAM401 carrying the IC mutant of the RNA I gene | Cmr | This work |
| pDAK901 | pGEM-T containing the RNA I gene for in vitro transcription | Ampr | 21 |
| pDAK902 | pGEM-T containing the RNA II gene for in vitro transcription | Ampr | 21 |
| pAD1 minireplicons | |||
| pDAK102 | pAD1 minireplicon in pAM201 replicon probe vector with pACYC184 replicon | Cmr | 47 |
| pDAK102Δ | pDAK102 deleted for the RNA I gene | Cmr | This work |
| pDAK2300K | pAD1 minireplicon marked with Tn917 | Emr | 48 |
TABLE 2.
Oligonucleotide probes and primers
| Primer | Sequencea |
|---|---|
| RNA I-specific probe | 5′-ATAACCAACGACATTAAATCTTTCAC-3′ |
| RNA II-specific probe | 5′-TGTGTTATCTGTACGATTTAATGTCG-3′ |
| Ef 5SrRNA-specific probe | 5′-AACAGGTGTATCCTTCTCGCTAT-3′ |
| 5′ RNA I EP-PCR | 5′-CGGGATCCAAATTGTTGAAGGTTTTATTATTAAATTGGCAGAATTTCAAATTATGATATAATTAAT-3′ |
| 3′ RNA I EP-PCR | 5′-GCTCTAGAAAAAAGCAATCCTACGGCGAATAGGATTGCTTTTTTTTACACTAGTTGCGACTTC-3′ |
| 19-stop | 5′-CCAATCTTTGTAGGATTGGTTCTGTAAATGATTTCTCG-3′ |
| 3′ RNA I-Xba | 5′-CGTCTAGATGAAAAAGCAATCCTACGGCGA-3′ |
| RNA I Ext #2 | 5′-CGCCTCGATTGGAGGTGTGTTATTTGTGAAAGATTTAATGTCGTTGGTTATCGCACCAATCTTTGTAGGAT-3′ |
| RNA I Pro | 5′-ATTGGCAGAATTTCAAATTATGATATAATTAATGCGGCAGCTCGCCTCGATTGGAGG-3′ |
| RNA I Ext | 5′-AAATTGTTGAAGGTTTTATTATTAAATTGGCAGAATTTC-3′ |
| RNAI-Sal-475 | 5′-GTCGACGTGTAAACGAGTTTACACGCTTAGAAAAGATCATAAAACAGCTAGATAAATTGTTGAAGGTTTTAT-3′ |
Restriction enzyme recognition sites are underlined.
Plasmid pDAK102Δ was constructed from pDAK102 by replacing a KpnI-SpeI fragment with the KpnI-SpeI fragment from the multiple-cloning site of pSPORT2. The pDAK102 KpnI site is upstream of the RNA I promoter and the SpeI site is immediately upstream of the par terminator, so deletion of the KpnI-SpeI fragment removes the RNA I gene without affecting RNA II expression. Approximately equal amounts of pDAK102 and pSPORT2 were cut, ligated, and then used to transform competent E. coli cells. Transformants were screened by colony blotting (3) with the RNA I-specific probe (Table 2), and colonies producing no signal were selected. The expected structure of the construct was confirmed by restriction analysis. pDAK102 was used instead of pDAK2300K because pDAK102 contains a pACYC184 E. coli replicon in addition to the pAD1 replicon, making genetic manipulation easier. pDAK2300K contains only the pAD1 replicon marked with Tn917. In E. faecalis, both plasmids depend on the pAD1 replicon for replication and therefore have similar copy numbers of 2 to 4 per cell.
The nontoxic derivatives of RNA I were isolated by two different approaches. The initiation codon (IC) mutant, was fortuitously isolated from a random mutagenesis procedure by error-prone PCR (EP-PCR). PCR was performed with RNA I primers 5′ RNA I EP-PCR and 3′ RNA I EP-PCR (Table 2) by the EP-PCR protocol previously described (3) for 22 cycles. pDAK606, a pDL278 derivative containing the wild-type par locus from pDAK607 cloned into the BamHI-SalI site, was used as a template. PCR products were cloned into pGEMT-Easy and transformed into E. coli. Transformants were washed from plates, and plasmid DNA was isolated from the pool. The amplified RNA I gene was then removed from pGEMT-Easy with restriction sites BamHI and XbaI introduced from the primers and cloned into similarly cut pAM401. pAM401 DNA was introduced into E. coli, and chloramphenicol-resistant transformants were selected and then screened for inactivation of the tetracycline resistance gene. Plasmid DNA was purified from individual tetracycline-sensitive transformants and introduced into E. faecalis to select constructs with mutated RNA I genes. The RNA I genes from successful transformations were sequenced (Lone Star Laboratories), and two were found to have a single mutation that changed the GUG start codon to GCG. This mutant plasmid was designated pDAK752.
The 19-stop mutation was constructed by PCR site-specific mutagenesis with pDAK606 as a template. In the first reaction, the mutagenic 19-stop primer was used as the 5′ primer, and the 3′ RNA I-Xba primer was used as the 3′ primer. The latter primer contains the 3′ end of RNA I and an XbaI recognition site for cloning. PCR was performed with PCR SuperMix High Fidelity (Invitrogen) according to the manufacturer's instructions. The PCR product from this reaction was gel purified with the Qiaex II gel extraction kit (QIAGEN) according to the manufacturer's instructions for removing the template. The remainder of the par sequence upstream of the fst coding sequence was then added with successive 5′ primers with extensions containing upstream sequences, each using the product of the previous reaction as a template. The 3′ RNA I-Xba primer was used as the 3′ primer in all reaction mixtures. The 5′ primers were used in the following sequence: RNA I Ext #2, RNA I Pro, RNA I Ext, and RNAI-Sal-475 (Table 2). The RNAI-Sal-475 primer contains a SalI recognition site resulting in a product flanked by SalI and XbaI sites at its 5′ and 3′ ends, respectively. The final product was cloned into pGEMT-Easy, sequenced to confirm that only the 19-stop mutation was present, and then transferred to pAM401 with the SalI-XbaI sites to create pDAK734.
Determination of RNA stability.
E. faecalis strains containing relevant plasmids were cultured overnight under appropriate selective conditions. Cultures were then diluted 1:10 without antibiotics and grown for an additional 2 h at 37°C to mid-exponential phase. Rifampin was added to the culture at a final concentration of 350 μg ml−1. Samples (each, 10 ml) were taken immediately after rifampin addition (time zero) and after various intervals thereafter, depending on the relative stability of the RNA being measured. Samples were immediately poured onto frozen 50 mM sodium azide in phosphate-buffered saline and stored on ice until the end of the experiment. RNA was then purified with the FastRNA Pro Blue kit (Qbiogene) according to the manufacturer's instructions. Two micrograms of total cellular RNA was fractionated on a 5% polyacrylamide-7 M urea gel with 1× TBE running buffer (89 mM Tris [pH 8.3], 2 mM EDTA, and 89 mM boric acid) at 325 V for 45 min. The RNA was transferred to a Nytran Supercharge nylon membrane (Schleicher & Schuell Bioscience) with the Transblot SD Semi-Dry transfer cell (Bio-Rad) at 15 V for 15 min. The membrane was blocked with Rapid-hyb buffer (Amersham Biosciences) at 42°C for 1 h. RNA I-, RNA II-, and E. faecalis 5S rRNA-specific probes were 5′ end labeled with 32P with T4 polynucleotide kinase and [γ-32P]ATP (ICN) as a substrate. End-labeled probes were separated from residual [γ-32P]ATP with a NucAway (Ambion) spin column. A total of 1 to 4 μl of each probe was added to the hybridization buffer and incubated for 1 h at 42°C. Blots were washed twice at room temperature with 1× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) and 0.1% sodium dodecyl sulfate for 10 min. Blots were imaged with an Amersham Biosciences storage phosphor screen and Typhoon imager, model 9410. Quantitation was performed with ImageQuant software. All counts were normalized to the 5S rRNA loading control. Half-lives were calculated from at least three independent experiments.
Quantitative Northern blotting and RNase protection assays (RPAs).
Quantitative Northern blotting was performed essentially as described above with the following exceptions: RNA was purified immediately after the 2-h incubation period without the addition of rifampin, and RNA I- and RNA II-specific probes were labeled to equal specific activity. The probes used have nearly equal melting temperatures for RNA I- and RNA II-specific probes, respectively, of 52.2 and 52.4°C at 50 mM salt concentration.
For RPA experiments, RNA was purified from OG1X(pDAK2300K) as described above (without rifampin). The antisense probe for detection of RNA I was created by an SP6 transcription of pDAK901 plasmid DNA digested with SphI, yielding a transcript of 339 nucleotides (nt). To construct an antisense probe for RNA II, the RNA II gene was removed from pDAK608 with SalI-NotI restriction enzymes and cloned into similarly cut pSPORT2 to construct pDAK616. This procedure places the RNA II gene antisense to the pSPORT2 T7 RNA polymerase start site. The RNA II probe was created by a T7 transcription of plasmid pDAK616 digested with BamHI (192 nt). The transcription reaction was performed using Ambion's MAXIscript in vitro transcription kit according to the manufacturer's instructions. The probes were random labeled with [α-32P]UTP (ICN) to a specific activity of approximately 5.3 × 108 cpm/μg. The synthetic standard for RNA I used to calibrate the system was created using a T7 transcription of pDAK901 plasmid DNA digested with BamHI and XbaI, yielding a wild-type RNA I transcript (215 nt). The synthetic standard for RNA II was created using T7 polymerase for the transcription of pDAK902, yielding a wild-type RNA II transcript (65 nt). The concentrations of the synthetic standard RNAs were determined by trace labeling with [32P]UTP. The antisense probes and synthetic standards were gel purified overnight at 37°C with the probe elution buffer (350 μl) supplied with the RPA III kit. The RPA was performed with Ambion's RPA III kit according to the manufacturer's instructions. Antisense probe and target RNA were hybridized overnight at 56°C. RNase T1 (1:50) was used to degrade single-stranded RNA, and protected RNA fragments were fractionated on a denaturing 5% polyacrylamide-7 M urea gel with 1× TBE running buffer (89 mM Tris [pH 8.3], 2 mM EDTA, and 89 mM boric acid) at 325 V for 45 min. The gel was dried on filter paper with the Bio-Rad model 583 gel dryer. RNA was quantified by comparing the net counts per minute of protected probe to the standard curve with synthetic RNA. The densitometry of the protected fragment bands was analyzed with an InstantImager (Packard).
To determine the number of molecules per cell from the RPA results, it was necessary to obtain an accurate viable count from the culture used. Since E. faecalis grows in chains, sonication was necessary to break the chains into individual cells prior to plating. For this purpose, aliquots of cells were removed from the culture before RNA preparation and sonicated for 5 s at different output settings on a Sonifier cell disrupter(model W185D; Heat Systems-Ultrasonics, Inc.) with a microtip. Cells were then serially diluted and plated on appropriate solid medium. Results indicated that viable counts increased to a peak at setting 3 of the cell disrupter, where counts were ∼15-fold higher than in unsonicated samples. Sonication at higher settings resulted in progressively lower cell counts, presumably due to cell disruption. Microscopic examination revealed that while most cells were singlets, some doublets remained even at the highest levels of sonication.
To estimate what proportion of RNA is lost during purification, E. faecalis strain OG1X containing the pAM401 vector was grown and harvested as if for RNA preparation. Prior to cell disruption, 100 fmol of RNA I and RNA II, purified in vitro from T7 promoter-containing constructs as previously described (23), was added to the samples. RNA was then purified and subjected to Northern blotting as described above with the equivalent amount of in vitro RNA expected to be in the sample if recovery was 100% in an adjacent lane. Results indicated that generally 60 to 75% of RNA is lost during RNA purification.
Dissociation experiments.
RNA I and RNA II were synthesized in vitro from pDAK901 and pDAK902 and gel purified as previously described (22), except that transcription was performed with the T7-MEGAshortscript high-yield transcription kit (Ambion). Purified RNA II was then end labeled as previously described (22). Duplex formation between RNA I (8 pmol) and end-labeled RNA II (0.4 pmol) was carried out in vitro at 37°C in 1× TMN buffer (20 mM Tris acetate [pH 7.5], 10 mM Mg acetate, 100 mM NaCl) for 30 min. Unlabeled RNA I was added at a 20-fold molar excess to ensure that all RNA II was in complex. The complex was challenged with increasing amounts of unlabeled RNA II (relative to end-labeled RNA II) in 1× TMN at 37°C for 1 h. The RNAs were electrophoresed on a native 5% polyacrylamide gel in 1× TBE at 140 V for 3.5 h. The lower voltage used was to prevent heating that might denature the complex. The gel was dried on filter paper and analyzed with the Instant Imager (Packard).
In the second approach, unlabeled RNA I and RNA II, synthesized and purified as described above, were mixed in various ratios in 1× TMN buffer at 37°C for 30 min to form a duplex. Duplex formation was carried out with 0.1 pmol of RNA I and a 1-, 2.5-, 5-, or 10-fold molar excess of RNA II. The preformed complex was then challenged with a 10- or 100-fold molar excess of end-labeled RNA II, prepared as above, relative to RNA I for 1 h at 37°C. The RNAs were fractionated and dried on filter paper as described above. The dried gel was imaged with the Amersham Biosciences storage phosphor screen and Typhoon imager, model 9410, and analyzed with ImageQuant software.
RESULTS
The presence of RNA I stabilizes RNA II.
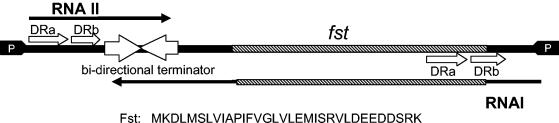
To determine if RNA II stability was affected by complex formation, persistence of RNA II after the addition of rifampin in the presence and absence of RNA I was determined. Stability of RNA II produced from pDAK611, containing the RNA II gene on vector pDL278, was measured in the presence of pDAK704, a pAM401 derivative carrying the RNA I gene, or the empty pAM401 vector. Representative results are shown in Fig. 2A. The average half-life of RNA II in the absence of RNA I was 8.1 ± 0.8 min. The half-life of RNA II in the presence of RNA I was consistently >40 min.
FIG. 2.
Stabilization of RNA II by RNA I. Samples were taken from cultures at time intervals following the addition of rifampin to stop transcription initiation. RNA was purified, fractionated, transferred to nylon membranes, and probed with radiolabeled oligonucleotides specific for RNA I, RNA II, and E. faecalis 5S rRNA, as described in Materials and Methods. The 5S rRNA probe was added as a loading control. Time points are 0, 5, 10, 20, and 40 min after the addition of rifampin, shown from left to right on all gels. (A) Stabilization of RNA II in trans. Lanes 1 to 5, OG1X(pDAK611, pAM401); lanes 6 to 10, OG1X(pDAK611, pDAK704). The lower panel shows an overexposure of the RNA II bands. (B) Stabilization of RNA II in cis. Lanes 1 to 5, OG1X(pDAK102); lanes 6 to 10, OG1X(pDAK102Δ).
These results showed that RNA II was stabilized by RNA I provided in trans. However, previous results with the par RNAs in their natural configuration on the pAD1 miniplasmid pDAK2300K showed a half-life of only ∼15 min (21), suggesting that vector context affected par RNA half-life. To ensure that the stabilization of RNA II observed with RNA I supplied in trans under high-copy-number conditions was relevant to par RNAs in their natural context, a derivative of pAD1 miniplasmid pDAK102 deleted for the RNA I gene, pDAK102Δ, was constructed. The stability of RNA II in OG1X cells containing either pDAK102 or pDAK102Δ was compared. Representative results are shown in Fig. 2B. The average half-life of RNA II was 4.1 ± 0.4 min in OG1X(pDAK102Δ) and 16.2 ± 1.5 min in OG1X(pDAK102), showing a fourfold stabilization of RNA II in the presence of RNA I.
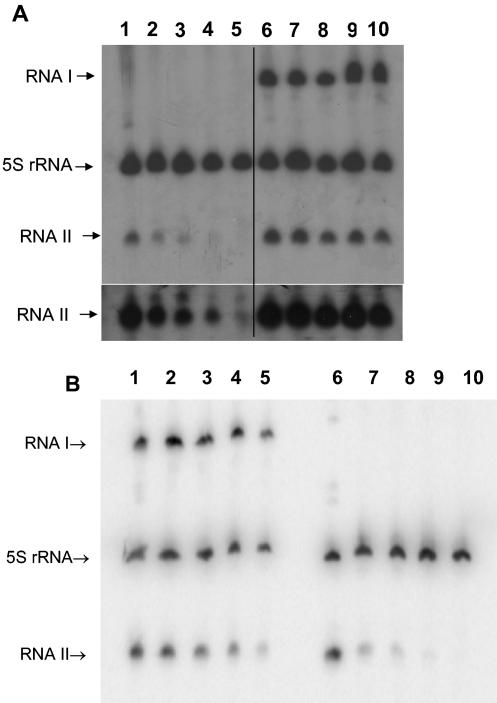
The stability of RNA II produced from pDAK611 was greater than that produced from pDAK102 in both the presence and absence of RNA I, although the difference was somewhat greater in the presence of RNA I (Table 3). Thus, RNA II stability was greater when produced on a high-copy-number vector with RNA I supplied in trans than when it was produced on the low-copy-number pAD1 miniplasmids with RNA I supplied in cis. To determine whether this effect was due to cis or trans presentation or to copy number differences, RNA II stability was measured when produced from pDAK607, which contains wild-type par on the high-copy-number pAM401 vector. Under these conditions, the half-life of RNA II was >40 min, ruling out a cis versus trans effect and implicating vector copy number as the cause of the stability difference. To determine if increased gene dosage due to higher copy number resulted in increased production of par RNAs, RNA was purified from cells carrying the par genes in various vector contexts. It was observed that RNA II levels were consistently 5- to 10-fold higher when produced from high-copy-number constructs in cis or in trans than when produced from a low-copy-number pAD1 miniplasmid (Fig. 3). Even in the absence of RNA I, cells containing pDAK611 produced over twice as much RNA II as cells containing pDAK2300K, in spite of the fact that the RNA II in OG1X (pDAK2300K) is twice as stable, due to the presence of RNA I. These results are consistent with a predominant effect of gene dosage on steady-state levels of the par RNAs. Therefore, it appears that higher levels of RNA result in greater stability, suggesting that some activity required for RNA II processing, particularly from the RNA I-RNA II complex, is limiting.
TABLE 3.
Stability of RNA II in the presence and absence of RNA I under various vector contexts
| RNA, and context (plasmid[s])a | Half-life (min)b
|
|
|---|---|---|
| RNA I | RNA II | |
| RNA II, high (pDAK611, pAM401) | — | 8.1 ± 0.8 |
| RNA I and II, high, in trans (pDAK611, pDAK704) | >40 | >40 |
| RNA I and II, low, in cis (pDAK2300K) | 60 ± 7 | 15.7 ± 2.6 |
| RNA I and II, low, in cis (pDAK102) | >40 | 16.2 ± 1.5 |
| RNA II, low, in cis (pDAK102Δ) | — | 4.1 ± 0.4 |
| RNA I and II, high, in cis (pDAK607) | >40 | >40 |
High, high copy number; low, low copy number.
Half-lives shown are an average of three independent experiments. —, not applicable.
FIG. 3.
Gene dosage effects of par RNA levels. RNA was purified from logarithmic-phase OG1X cells containing the par RNAs in various contexts, fractionated, transferred to nylon membranes, and probed with radiolabeled oligonucleotides specific for RNA I, RNA II, and E. faecalis 5S rRNA, as described in Materials and Methods. Lanes: 1, OG1X(pDAK611); 2, OG1X(pDAK704, pDAK611); 3, OG1X(pDAK2300K); 4, OG1X(pDAK607).
RNA I is exceptionally stable and is not destabilized by binding RNA II.
To determine if RNA I was targeted for degradation by RNA II binding, two RNA I mutants defective in production of the Fst toxin but not in interaction with RNA II were constructed. In the IC mutant, the IC for fst was changed from GUG to GCG. This mutation would be expected to inhibit ribosome binding to RNA I. In the 19-stop mutation, the 19th codon of the fst gene was changed from GAA to UAA, resulting in premature termination. This mutation was constructed to determine if ribosome binding affected stability with or without RNA II. Both mutations were within regions not predicted to interact with RNA II. The mutant genes under control of their native promoters were cloned into pAM401 and introduced into OG1X cells containing the pDL278 vector or the RNA II-encoding pDAK611. The IC and 19-stop constructs were designated pDAK752 and pDAK734, respectively.
Half-lives determined for RNA I in these experiments were much more variable than those observed for RNA II, probably due to secondary effects of rifampin exposure over the long incubation times required to determine the half-lives of the highly stable RNA I. However, it was clear that no detectable destabilization of RNA I occurred in the presence of RNA II. In fact, in parallel experiments RNA I was 1.6- to 2.2-fold more stable in the presence of RNA II than in its absence (Table 4). Half-lives of RNA II in the presence of the RNA I mutants were consistently >40 min, indicating that the introduced mutations were still capable of interacting with RNA II. The IC mutant RNA I was no less stable than the 19-stop mutation, suggesting that ribosome binding is not necessary for the stability of free RNA I or RNA I in complex with RNA II.
TABLE 4.
Stability of RNA I in the presence and absence of RNA II
| Expta | With RNA II (min)b | Without RNA II (min)c | Fold difference | RNA II half-life (min) |
|---|---|---|---|---|
| 19 stop #1 | 120 | 71 | 1.7 | 53 |
| 19 stop #2 | 102 | 46 | 2.2 | 44 |
| 19 stop #3 | 71 | 43 | 1.6 | 53 |
| IC #1 | 151 | 86 | 1.8 | 47 |
| IC #2 | 196 | 105 | 1.9 | NDd |
Each experiment represents results obtained with a different set of RNA preparations. RNA I genes were present on pAM401. The 19 stop construct contains a termination codon at the 19th codon of tst. In the IC mutant, the IC is changed from GUG to GCG. Time points used were 0, 60, 120, and 240 min.
RNA II was provided on pDAK611.
Strains contained pDL278.
ND, not done.
The half-life of RNA I on the pAD1 minireplicon pDAK2300K containing wild-type par was determined to be 60 ± 7 min (Table 3), close to what was observed for the 19-stop mutation and somewhat less than that for the IC mutation in the absence of RNA II. This is probably due to the fact that RNA II is degraded relatively rapidly in cells containing pDAK2300K and that RNA I is relatively free of RNA II for the majority of the time required for determining its half-life.
par-Encoded RNAs are present at an approximately 1:1 ratio at steady state.
The stabilization of both par RNAs in complex suggested a model of par function where most of the RNA I and RNA II in plasmid-containing cells was in complex. In other words, even though RNA II might be expected to be transcribed in excess over RNA I to ensure suppression of Fst toxin translation, at steady state the ratio of RNA I:RNA II should be close to 1:1, since most of the excess RNA II would be degraded. Evidence of a substantial excess of either RNA would suggest that some factor other than RNA stability controls Fst toxin expression, e.g., formation of a translationally inactive pool of the toxin message as in the hok/sok system (15, 40). To test this hypothesis, the relative levels of RNA I and RNA II were determined by quantitative Northern blotting and RPA.
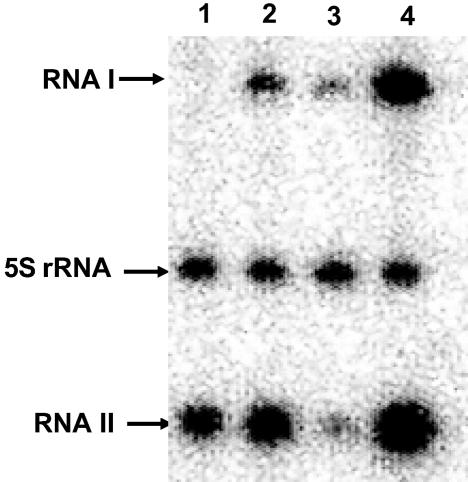
For quantitative Northern blots, RNA samples were prepared from OG1X cells containing the pAD1 miniplasmid pDAK2300K, which provides par at the wild-type copy number. Blots were probed with oligonucleotides specific for RNA I and RNA II that were designed to have equivalent melting temperatures and labeled to equal specific activities. The results are shown in Fig. 4. The ratio of RNA I to RNA II ranged from 1:1.01 to 1:1.08. Similar results were obtained in RPA experiments. With standard curves generated with known amounts of in vitro-produced RNA I and RNA II, the amount of RNA I in OG1X(pDAK2300K) was determined to be 1.22 × 10−9± 2.68 × 10−10 mol of RNA I/g of total RNA, and the amount of RNA II was determined to be 1.33 × 10−9± 4.97 × 10−10 mol of RNA II/g of total RNA purified from OG1X(pDAK2300K), for a ratio of 1:1.09 RNA I to RNA II. Measuring the number of colony-forming units per milliliter derived from the culture prior to RNA purification as described in Materials and Methods and the known concentration of the RNA preparation used for RPA analysis, a concentration of ∼40 molecules/cell was derived. This assumes that 100% of cellular RNA was recovered during purification and therefore represents a lower limit of RNA content. Repeated experiments in which harvested vector-containing cells were spiked with known concentrations of RNA I and RNA II prior to RNA purification showed that 60 to 75% of RNA was routinely lost during the purification process, suggesting a range of 100 to 150 molecules per cell.
FIG. 4.
Quantitative Northern blot analysis of par RNAs. Total RNA harvested from OG1X(pDAK2300K) (1.5 μg) was fractionated and transferred to a nylon membrane. Lane 1 was probed with oligonucleotides complementary to RNA I and RNA II. Lane 2 was probed with RNA I only. Lane 3 was probed with RNA II only. Densitometry revealed the following intensities for each lane. Lane 1, 2,231 net counts for RNA I and 2,253 net counts for RNA II; lane 2, 2,801 net counts for RNA I; lane 3, 3,041 net counts for RNA II.
The RNA I-RNA II complex does not spontaneously dissociate in vitro.
The results presented above indicated that most of the par-encoded RNAs exist in a rather stable complex in plasmid-containing cells. However, when the plasmid is lost RNA II must be removed from the complex to allow translation of fst from RNA I. Two alternative mechanisms of RNA II removal are possible. Either RNA II passively dissociates from the complex at a defined rate or RNA II is actively removed from the complex, perhaps by a specific cellular RNase(s). To test the former hypothesis, we attempted to determine the dissociation rate of preformed RNA I:RNA II complex in vitro. First, preformed complex made with a 20-fold molar excess of unlabeled RNA I relative to labeled RNA II was challenged with increasing concentrations of unlabeled RNA II. Dissociation was to be measured as the displacement of labeled RNA II from the preformed complex. However, even at a 1,000-fold molar excess of unlabeled RNA II, no dissociation of labeled RNA II from the preformed complex was observed (data not shown).
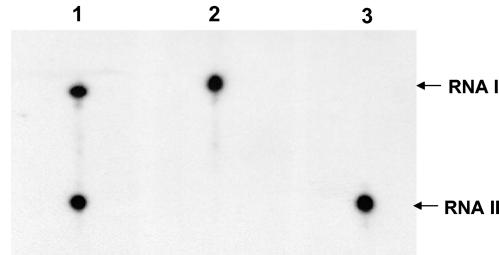
To further examine the resistance of the complex to dissociation, preformed complex was produced with unlabeled RNA II and RNA I at various molar ratios ranging from 1:1 to 10:1. The complexes were then challenged with an excess of labeled RNA II. It was reasoned that at the lower ratios of RNA II to RNA I, not all of the RNA I would be in complex and that some shifting of labeled challenge RNA II would be observed. The amount of shifted RNA would decrease with increasing RNA II:RNA I ratios in complex. If dissociation of the complex occurred at an appreciable rate, increasing the amount of challenge RNA II would result in a proportional increase in the amount of shifted RNA, due to greater competition. However, if there was no appreciable dissociation, challenge with higher concentrations of labeled RNA would not result in more shifting because the amount of available free RNA I would be the same. The results of this experiment are shown in Fig. 5. As expected, a decrease in shifting of challenge RNA II was observed with increasing RNA II:RNA I ratios in preformed complex. However, when the amount of labeled challenge RNA II was increased from a 10-fold molar excess to a 100-fold molar excess relative to RNA I, no increase in shifting occurred, suggesting that challenge RNA II could not displace unlabeled RNA II already in complex with RNA I. Therefore, there was no detectable dissociation of preformed RNA I-RNA II complex in vitro.
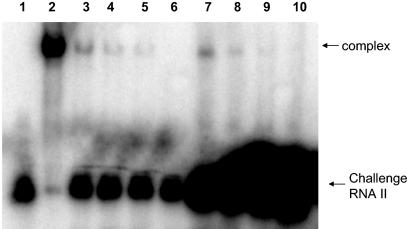
FIG. 5.
Dissociation of RNA I-RNA II complex in vitro. In vitro-produced RNA I and RNA II were allowed to form complexes at various ratios. The preformed complexes were then challenged with a 10-fold (lanes 3 to 6) or 100-fold (lanes 7 to 10) molar excess of labeled RNA II relative to RNA I. Lane 1, labeled RNA II; lane 2, labeled RNA II with unlabeled RNA I at a 1:1 ratio; lanes 3 to 6, unlabeled preformed RNA I-RNA II complex at 1:1, 1:2.5, 1:5, and 1:10 ratios, respectively, challenged with a 10-fold molar excess of labeled RNA II relative to RNA I; lanes 7 to 10, unlabeled preformed RNA I-RNA II complex at 1:1, 1:2.5, 1:5, and 1:10 ratios, respectively, challenged with 100-fold molar excess of labeled RNA II relative to RNA I. The large black spot at the bottom of lanes 7 to 10 is due to the 10-fold-larger amounts of labeled challenge RNA II used in these assays than in those shown in lanes 3 to 6. Counts from the shifted bands as determined by ImageQuant software are as follows: lane 3, 518,812; lane 4, 110,869; lane 5, 47,118; lane 7, 422,190; lane 8, 132,348; lane 9, 51,742.
DISCUSSION
The results presented in this report allowed the following conclusions. (i) The antisense RNA of the par addiction system, RNA II, was stabilized fourfold in the presence of RNA I, suggesting that the RNA I-RNA II complex has a half-life of at least 15 to 16 min. (ii) RNA I was not specifically targeted for degradation by complex formation as is observed with most other negatively regulated antisense RNA systems. (iii) Increasing expression of the par RNAs by increasing gene dosage resulted in an increased half-life of the RNA I-RNA II complex, suggesting that some factor required for complex degradation is limiting. (iv) RNA I and RNA II were present at a 1:1 ratio under natural conditions, consistent with the hypothesis that most par RNAs in the cell exist in complex and that any excess RNA II transcribed is rapidly degraded. (v) RNA II did not spontaneously dissociate from the par RNA complex at a detectable rate in vitro.
The requirements for antisense RNA-regulated postsegregational killing (PSK) systems are somewhat different than other negatively regulated antisense RNA systems. Translational repression of the toxin gene is required, but the antisense RNA cannot rapidly bind and target the toxin message for degradation because no mRNA would be left to kill plasmid-free segregants. In the well-studied hok/sok system, this problem is solved by separating translational inhibition and mRNA degradation functions. Translational inhibition is accomplished by hok RNA intramolecular structures that also inhibit sok binding, allowing a pool of hok to form in plasmid-containing cells (40). Slow processing of hok leads to isomerization of the message that allows sok binding and targeted degradation by RNase III. Interestingly, the sok antisense RNA is stabilized in the presence of its hok target but only in an rnc (RNase III) mutant (18).
Our results point to a different solution to this problem for the pAD1-encoded par PSK. A working model for the mechanism of par-mediated PSK based on these results is presented in Fig. 6. As RNA I is transcribed, the 5′ stem-loop sequesters the fst Shine-Dalgarno sequence, temporarily preventing translation of fst (21). fst translation is further repressed by the binding of RNA II (21), but instead of targeting RNA I for RNase degradation, RNA II and RNA I form a persistent complex that allows a pool of par RNAs to accumulate in the cell. Excess RNA II is rapidly degraded by a cellular RNase. The RNA I-RNA II complex is attacked more slowly, either by a different RNase or by the same RNase acting less efficiently. The removed RNA II is degraded, but in plasmid-containing cells a small excess of RNA II is maintained to replace the degraded RNA. The surplus of RNA II in plasmid-containing cells is small but may be sufficient given the inefficiency of RNA I translation in the presence of the 5′ stem-loop (21). In plasmid-free segregants, the surplus RNA II is rapidly depleted, and RNA II removed from the complex cannot be replaced. The resulting translation of fst from the released RNA I kills the cell by disrupting the cell membrane (52).
FIG. 6.
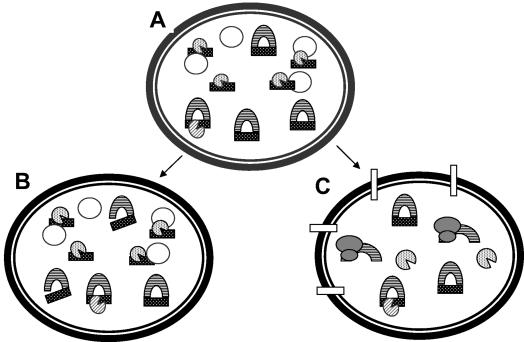
Model of the regulation of par PSK. In plasmid-containing cells (A) most RNA I (lined crescent) and RNA II (hatched rectangle) are in complex, as suggested by their 1:1 ratio observed in cells carrying wild-type par. Excess RNA II may be transcribed in order to ensure translational repression of RNA I, but any RNA II not in complex is rapidly degraded by a cellular RNase (hatched Pac-Man). RNA II is more slowly removed from the RNA I-RNA II complex, perhaps by a different RNase (lined Pac-Man). In cells that retain the plasmid (B), RNA II removed from the complex is rapidly replaced by newly synthesized RNA II. However, in cells that lose the plasmid (C), RNA II cannot be replaced, RNA I is translated by ribosomes (bilobed shaded shapes), and the toxin kills the cell (rectangles in the membrane). Empty circles represent pAD1 DNA.
The postulated role of a cellular RNase— that it removes RNA II to activate fst translation—is based on the in vitro dissociation experiments. Since we cannot prove at this time that the conditions prevailing in vivo are sufficiently similar to those used in vitro, we cannot rule out spontaneous dissociation of RNA II from the complex. Therefore, this part of the model should be considered tentative. However, the requirement of some cellular activity which, perhaps inefficiently, removes RNA II from the par RNA complex would explain the increased stability of the complex with increasing intracellular concentration. It should be noted that in spite of the prolonged stability of RNA II, par still functions to segregationally stabilize pAM401 in OG1X cultures with a concomitant loss of culture growth rate (50). Whether this is due to cell killing several generations after plasmid loss or activation of a small subset of total RNA I present is currently unclear.
Another possible explanation for the lower stability of RNA II produced from the pAD1 miniplasmids is that some pAD1 replicon-encoded function outside of par facilitates its degradation. The smallest pAD1 miniplasmid used in this study, pDAK2300K, encodes only five genes other than par, a replication initiator, a ParAB-type partition system, and two open reading frames (ORFs) of unknown function (51). An insertional mutation in the ORF closest to par, orfD, does not affect segregational stability of the replicon (51) or decay of RNA II (data not shown). prgN on pCF10, a homolog of the remaining ORF, orfE, has been implicated in negative control of conjugation (26). While the possibility that one or more of these genes might act on the par RNAs cannot be ruled out, considering the known functions of the gene products, it seems unlikely that they would play such a role.
RNA I is an extraordinarily stable RNA, with a minimum half-life of 45 min in the absence of RNA II and a half-life of around 1 h in its natural par context. Examination of the previously determined RNA I structure suggests possible reasons for its unusual stability (22). At the 3′ end, the complementarity within the terminator stem-loop extends to include the entire polyuridine stretch required for rho-independent termination. The result is a flush double-stranded RNA 3′ end that may be difficult for 3′-to-5′ exoribonucleases or poly-A polymerase to access. In contrast, RNA II has a three-uridine extension at the 3′ end that is not sequestered within the terminator stem. The 5′ end of RNA I is also tightly constrained within structural elements, including a 7-bp helix that may stack on the 5′ stem-loop that sequesters the ribosome-binding site. This structure leaves only two free nucleotides at the 5′ end. RNA II also has only two free nucleotides at its 5′ end, but in this case the constraining helix is only 4 bp long and there is no potential stacking stem. When RNA II binds to RNA I, however, the 4-bp intramolecular helix is replaced by a 14-bp intermolecular helix that involves all but the terminal 5′ nucleotide. Structural elements at the 5′ and 3′ ends of RNAs have been shown to inhibit degradation in both E. coli and Bacillus subtilis (6, 24, 38, 54). The complex must also adopt a conformation that is resistant to RNase III degradation, perhaps due to the dispersed nature of the complementary regions. Inhibition of ribosome binding by mutation of the IC does not appear to affect the stability of RNA I. This is perhaps not surprising, since ribosome binding to RNA I is inefficient due to the 5′ stem-loop which sequesters the Shine-Dalgarno sequence (21) and since the functionality of RNA I depends on persistence in the absence of ribosome binding to kill cells only after the coding plasmid is lost. RNA decay in gram-positive bacteria appears to be significantly different than in E. coli (11), and further study of the determinants of stability of RNA I and the par RNA complex will help further elucidate these mechanisms.
Acknowledgments
We acknowledge the technical assistance of Shirisha Reddy, Tony Greenfield, and Richard Duman and the helpful discussions of David Bechhofer.
This work was supported by Public Health Service grant GM55544, the Parson's Fund, National Science Foundation/EPSCoR Grant EPS-0091948, and the State of South Dakota.
REFERENCES
- 1.Andersen, J., S. Forst, K. Zhao, M. Inouye, and N. Delihas. 1989. The function of micF RNA. micF RNA is a major factor in the thermal regulation of OmpF protein in Escherichia coli. J. Biol. Chem. 264:17961-17970. [PubMed] [Google Scholar]
- 2.Asano, K., and K. Mizobuchi. 1998. Copy number control of IncIα plasmid ColIb-P9 by competition between pseudoknot formation and antisense RNA binding at a specific RNA site. EMBO J. 17:5201-5213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl. 1998. Current protocols in molecular biology, vol. 1. John Wiley & Sons, Boston, Mass.
- 4.Bass, B. L. 2000. Double-stranded RNA as a template for gene silencing. Cell 101:235-238. [DOI] [PubMed] [Google Scholar]
- 5.Blomberg, P., E. G. H. Wagner, and K. Nordstrom. 1990. Control of replication of plasmid R1: the duplex between antisense RNA, CopA and its target, CopT, is processed specifically in vivo and in vitro by RNase III. EMBO J. 9:2331-2340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bouvet, P., and J. G. Belasco. 1992. Control of RNase E-mediated degradation by 5′-terminal base pairing in E. coli. Nature 360:488-491. [DOI] [PubMed] [Google Scholar]
- 7.Brantl, S. 2002. Antisense RNAs in plasmids: control of replication and maintenance. Plasmid 48:165-173. [DOI] [PubMed] [Google Scholar]
- 8.Brantl, S. 2002. Antisense-RNA regulation and RNA interference. Biochim. Biophys. Acta 1575:15-25. [DOI] [PubMed] [Google Scholar]
- 9.Brantl, S., E. Birch-Hirschfeld, and D. Behnke. 1993. RepR protein expression on plasmid pIP501 is controlled by an antisense RNA-mediated transcription attenuation mechanism. J. Bacteriol. 175:4052-4061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Case, C. C., E. L. Simons, and R. W. Simons. 1990. The IS10 transposase mRNA is destabilized during antisense RNA control. EMBO J. 9:1259-1266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Condon, C. 2003. RNA processing and degradation in Bacillus subtilis. Microbiol. Mol. Biol. Rev. 67:157-174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Delihas, N. 1995. Regulation of gene expression by trans-encoded antisense RNAs. Mol. Microbiol. 15:411-414. [DOI] [PubMed] [Google Scholar]
- 13.Dunny, G. M., B. L. Brown, and D. B. Clewell. 1978. Induced cell aggregation and mating in Streptococcus faecalis: evidence for a bacterial sex pheromone. Proc. Natl. Acad. Sci. USA 75:3479-3483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fire, A., S. Xu, M. K. Montgomery, S. A. Kostas, S. E. Driver, and C. C. Mello. 1998. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature 391:806-811. [DOI] [PubMed] [Google Scholar]
- 15.Franch, T., and K. Gerdes. 1996. Programmed cell death in bacteria: translational repression by mRNA end-pairing. Mol. Microbiol. 21:1049-1060. [DOI] [PubMed] [Google Scholar]
- 16.Franch, T., A. P. Gultyaev, and K. Gerdes. 1997. Programmed cell death by hok/sok of plasmid R1: processing at the hok mRNA 3′-end triggers structural rearrangements that allow translation and antisense RNA binding. J. Mol. Biol. 273:38-51. [DOI] [PubMed] [Google Scholar]
- 17.Franch, T., M. Petersen, E. G. H. Wagner, J. P. Jacobsen, and K. Gerdes. 1999. Antisense RNA regulation in prokaryotes: rapid RNA/RNA interaction facilitated by a general U-turn loop structure. J. Mol. Biol. 294:1115-1125. [DOI] [PubMed] [Google Scholar]
- 18.Gerdes, K., A. Nielsen, P. Thorsted, and E. G. Wagner. 1992. Mechanism of killer gene activation. Antisense RNA-dependent RNase III cleavage ensures rapid turn-over of the stable hok, srnB and pndA effector messenger RNAs. J. Mol. Biol. 226:637-649. [DOI] [PubMed] [Google Scholar]
- 19.Gerdes, K., L. K. Poulsen, T. Thisted, A. K. Nielsen, J. Martinussen, and P. H. Andreasen. 1990. The hok killer gene family in gram-negative bacteria. New Biol. 2:946-956. [PubMed] [Google Scholar]
- 20.Gerdes, K., P. B. Rasmussen, and S. Molin. 1986. Unique type of plasmid maintenance function: postsegregational killing of plasmid-free cells. Proc. Natl. Acad. Sci. USA 83:3116-3120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Greenfield, T. J., E. Ehli, T. Kirshenmann, T. Franch, K. Gerdes, and K. E. Weaver. 2000. The antisense RNA of the par locus of pAD1 regulates the expression of a 33-amino-acid toxic peptide by an unusual mechanism. Mol. Microbiol. 37:652-660. [DOI] [PubMed] [Google Scholar]
- 22.Greenfield, T. J., T. Franch, K. Gerdes, and K. E. Weaver. 2001. Antisense RNA regulation of the par post-segregational killing system: structural analysis and mechanism of binding of the antisense RNA, RNAII and its target, RNAI. Mol. Microbiol. 42:527-537. [DOI] [PubMed] [Google Scholar]
- 23.Greenfield, T. J., and K. E. Weaver. 2000. Antisense RNA regulation of the pAD1 par post-segregational killing system requires interaction at the 5′ and 3′ ends of the RNAs. Mol. Microbiol. 37:661-670. [DOI] [PubMed] [Google Scholar]
- 24.Hambraeus, G., M. Persson, and B. Rutberg. 2000. The aprE leader is a determinant of extreme mRNA stability in Bacillus subtilis. Microbiology 146:3051-3059. [DOI] [PubMed] [Google Scholar]
- 25.Hayes, F. 2003. Toxins-antitoxins: plasmid maintenance, programmed cell death, and cell cycle arrest. Science 301:1496-1499. [DOI] [PubMed] [Google Scholar]
- 26.Hedberg, P. J., B. A. B. Leonard, R. E. Ruhfel, and G. M. Dunny. 1996. Identification and characterization of the genes of Enterococcus faecalis plasmid pCF10 involved in replication and in negative control of pheromone-inducible conjugation. Plasmid 35:46-57. [DOI] [PubMed] [Google Scholar]
- 27.Hershberg, R., S. Altuvia, and H. Margalit. 2003. A survey of small RNA-encoding genes in Escherichia coli. Nucleic Acids Res. 31:1813-1820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Holmes, D. S., and M. Quigley. 1981. A rapid boiling method for the preparation of bacterial plasmids. Anal. Biochem. 114:193-197. [DOI] [PubMed] [Google Scholar]
- 29.Ike, Y., R. A. Craig, B. A. White, Y. Yagi, and D. B. Clewell. 1983. Modification of Streptococcus faecalis sex pheromones after acquisition of plasmid DNA. Proc. Natl. Acad. Sci. USA 80:5369-5373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Krinke, L., and D. L. Wulff. 1990. RNase III-dependent hydrolysis of lambda cII-O gene mRNA mediated by lambda OOP antisense RNA. Genes Dev. 4:2223-2233. [DOI] [PubMed] [Google Scholar]
- 31.Lease, R. A., and M. Belfort. 2000. A trans-acting RNA as a control switch in Escherichia coli: DsrA modulates function by forming alternative structures. Proc. Natl. Acad. Sci. USA 97:9919-9924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.LeBlanc, D. J., L. N. Lee, and A. Abu-Al-Jaibat. 1992. Molecular, genetic, and functional analysis of the basic replicon of pVA380-1, a plasmid of oral streptococcal origin. Plasmid 28:130-145. [DOI] [PubMed] [Google Scholar]
- 33.Malmgren, C., H. M. Engdahl, P. Romby, and E. G. H. Wagner. 1996. An antisense/target RNA duplex or a strong intramolecular RNA structure 5′ of a translation initiation signal blocks ribosome binding: the case of plasmid R1. RNA 2:1022-1032. [PMC free article] [PubMed] [Google Scholar]
- 34.Masse, E., F. E. Escorcia, and S. Gottesman. 2003. Coupled degradation of a small regulatory RNA and its mRNA targets in Escherichia coli. Genes Dev. 17:2374-2383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Morfeldt, E., D. Taylor, A. von Gabain, and S. Arvidson. 1995. Activation of alpha-toxin translation in Staphylococcus aureus by the trans-encoded antisense RNA, RNAIII. EMBO J. 14:4569-4577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Novick, R. P., S. Iordanescu, S. J. Projan, J. Kornblum, and I. Edelman. 1989. pT181 plasmid replication is regulated by a countertranscript-driven transcriptional attenuator. Cell 59:395-404. [DOI] [PubMed] [Google Scholar]
- 37.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 38.Spickler, C., and G. A. Mackie. 2000. Action of RNase II and polynucleotide phosphorylase against RNAs containing stem-loops of defined structure. J. Bacteriol. 182:2422-2427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stougarrd, P., S. Molin, and K. Nordstrom. 1981. RNAs involved in copy-number control and incompatibility of plasmid R1. Proc. Natl. Acad. Sci. USA 78:6008-6012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Thisted, T., N. S. Sorensen, and K. Gerdes. 1995. Mechanism of post-segregational killing: secondary structure analysis of the entire Hok mRNA from plasmid R1 suggests a fold-back structure that prevents translation and antisense RNA binding. J. Mol. Biol. 247:859-873. [DOI] [PubMed] [Google Scholar]
- 41.Tomizawa, J. 1986. Control of ColE1 plasmid replication: binding of RNA I to RNA II and inhibition of primer formation. Cell 47:89-97. [DOI] [PubMed] [Google Scholar]
- 42.Tomizawa, J., T. Itoh, G. Selzer, and T. Som. 1981. Inhibition of ColE1 RNA primer formation by a plasmid-specified small RNA. Proc. Natl. Acad. Sci. USA 78:1421-1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.van Biesen, T., and L. S. Frost. 1994. The FinO protein of IncF plasmid binding FinP antisense RNA and its target, traJ mRNA, and promotes duplex formation. Mol. Microbiol. 14:427-436. [DOI] [PubMed] [Google Scholar]
- 44.Vogel, J., V. Bartels, T. H. Tang, G. Churakov, J. G. Slagter-Jager, A. Huttenhofer, and E. G. H. Wagner. 2003. RNomics in Escherichia coli detects new sRNA species and indicates parallel transcriptional output in bacteria. Nucleic Acids Res. 31:6435-6443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wagner, E. G. H., S. Altuvia, and P. Romby. 2002. Antisense RNAs in bacteria and their genetic elements. Adv. Genet. 46:361-398. [DOI] [PubMed] [Google Scholar]
- 46.Waldbeser, L. S., Q. Chen, and J. H. Crosa. 1995. Antisense RNA regulation of the fatB iron transport protein gene in Vibrio anguillarum. Mol. Microbiol. 17:747-756. [DOI] [PubMed] [Google Scholar]
- 47.Weaver, K. E., F. An, and D. B. Clewell. 1993. Identification, characterization, and nucleotide sequence of a region of Enterococcus faecalis pheromone-responsive plasmid pAD1 capable of autonomous replication. J. Bacteriol. 175:1900-1909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Weaver, K. E., and D. B. Clewell. 1991. Control of Enterococcus faecalis sex pheromone cAD1 elaboration: effects of culture aeration and pAD1 plasmid-encoded determinants. Plasmid 25:177-189. [DOI] [PubMed] [Google Scholar]
- 49.Weaver, K. E., and D. B. Clewell. 1988. Regulation of the pAD1 sex pheromone response in Enterococcus faecalis: construction and characterization of lacZ transcriptional fusions in a key control region of the plasmid. J. Bacteriol. 170:4343-4352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Weaver, K. E., K. D. Jensen, A. Colwell, and S. I. Sriram. 1996. Functional analysis of the Enterococcus faecalis plasmid pAD1-encoded stability determinant par. Mol. Microbiol. 20:53-63. [DOI] [PubMed] [Google Scholar]
- 51.Weaver, K. E., and D. J. Tritle. 1994. Identification and characterization of an Enterococcus faecalis plasmid pAD1-encoded stability determinant which produces two small RNA molecules necessary for its function. Plasmid 32:168-181. [DOI] [PubMed] [Google Scholar]
- 52.Weaver, K. E., D. M. Weaver, C. L. Wells, C. M. Waters, M. E. Gardner, and E. A. Ehli. 2003. Enterococcus faecalis plasmid pAD1-encoded Fst toxin affects membrane permeability and alters cellular responses to lantibiotics. J. Bacteriol. 185:2169-2177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wirth, R., F. An, and D. B. Clewell. 1987. Highly efficient cloning system for Streptococcus faecalis protoplast transformation, shuttle vectors, and applications, p. 25-27. In J. J. Ferretti and R. Curtiss III (ed.), Streptococcal genetics. American Society for Microbiology, Washington, D.C.
- 54.Wong, H. C., and S. Chang. 1986. Identification of a positive retroregulator that stabilizes mRNAs in bacteria. Proc. Natl. Acad. Sci. USA 83:3233-3237. [DOI] [PMC free article] [PubMed] [Google Scholar]