Abstract
In Pseudomonas aeruginosa, the antibiotic dihydroaeruginoate (Dha) and the siderophore pyochelin are produced from salicylate and cysteine by a thiotemplate mechanism involving the peptide synthetases PchE and PchF. A thioesterase encoded by the pchC gene was found to be necessary for maximal production of both Dha and pyochelin, but it was not required for Dha release from PchE and could not replace the thioesterase function specified by the C-terminal domain of PchF. In vitro, 2-aminobutyrate, a cysteine analog, was adenylated by purified PchE and PchF proteins. In vivo, this analog strongly interfered with Dha and pyochelin formation in a pchC deletion mutant but affected production of these metabolites only slightly in the wild type. Exogenously supplied cysteine overcame the negative effect of a pchC mutation to a large extent, whereas addition of salicylate did not. These data are in agreement with a role for PchC as an editing enzyme that removes wrongly charged molecules from the peptidyl carrier protein domains of PchE and PchF.
Under iron-limiting growth conditions, aerobic bacteria produce siderophores which form complexes with Fe3+ in the environment and deliver it, via specific outer membrane receptors, to the bacterial cytoplasm. The two major siderophores made by Pseudomonas aeruginosa are pyoverdin (10, 23) and pyochelin (8, 34), both of which contribute to the virulence of this opportunistic human pathogen (9, 24, 43). The biosynthetic genes of the aryl-peptide pyochelin (32, 33, 39, 40) are clustered with the pyochelin receptor gene fptA (1) and the regulatory gene pchR (21) on the P. aeruginosa chromosome. The biosynthetic genes are organized in two divergent operons, pchDCBA and pchEFGHI. In the presence of iron, pchDCBA, pchEFGHI, and fptA are repressed by the Fur protein (1, 32, 40). During iron limitation, the biosynthesis of pyochelin and its receptor protein is induced by a positive feedback loop involving pyochelin and the transcriptional regulator PchR (17, 21, 32).
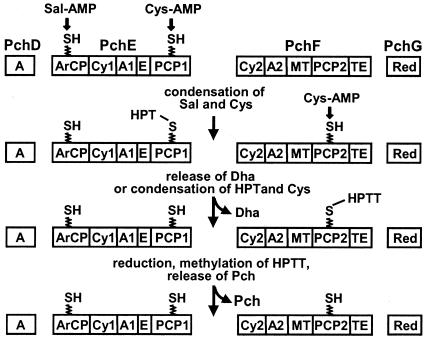
Pyochelin is made from salicylate and two molecules of cysteine by a thiotemplate mechanism on a four-protein, 12-domain assembly line (11, 27) (Fig. 1). The salicylate moiety is generated from chorismate by PchA (isochorismate synthase) (15) and PchB (isochorismate pyruvate-lyase) (14, 39) and is subsequently adenylated by PchD (31). Adenylation of l-cysteine is catalyzed by the nonribosomal peptide synthetase PchE, which then covalently links both activated substrates, as thioesters, to two posttranslationally added 4′-phosphopantetheine prosthetic groups on PchE. Condensation, epimerization, and cyclization reactions, also catalyzed by PchE, generate the enzyme-bound intermediate hydroxyphenyl-thiazoline (HPT), which can be released by slow hydrolysis of the thioester bond, to produce dihydroaeruginoate (Dha) (28, 31). This metabolite is found in small amounts in culture supernatants of P. aeruginosa (40). The 4′-phosphopantetheinylated (primed) form of the peptide synthetase PchF adenylates a second molecule of l-cysteine and, like PchE, links it covalently via a thioester. PchF subsequently catalyzes the condensation of the PchE-bound intermediate HPT with cysteinyl-PchF and thereby generates the second thiazoline ring, which is reduced by the PchG reductase and N-methylated by a tailoring domain of PchF. Hydrolysis of the remaining thioester bond finally releases pyochelin from PchF (27, 33).
FIG. 1.
Model for PchDEFG-dependent biosynthesis of pyochelin from salicylate in P. aeruginosa (adapted from references 27, 28, 31, and 33). Adenylated forms of salicylate (Sal) and l-cysteine (Cys) are loaded onto the peptide synthetases PchE and PchF via covalent thioester linkages. PchE-dependent condensation of salicylate with l-cysteine, followed by epimerization and cyclization reactions, generates the intermediate HPT, which can be released from PchE to give Dha or can be coupled to PchF-bound l-cysteine to give hydroxyphenyl-bis-thiazoline (HPTT). After reduction of HPTT by PchG and methylation by PchF, pyochelin (Pch) is released by thioester cleavage. Functional domains are indicated for adenylation (A, A1, and A2), cyclization (Cy1 and Cy2), epimerization (E), thiolation (ArCP for the aryl carrier protein domain and PCP1 and PCP2 for peptidyl carrier protein domains), reduction (Red), methyl transfer (MT) and thioesterase (TE) activity.
The pch gene cluster encodes two thioesterases. One of these thioesterases resides in the C-terminal thioesterase domain of PchF (internal thioesterase) (Fig. 1) and releases pyochelin from PchF (31). The other, a type II thioesterase (external thioesterase), is encoded by the pchC gene (40), and its function has not been studied previously. Thioesterases carry two conserved sequences: GXSXG, a motif also present in the active sites of serine proteases, lipases, and acyltransferases; and GXHF, which is located approximately 140 amino acids further downstream and whose His residue is essential for catalytic activity (29, 36, 48). Both motifs occur in the thioesterase domain of PchF (GYCSGX176GGHF [31]) and in PchC (GHSLGX126GGHF [40]). Mutations in the first, integrated thioesterase motif of PchF prevent pyochelin release from PchF in vitro (31). The purpose of the present study was to examine the role of the second, external thioesterase, PchC.
In prokaryotic polyketide or nonribosomal peptide syntheses, it is common to find a distinct, external type II thioesterase. Mutational loss of the external thioesterase generally results in a reduced amount of product, and it has been suggested that such enzymes could either assist in product release from the thiotemplate or play a role as editing enzymes that remove wrongly charged substrates or aberrant intermediates from the enzyme complex (19, 36, 37). In vitro, type II thioesterases can regenerate misacylated peptide synthetases resulting from erroneous priming or from loading of an amino acid which cannot be processed (37, 51). Here we provide evidence for such a proofreading role for the type II PchC thioesterase of P. aeruginosa in vivo, which results in optimized Dha and pyochelin production.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
Bacterial strains used in this study are listed in Table 1. Bacteria were usually grown on nutrient agar and in nutrient yeast broth (42) at 37°C. To quantify salicylate, Dha, and pyochelin in culture supernatants of P. aeruginosa, strains were grown in GGP medium (6). Antibiotics, when required, were added to the growth media at the following concentrations: tetracycline, 25 μg/ml for Escherichia coli and 100 μg/ml for P. aeruginosa; ampicillin, 100 μg/ml for E. coli; carbenicillin, 250 μg/ml for P. aeruginosa; kanamycin, 25 μg/ml for E. coli; and gentamicin, 10 μg/ml for E. coli and P. aeruginosa. To counterselect E. coli donor cells in matings with P. aeruginosa, chloramphenicol was used at a concentration of 10 μg/ml. 2-Aminobutyrate was purchased from Acros Organics.
TABLE 1.
Strains and plasmids
| Strain or plasmid | Relevant characteristics | Reference or source |
|---|---|---|
| E. coli S17-1 | thi pro hsdR recA; chromosomal RP4 (Tra+ Tcs Kms Aps) | 41 |
| P. aeruginosa strains | ||
| PALS128 | pvdB | 45 |
| PALS128-6 | pvdB pchA::ω (insertion of 35 bp) | 39 |
| PAO1 | Wild type | ATCC 15692 |
| PAO6331 | Δpch (= ΔpchDCBA ΔpchR ΔpchEFGHI) | 33 |
| PAO6339 | ΔpchC | This study |
| PAO6342 | pvdB ΔpchC | This study |
| PAO6357 | pvdB pchA::ω ΔpchC | This study |
| Plasmids | ||
| pBLS II KS | Cloning vector, Apr | Stratagene |
| pPchE | pchE overexpression construct; Kmr | 31 |
| pPchF | pchF overexpression construct; Kmr | 31 |
| pPchFTE | pchF overexpression construct with C1606A/S1607A mutations; Kmr | 31 |
| pME3087 | Suicide vector; ColE1 replicon; Tcr; EcoRI-KpnI-BamHI-XbaI-PstI-SphI-HindIII polylinker | 46 |
| pME3318 | pQF10 derivative carrying pchDCBA | 40 |
| pME6001 | pBBR1-based cloning vector; Gmr | 4 |
| pME6012 | pVS1-p15A shuttle vector; Tcr | 20 |
| pME6178 | pME6001 carrying entD pchDCBA under Plac control | 33 |
| pME6488 | pME6012 carrying pchEFG under Pkan control | 33 |
| pME6815 | Suicide plasmid for construction of pchC deletion; derivative of pME3087 containing the 3′ part of pchD, pchC with an in-frame deletion, and the 5′ part of pchB on a 1.0-kb BamHI-HindIII fragment | This study |
| pME6831 | pUCPSK carrying pchDC on a 2.8-kb EcoRI-ScaI fragment from pME3318 | This study |
| pME6852 | pME6001 carrying entD pchDBA under Plac control (same as pME6178 but with in-frame deletion in pchC) | This study |
| pME6886 | pME6012 carrying pchEFC1606A/S1607AG under Pkan control (same as pME6488 but with mutations affecting the PchF thioesterase motif) | This study |
| pME7538 | pUCPSK carrying pchDCS90A (same as pME6831 but with mutation affecting the thioesterase motif) | This study |
| pUCSK | ColE1-pRO1600 shuttle vector; Apr | 47 |
| pUK21 | Cloning vector; Kmr | 44 |
Construction of plasmids and gene replacement mutants.
All plasmids used in this study are listed in Table 1. The suicide plasmid pME6815 used to create an in-frame deletion in the chromosomal pchC gene was constructed as follows. A 0.9-kb XmaI-ScaI fragment carrying most of the pchC gene and the 5′ part of pchB was cloned into pBluescript (pBLS II KS) between XmaI and EcoRV sites. This plasmid was used as a template in an inverse PCR performed with PchC1 (5′-CTGACCCGGGCCTCGCACATCCCGAC; XmaI site underlined) and PchC2 (5′-GAAGCGGTCCTCGCGACCCGGG; XmaI site underlined) as the primers to amplify a 3.3-kb fragment. The PCR product was cleaved with XmaI and religated. From the resulting plasmid, a 0.32-kb XmaI-HindIII fragment was excised, ligated to a 0.65-kb fragment carrying the 5′ part of pchC (upstream of the XmaI site) and part of the flanking pchD gene, and cloned into the suicide vector pME3087 between BamHI and HindIII sites. The resulting suicide plasmid, pME6815, carried the flanking regions of pchC and a mutated pchC gene which lacked codons 53 to 218. For construction of chromosomal pchC deletions in P. aeruginosa PAO1, PALS128, and PALS128-6, pME6815 was mobilized from E. coli S17-1 to the recipient P. aeruginosa strains and chromosomally integrated with selection for tetracycline resistance. Excision of the vector via a second crossing over was accomplished by enrichment for tetracycline-sensitive cells as described previously (50). The resulting pchC mutants, PAO6339, PAO6342, and PAO6357, were checked by Southern blot analysis (data not shown). To generate the same mutation in the Plac-entDpchDBA expression plasmid pME6852, the 671-bp XmaI-EcoRV fragment of pME6178 was replaced by the corresponding XmaI-EcoRV fragment of pME6815 (173 bp) carrying the pchC deletion.
The thioesterase motif in the pchC gene of plasmid pME7538 was mutated by overlap extension (25) as follows. Two 0.3-kb fragments, fragments A and B, were amplified by PCR from chromosomal DNA of PAO1 by using primer PchCmut1 (5′-CTGCGCGAGCGCCTGCGCCGCGAGC) plus primer PchCmut6 (5′-CAGCGCCGCGCCGAGGGCGTGGCCGAACAG) and primer PchCmut5 (5′-GCGCTGTTCGGCCACGCCCTCGGCGCGGCG) plusprimer PchCmut4 (5′-GACTTCCTCGTCGTGCTCGCCGAGG), respectively. PchCmut6 and PchCmut5 are complementary to each other to a large extent and specify a serine-to-alanine mutation in codon 90 (underlined). Equimolar amounts of PCR fragments A and B served as templates for the subsequent PCR amplification with primers PchCmut1 and PchCmut4. From the 0.6-kb PCR product obtained, an internal 0.3-kb XmaI-XhoI fragment was excised and used to replace the corresponding wild-type fragment in pME6831, resulting in the Plac-pchDCS90A expression plasmid pME7538.
Plasmid pME6886, which expresses pchEFC1606A/S1607AG under Pkan control and carries a mutation in the PchF thioesterase motif, was constructed by replacing the 311-bp SalI-NotI fragment of pME6488 with the corresponding fragment from pPchFTE. All constructs carrying pchC in-frame deletions or point mutations in pchC or pchF were verified by sequence analysis.
DNA manipulation and cloning procedures.
DNA manipulations were carried out as described by Sambrook et al. (35). Small-scale preparation of plasmid DNA was carried out by the cetyltrimethylammonium bromide method (12), and large-scale preparation was performed by using JetStar-Tips (Genomed, Basel, Switzerland). Chromosomal DNA was prepared by the method of Gamper et al. (16). DNA fragments were purified from agarose gels with a Gene Clean DNA extraction kit (Bio 101, La Jolla, Calif.). Transformation of E. coli and P. aeruginosa was carried out by electroporation (13). Nucleotide sequences were determined with a Big Dye terminator cycle sequencing kit and an ABI-PRISM 373 automatic sequencer (Applied Biosystems). Nucleotide sequences were analyzed with programs of the University of Wisconsin Genetics Computer Group package (version 9.1).
Identification of salicylate, Dha, and pyochelin in culture supernatants of P. aeruginosa.
P. aeruginosa strains were grown in GGP medium for the time indicated. For high-pressure liquid chromatography (HPLC) analysis, ethyl acetate extracts of acidified culture supernatants were dried by evaporation, dissolved in 60% (vol/vol) methanol-10 mM H3PO4, and injected into an HPLC system as described previously (32). Compounds were identified by their retention times and UV spectra. Dha and salicylate were quantified at 256 and 237 nm, respectively. Pyochelin, which exists as a mixture of two interconvertible isomers, pyochelin I and pyochelin II (34), was quantified at 258 and 254 nm, respectively.
ATP-32PPi exchange reactions.
PchE and PchF proteins used in the assays were overexpressed and purified in E. coli and were converted to their phosphopantetheinyl holoforms as described previously (30, 31). ATP-PPi exchange reactions were performed by using previously described procedures (31); the reaction mixtures contained 100 nM PchE and 100 nM PchF, respectively, and the amino acid tested at a concentration of 1 mM. Incubation was at 30°C for 20 min.
RESULTS
Inactivation of the PchC thioesterase decreases production of Dha and pyochelin in P. aeruginosa. To study the role of PchC, we constructed a chromosomal in-frame deletion in its gene and evaluated the impact of this on the production of Dha and pyochelin. As shown in Table 2, the production of both metabolites was significantly reduced in the pchC mutant PAO6339 compared to the production in the wild-type strain, PAO1, and as a consequence, large amounts of the precursor salicylate accumulated. When the PAO6339 mutant was complemented with plasmid pME6831 carrying pchDC, production of Dha and pyochelin was restored to the wild-type level. By contrast, plasmid pME7538, which is identical to pME6831 except for a point mutation (Cys90Ala) destroying the essential thioesterase motif of PchC, did not complement the PchC-negative phenotype of PAO6339 (Table 2). Note that, as reported previously, expression of pchC requires the presence of the upstream pchD gene in cis (40); pchD was therefore included in these plasmids.
TABLE 2.
Effects of a pchC mutation on the concentrations of salicylate, Dha, and pyochelin in culture supernatants of P. aeruginosaa
| Strain | Mutation | Plasmid | Genes carried | Salicylate concn (nmol/ml)b | Dha concn (nmol/ml)b | Pyochelin concn (nmol/ml)b |
|---|---|---|---|---|---|---|
| PAO1 | Wild type | <8 | 113 ± 14 | 1,047 ± 36 | ||
| PAO6339 | ΔpchC | 1,034 ± 263 | 45 ± 4 | 408 ± 13 | ||
| PAO6339 | ΔpchC | pME6831 | pchDC | <8 | 89 ± 4 | 1,010 ± 83 |
| PAO6339 | ΔpchC | pME7538 | pchDCS90A | 961 ± 218 | 10 ± 3 | 127 ± 41 |
GGP medium (30 ml) (6) was inoculated with 0.3-ml portions of cultures grown in the same medium. After incubation at 37°C and 220 rpm for 38 h, supernatants were extracted and analyzed for salicylate, Dha, and pyochelin by HPLC as described previously (33). The values are the means ± standard deviations for three parallel experiments.
Concentration in culture supernatant.
PchC cannot release pyochelin from its thiotemplate, PchF, and is not essential for Dha formation.
Since pyochelin is required for full transcriptional expression of the pchDCBA and pchEFGHI operons by positive autoregulation (32), it is important to express both operons under the control of constitutive promoters when the roles of individual genes in the pyochelin biosynthetic pathway are assessed. We therefore used a two-plasmid system described previously (32) with the deletion mutant PAO6331 lacking pchDCBA, pchR, and pchEFGHI. The E. coli entD gene encoding a phosphopantetheinyl transferase with relaxed specificity was coexpressed with the pchDCBA operon to ensure constitutive conversion of PchE and PchF to their phosphopantetheinyl holoforms (33). In this system pchDCBA and pchEFG are sufficient for pyochelin biosynthesis (33), but the amounts of pyochelin and Dha produced under these conditions are two- to threefold lower than the amounts in PAO1, probably because the kanamycin promoter used to drive pchEFG expression on pME6488 is weaker than the natural, pyochelin-inducible promoter of the pchEFGHI operon. As a consequence, salicylate is not quantitatively incorporated into Dha and pyochelin and accumulates to some extent in PAO6331 carrying pME6178 (entD pchDCBA) and pME6488 (pchEFG) (33) (Table 3). Pyochelin and Dha formation in PAO6331 carrying pME6852 (entD pchDBA) and pME6488 (pchEFG) was reduced threefold compared to the formation in PAO6331 carrying pME6178 and pME6488, confirming that pchC is important, but not essential, for Dha and pyochelin formation in P. aeruginosa. Increased amounts of salicylate accumulated in culture supernatants as a result of reduced incorporation into Dha and pyochelin.
TABLE 3.
Impact of thioesterases encoded by pchC and pchF on salicylate, Dha, and pyochelin formation in P. aeruginosaa
| Strain | Chromosomal mutation | Plasmids | Genes carried | Salicylate concn (nmol/ml)b | Dha concn (nmol/ml)b | Pyochelin concn (nmol/ml)b |
|---|---|---|---|---|---|---|
| PAO1 | Wild type | <8.0 | 60 ± 4.0 | 748 ± 25 | ||
| PAO6331 | Δpch | <8.0 | <1.2 | <2 | ||
| PAO6331 | Δpch | pME6178 + pME6488 | entD pchDCBA + pchEFG | 256 ± 29 | 18 ± 2.5 | 400 ± 74 |
| PAO6331 | Δpch | pME6852 + pME6488 | entD pchDBA + pchEFG | 636 ± 72 | 5 ± 0.3 | 120 ± 15 |
| PAO6331 | Δpch | pME6178 + pME6886 | entD pchDCBA + pchEFC1606A/S1607AG | 872 ± 194 | 81 ± 19.7 | <2 |
| PAO6331 | Δpch | pME6852 + pME6886 | entD pchDBA + pchEFC1606A/S1607AG | 757 ± 168 | 24 ± 1.9 | <2 |
GGP medium (30 ml) containing, when required, gentamicin (10 μg/ml) and tetracycline (100 μg/ml) was inoculated with 0.3-ml portions of cultures grown in the same medium. After incubation at 37°C and 220 rpm for 33 h, supernatants were extracted and analyzed for salicylate, Dha, and pyochelin by HPLC. The values are the means ± standard deviations for three parallel experiments.
Concentration in culture supernatant.
To investigate a potential role of PchC in the release of pyochelin from PchF, we tested whether the pchC gene was able to functionally complement a double point mutation (Cys1606Ala/Ser1607Ala) blocking the thioesterase function of pchF (31). As illustrated in Table 3, strain PAO6331 carrying pME6852 (entD pchDBA) and pME6886 (pchEFC1606A/S1607AG) was unable to produce pyochelin because of a lack of both thioesterases. When the entD pchDCBA construct pME6178 was used instead of pME6852, pyochelin was not produced either, indicating that PchC could not substitute for the mutated thioesterase function of PchF. Small amounts of Dha were detected in culture supernatants of the wild-type strain, PAO1, as well as in the recombinant strain lacking both thioesterases, probably as a result of nonspecific hydrolysis of the PchE-HPT thioester bond (31).
The recombinant strain lacking the pchF-encoded thioesterase (PAO6331 with pME6178 and pME6886) produced more Dha than the pyochelin-producing pchF+ strain (PAO6331 carrying pME6178 and pME6488) produced (Table 3). Similarly, PAO6331 expressing entD pchDCBA and pchE produces more Dha than its pyochelin-producing counterpart produces (33). It seems that nonspecific hydrolysis of the PchE-HPT thioester bond increases when the pyochelin assembly line is interrupted because of mutations in pchF. The fact that more Dha was excreted in the presence of PchC than in the absence of PchC may be explained by a positive effect of PchC on the catalytic efficiency of PchE (see below). In conclusion, PchC does not seem to be essential for releasing Dha from its thiotemplate, PchE.
The PchC-negative phenotype is suppressed by addition of cysteine.
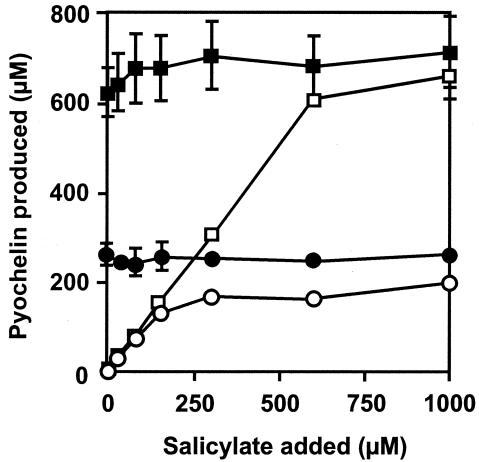
We next evaluated a potential role of PchC as an editing enzyme in case of mischarging of PchE and/or PchF during Dha and pyochelin production. Two sources of mischarging were considered: (i) PchD could activate endogenous or exogenously added salicylate analogs and deliver these molecules to the aryl carrier protein domain of PchE; (ii) PchE and PchF could adenylate and subsequently load amino acids other than l-cysteine onto their peptidyl carrier protein (PCP) domains. In both cases, correct assembly of Dha and pyochelin would be hampered, and thus the role of PchC could be to recognize and eliminate wrongly charged molecules by thioester cleavage. The first hypothesis was tested by measuring the influence of PchC on pyochelin formation in a salicylate-requiring mutant to which salicylate was added at different concentrations (10 μM to 1 mM) together with a large excess of various salicylate analogs, such as p-aminosalicylate, benzoate, p-hydroxybenzoate, p-aminobenzoate, or anthranilate. Pyochelin formation was measured semiquantitatively on CAS agar, which allows the production of siderophores to be visualized by the formation of an orange halo around the colonies (38). Since the siderophore pyoverdin would interfere in this assay, we used a pvdB (pyoverdin-negative) pchA strain, PALS128-6, and its ΔpchC derivative PAO6357. None of the salicylate analogs tested reduced the diameter of the halo around the bacterial colonies, regardless of the presence of pchC, suggesting that pyochelin formation was not inhibited (data not shown). If the role of PchC were indeed to remove an endogenous mischarged salicylate analog from the aryl carrier protein domain of PchE, then an excess of salicylate should competitively inhibit mischarging and thereby suppress the negative effect of a pchC mutation. We therefore tested whether the PchC-negative phenotype could be suppressed by salicylate addition. Whereas pyochelin production was stimulated somewhat in the PchC-positive strain PALS128, this was not the case in the pchC mutant PAO6357 (Fig. 2). Pyochelin formation in the corresponding pchA mutants depended on salicylate addition, confirming that salicylate was readily taken up (Fig. 2). In the salicylate-requiring pchC mutant PAO6357 the concentration of pyochelin formed remained below 200 μM, even when the culture medium was amended with 1 mM salicylate. Taken together, these results do not support the first hypothesis.
FIG. 2.
Pyochelin formation by the pyoverdin-negative P. aeruginosa strains PALS128 (▪), PAO6342 (pchC) (•), PALS128-6 (pchA) (□), and PAO6357 (pchA pchC) (○) in GGP medium amended with different amounts of salicylate. The growth conditions, extraction, and analysis of pyochelin were as described in Table 2, footnote a. Means ± standard deviations for three parallel experiments are shown.
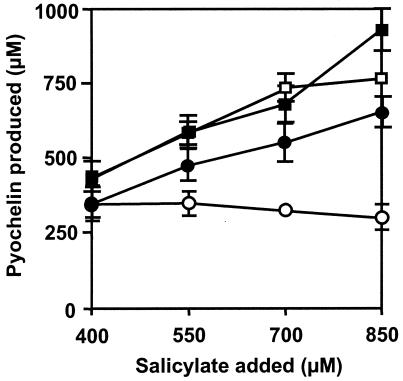
To evaluate the second hypothesis, we tested whether the PchC-negative phenotype could be suppressed by addition of l-cysteine to GGP medium, which is rich in amino acids except for l-cysteine (15). As illustrated in Fig. 3, addition of 2 mM l-cysteine to a medium containing salicylate at concentrations between 400 and 850 μM enabled strain PAO6357 (pvdB pchA pchC) to produce almost the same amounts of pyochelin as its pchC+ parent PALS128-6 produced. By contrast, we confirmed that in the absence of l-cysteine, the pchC-negative mutant produced significantly less siderophore than the pchC-positive strain produced. Similar results were obtained when the growth medium was amended with 5 mM l-cysteine (data not shown). These data are in agreement with a role of PchC in the removal of wrongly charged substrates from the PCP domains of PchE and/or PchF.
FIG. 3.
Effect of cysteine on pyochelin formation by P. aeruginosa strains. In GGP medium amended with different amounts of salicylate, pyochelin formation was measured in strain PALS128-6 (pvdB pchA) grown without cysteine (□) and with 2 mM l-cysteine (▪), as well as in strain PAO6357 (pvdB pchA pchC) grown without cysteine (○) and with 2 mM l-cysteine (•). The growth conditions, extraction, and analysis of pyochelin were as described in Table 2, footnote a. Means ± standard deviations are shown.
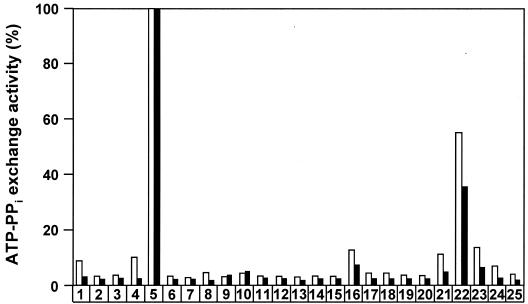
Which amino acid might be misloaded onto these enzymes in vivo? ATP-PPi exchange assays were performed to test whether purified PchE and PchF could adenylate amino acids other than l-cysteine. Both enzymes were highly specific for l-cysteine; of all the natural amino acids tested, only l-serine was activated by PchE and PchF at levels above background levels (Fig. 4). PchE was found to load radiolabeled l-serine instead of l-cysteine onto the PCP1 domain in vitro, but no subsequent condensation products could be detected (data not shown).
FIG. 4.
Relative activation of amino acids by PchE (open bars) and PchF (solid bars). ATP-PPi exchange activities were measured with proteinogenic amino acids (bars 1 to 20) and nonproteinogenic amino acids (bars 21 to 24) at a concentration of 1 mM. The highest exchange activity measured with l-cysteine was defined as 100%. The standard deviations of values in this assay were ≤20%. Bars 1, l-alanine; bars 2, l-arginine; bars 3, l-asparagine; bars 4, l-aspartate; bars 5, l-cysteine; bars 6, l-glutamate; bars 7, l-glutamine; bars 8, l-glycine; bars 9, l-histidine; bars 10, l-isoleucine; bars 11, l-leucine; bars 12, l-lysine; bars 13, l-methionine; bars 14, l-phenylalanine; bars 15, l-proline; bars 16, l-serine; bars 17, l-threonine; bars 18, l-tryptophan; bars 19, l-tyrosine; bars 20, l-valine; bars 21, l-homoserine; bars 22, l-2-aminobutyrate; bars 23, S-methyl-l-cysteine; bars 24, dl-allylglycine; bars 25, no amino acid added.
2-Aminobutyrate strongly inhibits Dha and pyochelin formation in a pchC mutant.
In the ATP-PPi exchange assays for PchE and PchF, the cysteine analog 2-aminobutyrate was activated with good efficiency (Fig. 4). Previously, 2-aminobutyrate had been shown to be activated and loaded instead of l-cysteine onto the PCP domain of HMWP1 (high-molecular-weight protein 1), a PchE-like nonribosomal peptide synthetase involved in the biosynthesis of the siderophore yersiniabactin in Yersinia pestis (22). We therefore tested whether this analog interfered with Dha and pyochelin formation by P. aeruginosa in vivo. When 2-aminobutyrate was added to the growth medium at a final concentration of 5 mM, pyochelin formation was very strongly decreased in pchC mutant PAO6339, and no Dha was detected (Table 4). In the absence of pyochelin, the expression of the pchDCBA and pchEFG genes required for salicylate, Dha, and pyochelin formation is low because the positive feedback regulation operating in this biosynthetic pathway is disrupted (32). It is therefore not surprising that addition of 2-aminobutyrate to the pchC mutant also reduced the production of salicylate (Table 4) as the amount of pyochelin produced under these conditions was not sufficient to allow high-level expression of the pch biosynthetic genes. By contrast, addition of 5 mM 2-aminobutyrate to a culture of the wild-type strain, PAO1, had only a modest effect on the production of pyochelin and Dha (Table 4), supporting the hypothesis that the PchC type II thioesterase is able to remove 2-aminobutyrate from PchE and PchF.
TABLE 4.
Impact of 2-aminobutyrate on salicylate, Dha, and pyochelin formation
| Strain | Genotype | Growth mediuma | Salicylate concn (nmol/ml)b | Dha concn (nmol/ml)b | Pyochelin concn (nmol/ml)b |
|---|---|---|---|---|---|
| PAO1 | Wild type | GGP | <8.0 | 87 ± 16 | 869 ± 111 |
| PAO1 | Wild type | GGP + 2-ABA | <8.0 | 62 ± 10 | 552 ± 28 |
| PAO6339 | ΔpchC | GGP | 708 ± 100 | 70 ± 4 | 422 ± 57 |
| PAO6339 | ΔpchC | GGP + 2-ABA | 87 ± 40 | <1.2 | 40 ± 22 |
GGP medium (30 ml) containing, where indicated, dl-2-aminobutyrate (2-ABA) at a final concentration of 5 mM, was inoculated with 0.1-ml portions of cultures grown in GGP medium. After incubation at 37°C and 220 rpm for 38 h, supernatants were extracted and analyzed for salicylate, Dha, and pyochelin by HPLC. The values are the means ± standard deviations for three parallel experiments.
Concentration in culture supernatant.
DISCUSSION
In this study, we demonstrated that PchC, an external type II thioesterase encoded by the pyochelin gene cluster, is important for optimal production of the siderophore pyochelin and of its precursor, the antibiotic Dha. Type II thioesterase genes have been identified in many gene clusters specifying the biosynthesis of nonribosomal peptides (3, 7, 26, 40) and polyketides (5, 18, 49). Mutational loss of these genes generally results in a decreased amount of product formed, and in vitro, type II thioesterases can regenerate misacylated thiol groups of the 4′-phosphopantetheine cofactors attached to the PCP domains of nonribosomal peptide synthetases (37). In vivo, misacylated enzymes may be formed either when a wrong amino acid is activated and loaded onto the PCP domain or when the 4′-phosphopantetheinyl transferase uses acyl coenzyme A instead of free coenzyme A as the 4′-phosphopantetheine donor (37). Similarly, during polyketide biosynthesis, type II thioesterases are thought to be responsible for hydrolyzing aberrant cofactor-bound acyl groups (19).
Our data support such a proofreading role for PchC in P. aeruginosa. The production of Dha and pyochelin was strongly decreased in the pchC-negative mutant but not in the wild type, especially when the growth medium was amended with 2-aminobutyrate (Table 4). This cysteine analog was found to be activated by both PchE and PchF and thus could compete with cysteine for loading onto the PCP domains of these nonribosomal peptide synthetases. If 2-aminobutyrate instead of l-cysteine were attached to PchE and PchF, the formation of thiazoline rings would not occur and Dha and pyochelin assembly would be prevented. In the wild type, the most likely function of the PchC enzyme is to remove the wrongly charged substrate, thereby regenerating peptide synthetase activity. To our knowledge, this is the first evidence for a proofreading role of a type II thioesterase in vivo.
There are two indications that a wrong substrate loaded onto PchE causes the enzyme to stall rather than to form aberrant products. (i) Under the conditions used in this study, strain PAO1 converted salicylate quantitatively to Dha and pyochelin. In the pchC mutant, however, which produced less Dha and pyochelin, large amounts of salicylate accumulated (Tables 2 to 4), indicating that little, if any, salicylate is coupled to substrates other than l-cysteine by PchE. (ii) Although PchE was able to load l-serine instead of l-cysteine onto the PCP1 domain, no subsequent condensation products were observed in vitro. In addition, HPLC analysis of culture supernatants of the pchC mutant (Tables 2 to 4) did not reveal aberrant condensation products of salicylate.
The role of PchC in removing wrongly charged molecules from the PCP domain of PchE and possibly the PCP domain of PchF is corroborated by the fact that addition of l-cysteine to the growth medium largely suppressed the adverse effect of a pchC mutation, probably by ensuring that l-cysteine rather than its potential natural competitors (e.g., l-serine) is charged on PchE (and PchF). A stimulating effect of l-cysteine on pyochelin biosynthesis has been noticed previously (2, 15) and may also be important in the wild type when the amount of salicylate available is greater than the amount of l-cysteine. In the experiments shown in Fig. 2 and 3, the amount of salicylate was controlled by exogenous salicylate added to a salicylate-requiring mutant.
Given the fact that in vitro some type II thioesterases hydrolyze PCP-bound peptides (37), we considered the possibility that PchC might detach the biosynthetic intermediate Dha and/or participate in the release of the final product, pyochelin, from the thiotemplate. Whereas it is clear from the results shown in Table 3 that PchC does not cleave the pyochelin-PchF thioester bond, we cannot entirely rule out involvement of PchC in Dha release from PchE since the production of this antibiotic compound was greater in a pchC-positive background than in the absence of pchC. However, it seems more likely that the editing function of PchC accounts for this effect, as some Dha was formed even in the absence of thioesterase activities encoded by both pchC and pchF (Table 3).
In conclusion, we showed that the PchC thioesterase is not essential for the release of Dha from PchE and does not participate in the release of pyochelin from PchF. Our data are consistent with PchC having a quality control function in pyochelin biosynthesis by removing wrongly charged molecules from the PCP domains of PchE and PchF. Further support for this conclusion comes from a recent study (51) which demonstrated that a type II thioesterase removes misloaded amino acids from tyrocidine synthetase prior to peptide bond formation and thereby restores the activity of enzyme modules stalled with unprocessed aminoacyl intermediates.
Acknowledgments
We thank Thomas A. Keating for providing pPchFTE, Jean-Pierre Rey for technical assistance, Patrick Michaux for help with HPLC analyses, and GlaxoSmithKline for a generous gift of carbenicillin.
This work was supported by the Swiss National Foundation for Scientific Research (projects 31-56608.99 and 31-102174).
REFERENCES
- 1.Ankenbauer, R. G., and H. N. Quan. 1994. FptA, the Fe(III)-pyochelin receptor of Pseudomonas aeruginosa: a phenolate siderophore receptor homologous to hydroxamate siderophore receptors. J. Bacteriol. 176:307-319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Audenaert, K., T. Pattery, P. Cornelis, and M. Höfte. 2002. Induction of systemic resistance to Botrytis cinerea in tomato by Pseudomonas aeruginosa 7NSK2: role of salicylic acid, pyochelin, and pyocyanin. Mol. Plant-Microbe Interact. 15:1147-1156. [DOI] [PubMed] [Google Scholar]
- 3.Bearden, S. W., J. D. Fetherston, and R. D. Perry. 1997. Genetic organization of the yersiniabactin biosynthetic region and construction of avirulent mutants in Yersinia pestis. Infect. Immun. 65:1659-1668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blumer, C., S. Heeb, G. Pessi, and D. Haas. 1999. Global GacA-steered control of cyanide and exoprotease production in Pseudomonas fluorescens involves specific ribosome binding sites. Proc. Natl. Acad. Sci. USA 96:14073-14078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Butler, A. R., N. Bate, and E. Cundliffe. 1999. Impact of thioesterase activity on tylosin biosynthesis in Streptomyces fradiae. Chem. Biol. 6:287-292. [DOI] [PubMed] [Google Scholar]
- 6.Carmi, R., S. Carmeli, E. Levy, and F. J. Gough. 1994. (+)-(S)-Dihydro-aeruginoic acid, an inhibitor of Septoria tritici and other phytopathogenic fungi and bacteria, produced by Pseudomonas fluorescens. J. Nat. Prod. 57:1200-1205. [DOI] [PubMed] [Google Scholar]
- 7.Cosmina, P., F. Rodriguez, F. de Ferra, G. Grandi, M. Perego, G. Venema, and G. van Sinderen. 1993. Sequence and analysis of the genetic locus responsible for surfactin synthesis in Bacillus subtilis. Mol. Microbiol. 8:821-831. [DOI] [PubMed] [Google Scholar]
- 8.Cox, C. D. 1980. Iron uptake with ferripyochelin and ferric citrate by Pseudomonas aeruginosa. J. Bacteriol. 142:581-587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cox, C. D. 1982. Effect of pyochelin on the virulence of Pseudomonas aeruginosa. Infect. Immun. 36:17-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cox, C. D., and P. Adams. 1985. Siderophore activity of pyoverdin in Pseudomonas aeruginosa. Infect. Immun. 48:130-138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Crosa, J. H., and C. T. Walsh. 2002. Genetics and assembly line enzymology of siderophore biosynthesis in bacteria. Microbiol. Mol. Biol. Rev. 66:223-249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Del Sal, G., G. Manfioletti, and C. Schneider. 1988. A one-tube plasmid DNA mini-preparation suitable for sequencing. Nucleic Acids Res. 16:9878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Farinha, M. A., and A. M. Kropinski. 1990. High efficiency electroporation of Pseudomonas aeruginosa using frozen cell suspensions. FEMS Microbiol. Lett. 58:221-225. [DOI] [PubMed] [Google Scholar]
- 14.Gaille, C., P. Kast, and D. Haas. 2002. Salicylate biosynthesis in Pseudomonas aeruginosa. Purification and characterization of PchB, a novel bifunctional enzyme displaying isochorismate pyruvate-lyase and chorismate mutase activities. J. Biol. Chem. 277:21768-21775. [DOI] [PubMed] [Google Scholar]
- 15.Gaille, C., C. Reimmann, and D. Haas. 2003. Isochorismate synthase (PchA), the first and rate-limiting enzyme in salicylate biosynthesis of Pseudomonas aeruginosa. J. Biol. Chem. 278:16893-16898. [DOI] [PubMed] [Google Scholar]
- 16.Gamper, M., B. Ganter, M. Polito, and D. Haas. 1992. RNA processing modulates the expression of the arcDABC operon in Pseudomonas aeruginosa. J. Mol. Biol. 226:943-957. [DOI] [PubMed] [Google Scholar]
- 17.Gensberg, K., K. Hughes, and A. W. Smith. 1992. Siderophore-specific induction of iron uptake in Pseudomonas aeruginosa. J. Gen. Microbiol. 138:2381-2387. [DOI] [PubMed] [Google Scholar]
- 18.Haydock, S. F., J. A. Dowson, N. Dhillon, G. A. Roberts, J. Cortes, and P. F. Leadlay. 1991. Cloning and sequence analysis of genes involved in erythromycin biosynthesis in Saccharopolyspora erythraea: sequence similarities between EryG and a family of S-adenosylmethionine-dependent methyltransferases. Mol. Gen. Genet. 230:120-128. [DOI] [PubMed] [Google Scholar]
- 19.Heathcote, M. L., J. Staunton, and P. F. Leadlay. 2001. Role of type II thioesterases: evidence for removal of short acyl chains produced by aberrant decarboxylation of chain extender units. Chem. Biol. 8:207-220. [DOI] [PubMed] [Google Scholar]
- 20.Heeb, S., Y. Itoh, T. Nishijyo, U. Schnider, C. Keel, J. Wade, U. Walsh, F. O'Gara, and D. Haas. 2000. Small, stable shuttle vectors based on the minimal pVS1 replicon for use in Gram-negative, plant-associated bacteria. Mol. Plant-Microbe Interact. 13:232-237. [DOI] [PubMed] [Google Scholar]
- 21.Heinrichs, D. E., and K. Poole. 1993. Cloning and sequence analysis of a gene (pchR) encoding an AraC family activator of pyochelin and ferripyochelin receptor synthesis in Pseudomonas aeruginosa. J. Bacteriol. 175:5882-5889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Keating, T. A., Z. Suo, D. E. Ehmann, and C. T. Walsh. 2000. Selectivity of the yersiniabactin synthetase adenylation domain in the two-step process of amino acid activation and transfer to a holo-carrier protein domain. Biochemistry 39:2297-2306. [DOI] [PubMed] [Google Scholar]
- 23.Meyer, J. M., and M. A. Abdallah. 1978. The fluorescent pigment of Pseudomonas fluorescens: biosynthesis, purification and physicochemical properties. J. Gen. Microbiol. 107:319-328. [Google Scholar]
- 24.Meyer, J. M., A. Neely, A. Stintzi, C. Georges, and I. A. Holder. 1996. Pyoverdin is essential for virulence of Pseudomonas aeruginosa. Infect. Immun. 64:518-523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mikaelian, I., and A. Sergeant. 1996. Modification of the overlap extension method for extensive mutagenesis on the same template. Methods Mol. Biol. 57:193-202. [DOI] [PubMed] [Google Scholar]
- 26.Mootz, H. D., and M. A. Marahiel. 1997. The tyrocidine biosynthesis operon of Bacillus brevis: complete nucleotide sequence and biochemical characterization of functional internal adenylation domains. J. Bacteriol. 179:6843-6850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Patel, H., and C. T. Walsh. 2001. In vitro reconstitution of the Pseudomonas aeruginosa nonribosomal peptide synthesis of pyochelin: characterization of backbone tailoring thiazoline reductase and N-methyltransferase activities. Biochemistry 40:9023-9031. [DOI] [PubMed] [Google Scholar]
- 28.Patel, H. M., J. Tao, and C. T. Walsh. 2003. Epimerization of an l-cysteinyl to a d-cysteinyl residue during thiazoline ring formation in siderophore chain elongation by pyochelin synthetase from Pseudomonas aeruginosa. Biochemistry 42:10514-10527. [DOI] [PubMed] [Google Scholar]
- 29.Pazirandeh, M., S. S. Chirala, and S. J. Wakil. 1991. Site-directed mutagenesis studies on the recombinant thioesterase domain of chicken fatty acid synthase expressed in Escherichia coli. J. Biol. Chem. 266:20946-20952. [PubMed] [Google Scholar]
- 30.Quadri, L. E. N., P. H. Weinreb, M. Lei, M. M. Nakano, P. Zuber, and C. T. Walsh. 1998. Characterization of Sfp, a Bacillus subtilis phosphopantetheinyl transferase for peptidyl carrier protein domains in peptide synthetases. Biochemistry 37:1585-1595. [DOI] [PubMed] [Google Scholar]
- 31.Quadri, L. E. N., T. A. Keating, H. M. Patel, and C. T. Walsh. 1999. Assembly of the Pseudomonas aeruginosa nonribosomal peptide siderophore pyochelin: in vitro reconstitution of aryl-4,2-bisthiazoline synthetase activity from PchD, PchE, and PchF. Biochemistry 38:14941-14954. [DOI] [PubMed] [Google Scholar]
- 32.Reimmann, C., L. Serino, M. Beyeler, and D. Haas. 1998. Dihydroaeruginoic acid synthetase and pyochelin synthetase, products of the pchEF genes, are induced by extracellular pyochelin in Pseudomonas aeruginosa. Microbiology 144:3135-3148. [DOI] [PubMed] [Google Scholar]
- 33.Reimmann, C., H. M. Patel, L. Serino, M. Barone, C. T. Walsh, and D. Haas. 2001. Essential PchG-dependent reduction in pyochelin biosynthesis of Pseudomonas aeruginosa. J. Bacteriol. 183:813-820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rinehart, K. L., A. L. Staley, S. R. Wilson, R. G. Ankenbauer, and C. D. Cox. 1995. Stereochemical assignment of the pyochelins. J. Org. Chem. 60:2786-2791. [Google Scholar]
- 35.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Plainview, N.Y.
- 36.Schneider, A., and M. A. Marahiel. 1998. Genetic evidence for a role of thioesterase domains, integrated in or associated with peptide synthetases, in non-ribosomal peptide biosynthesis in Bacillus subtilis. Arch. Microbiol. 169:404-410. [DOI] [PubMed] [Google Scholar]
- 37.Schwarzer, D., H. D. Mootz, U. Linne, and M. A. Marahiel. 2002. Regeneration of misprimed nonribosomal peptide synthetases by type II thioesterases. Proc. Natl. Acad. Sci. USA 99:14083-14088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schwyn, B., and J. B. Neilands. 1987. Universal chemical assay for the detection and determination of siderophores. Anal. Biochem. 160:47-56. [DOI] [PubMed] [Google Scholar]
- 39.Serino, L., C. Reimmann, B. Baur, M. Beyeler, P. Visca, and D. Haas. 1995. Structural genes for salicylate biosynthesis from chorismate in Pseudomonas aeruginosa. Mol. Gen. Genet. 249:217-228. [DOI] [PubMed] [Google Scholar]
- 40.Serino, L., C. Reimmann, P. Visca, M. Beyeler, V. Della Chiesa, and D. Haas. 1997. Biosynthesis of pyochelin and dihydroaeruginoic acid requires the iron-regulated pchDCBA operon in Pseudomonas aeruginosa. J. Bacteriol. 179:248-257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Simon, R., U. Priefer, and A. Pühler. 1983. A broad host range mobilization system for in vitro genetic engineering: transposon mutagenesis in Gram negative bacteria. Bio/Technology 1:784-790. [Google Scholar]
- 42.Stanisich, V. A., and B. W. Holloway. 1972. A mutant sex factor of Pseudomonas aeruginosa. Genet. Res. Camb. 19:91-108. [DOI] [PubMed] [Google Scholar]
- 43.Takase, H., H. Nitanai, K. Hoshino, and T. Otani. 2000. Impact of siderophore production on Pseudomonas aeruginosa infections in immunosuppressed mice. Infect. Immun. 68:1834-1839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vieira, J., and J. Messing. 1991. New pUC-derived cloning vectors with different selectable markers and DNA replication origins. Gene 100:189-194. [DOI] [PubMed] [Google Scholar]
- 45.Visca, P., L. Serino, and N. Orsi. 1992. Isolation and characterization of Pseudomonas aeruginosa mutants blocked in the synthesis of pyoverdin. J. Bacteriol. 174:5727-5731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Voisard, C., C. T. Bull, C. Keel, J. Laville, M. Maurhofer, U. Schnider, G. Défago, and D. Haas. 1994. Biocontrol of root diseases by Pseudomonas fluorescens CHA0: current concepts and experimental approaches, p. 67-89. In F. O'Gara, D. Dowling, and B. Boesten (ed.), Molecular ecology of rhizosphere microorganisms. VCH Publishers, Weinheim, Germany.
- 47.Watson, A., R. A. Alm, and J. S. Mattick. 1996. Construction of improved vectors for protein production in Pseudomonas aeruginosa. Gene 172:163-164. [DOI] [PubMed] [Google Scholar]
- 48.Witkowski, A., H. E. Witkowski, and S. Smith. 1994. Reengineering the specificity of a serine active-site enzyme. Two active-site mutations convert a hydrolase to a transferase. J. Biol. Chem. 269:379-383. [PubMed] [Google Scholar]
- 49.Xue, Y., L. Zhao, H.-W. Liu, and D. H. Sherman. 1998. A gene cluster for macrolide antibiotic biosynthesis in Streptomyces venezuelae: architecture of metabolic diversity. Proc. Natl. Acad. Sci. USA 95:12111-12116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ye, R. W., D. Haas, J. O. Ka, V. Krishnapillai, A. Zimmermann, C. Baird, and J. M. Tiedje. 1995. Anaerobic activation of the entire denitrification pathway in Pseudomonas aeruginosa requires Anr, an anolog of Fnr. J. Bacteriol. 177:3606-3609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yeh, E., R. M. Kohli, S. D. Bruner, and C. T. Walsh. Type II thioesterase restores activity of a NRPS module stalled with an aminoacyl-S-enzyme that cannot be elongated. Chembiochem, in press. [DOI] [PubMed]