Abstract
Xenorhabdus nematophila is an insect pathogen and produces protein toxins which kill the larval host. Previously, we characterized an orally toxic, large, outer membrane-associated protein complex from the culture medium of X. nematophila. Here, we describe the cloning, expression, and characterization of a 17-kDa pilin subunit of X. nematophila isolated from that protein complex. The gene was amplified by PCR, cloned, and expressed in Escherichia coli. The recombinant protein was refolded in vitro in the absence of its cognate chaperone by using a urea gradient. The protein oligomerized during in vitro refolding, forming multimers. Point mutations in the conserved N-terminal residues of the pilin protein greatly destabilized its oligomeric organization, demonstrating the importance of the N terminus in refolding and oligomerization of the pilin subunit by donor strand complementation. The recombinant protein was cytotoxic to cultured Helicoverpa armigera larval hemocytes, causing agglutination and subsequent release of the cytoplasmic enzyme lactate dehydrogenase. The agglutination of larval cells by the 17-kDa protein was inhibited by several sugar derivatives. The biological activity of the purified recombinant protein indicated that it has a conformation similar to that of the native protein. The 17-kDa pilin subunit was found to be orally toxic to fourth- or fifth-instar larvae of an important crop pest, H. armigera, causing extensive damage to the midgut epithelial membrane. To our knowledge, this is first report describing an insecticidal pilin subunit of a bacterium.
Recent concerns about development of resistance to insecticidal proteins of Bacillus thuringiensis have directed the focus of research toward isolation of novel toxin molecules from other microorganisms in the soil. Xenorhabdus nematophila is an entomopathogenic gram-negative bacterium belonging to the family Enterobacteriaceae (4). It lives in symbiotic association with a soil nematode, Steinernema carpocapsae (2). An infective juvenile larva of the nematode harbors X. nematophila in its intestinal pouch and transports the bacteria to the gut or hemocoel of the target insect. Infestation of an insect larva by the nematode-bacterium complex leads to death of the insect host in a short time (1, 2). The main causes of larval death are thought to be a combination of septicemia and the lethal toxins produced by the bacterium. The presence of the bacterium is considered necessary for optimum growth and reproduction of the nematode in the larval prey (30). Formulations containing a Steinernema-X. nematophila complex are used as biological control agents against a wide range of insect pests (9, 10, 28, 35).
Several factors in X. nematophila that are directly responsible for the death of insects or assist in the pathogenic process have been described (6, 14, 16, 26). Outer membrane proteins of the pathogenic bacteria perform several critical functions in the host environment, such as recognition and interaction with the target host cells (3), transport of effector proteins (22), and secretion of virulence factors and antibacterial proteins (31). We are investigating the role of outer membrane and associated proteins of X. nematophila which are responsible for causing pathogenicity and death of the insect host. In a previous study oral insecticidal activity associated with outer membrane vesicles (OMVs) of X. nematophila, which were released into the extracellular medium, was demonstrated (19).
The N-terminal and internal sequences of a major band corresponding to the 17-kDa band in the OMV preparation showed homology with the PapA structural subunit of P pili of Escherichia coli (34). Subsequently, we purified the pilin subunit from the surface of X. nematophila cells and characterized its interaction with insect hemocytes. The pilin subunit was found to be cytotoxic to the cultured larval hemocytes of an important crop pest, Helicoverpa armigera (20). It has been suggested that fimbriae expressed on the cell surface aid in colonization of the nematode host by X. nematophila (27). In gram-negative pathogens, fimbriae are surface organelles which mediate host cell binding (21) and are important virulence factors in uropathogenic strains of E. coli, Proteus mirabilis, Salmonella, Serratia, Neisseria, etc. (15, 29, 38, 43).
The biogenesis of fimbriae or pili has been studied extensively in E. coli and other gram-negative bacteria (8, 34). In E. coli this biogenesis is known to occur through the highly conserved chaperone-usher pathway, in which individual pilin subunits interact with a specific periplasmic chaperone via a mechanism termed donor strand complementation (34). The pilin domains of all the subunits have immunoglobulin (Ig)-like folds. However, they lack the seventh C-terminal β-strand present in the canonical Ig fold. The absence of this strand produces a deep groove along the surface of the domain and exposes its hydrophobic core. The chaperone donates the missing strand to complement the incomplete Ig-like fold by transiently shielding the hydrophobic core and contributes to stabilization of the pilin subunit (8, 34). Assembly of subunits into the pilus fiber proceeds by a donor strand exchange mechanism in which the chaperone's donor strand is replaced by the N terminus of the next subunit (8). The structural basis of fiber formation has been revealed by high-resolution crystallography (33, 41).
To study the interaction of the 17-kDa pilin subunit of X. nematophila with the larval host, we cloned and expressed the protein in E. coli. The protein was produced as inclusion bodies, which were refolded in a biologically active, oligomeric form. Here we provide evidence that the recombinant structural subunit of the pilin of X. nematophila undergoes intermolecular donor strand complementation during in vitro folding and forms oligomers in the absence of its cognate chaperone. The 17-kDa protein showed oral larvicidal activity against H. armigera larvae. This is the first report demonstrating insecticidal activity in a pilin subunit.
MATERIALS AND METHODS
Bacteria and growth conditions.
X. nematophila strain ATCC 19061 was obtained from the American Type Culture Collection (Rockville, Md.). The X. nematophila culture was streaked on nutrient agar supplemented with 0.004% (wt/vol) triphenyl tetrazolium chloride and 0.025% (wt/vol) bromothymol blue (4). Broth cultures were grown from a single blue colony in Luria-Bertani (LB) medium at 28°C with shaking at 150 rpm. E. coli K-12 was used as a reference strain.
Preparation and fractionation of OMV proteins of X. nematophila.
OMVs were prepared from the culture supernatant as described previously by Khandelwal and Bhatnagar (19). The OMV proteins were solubilized in TENS buffer (50 mM Tris-HCl [pH 7.2], 5 mM EDTA, 400 mM NaCl, 1.0% sodium dodecyl sulfate [SDS]) at 37°C overnight and applied to a Sephacryl S-300 column; then the proteins were eluted with TENS buffer, and the fractions were examined by SDS-polyacrylamide gel electrophoresis (PAGE).
Isolation and purification of native pilin protein from X. nematophila.
The native pilin protein was obtained from the surface of X. nematophila cells and purified by sucrose density gradient centrifugation as described by Korhonen et al. (23).
Preparation of polyclonal antiserum.
The purified native pilin protein was emulsified with Freund's adjuvant and injected intramuscularly into a rabbit. Two booster doses were given at 2-week intervals. After 6 weeks, the rabbit was bled, and the antiserum was examined by Western blotting.
Amino-terminal and internal peptide sequencing.
The purified 17-kDa protein from X. nematophilus and the void volume fraction of the Sephacryl S-300 column loaded with OMV protein were transferred to a polyvinylidene difluoride membrane under standard conditions, stained with amido black, and excised for N-terminal peptide sequencing by Edman's method. To obtain an internal sequence of the protein, it was subjected to tryptic digestion, and one of the peptides was purified and sequenced. Since the peptide did not end with Arg or Lys (the terminal amino acid residues recognized by trypsin), it was the C terminus of the protein, which matched the C terminus of PapA. The N-terminal sequences of the purified recombinant 17-kDa proteins obtained from both pQE30 and pET23a plasmid constructs were also determined as described above.
Cloning of the gene encoding the 17-kDa protein.
Table 1 lists the strains and plasmids used in this study. Based on the N- and C-terminal sequences, degenerate primers were synthesized, and a 468-bp DNA fragment was amplified by using X. nematophila genomic DNA as the template. The PCR-amplified fragment was cloned in the PCR cloning vector pGEMT-easy (Promega, Madison, Wis.), which produced plasmid pPK1, which was transformed into E. coli DH5α. The sequence of clone PK1 was determined and confirmed.
TABLE 1.
Plasmids and strains used in this study
| Plasmid or strain | Relevent characteristics | Reference or Source |
|---|---|---|
| Plasmids | ||
| pPK1 | pGEMT-easy containing 468-bp fragment | This study |
| pPK2 | pGEMT-easy containing 1-kbp fragment with upstream sequence | This study |
| pPK3 | pGEMT-easy containing 537-bp pilus gene with BamHI and HindIII sites, with signal sequence | This study |
| pPK4 | pQE30 with 537-bp BamHI-HindIII fragment with His tag upstream of signal sequence | This study |
| pPK5 | pET23a with 537-bp BamHI-HindIII fragment with C-terminal His tag | This study |
| Strains | ||
| BL21(DE3) | F−ompT hsdSB(rB mB) gal dcm (DE3) | Novagen |
| M15 | NaIs Strs R Fs Thi− Lac− Ara+ Gal+ MtI− F RecA+ Uvr+ Lon+ | Qiagen |
| PK4 | E. coli M15 containing pPK4 plasmid | This study |
| PK5 | E. coli BL21(DE3) containing pPK5 plasmid | This study |
| PK6 | E. coli BL21(DE3) containing pET23a plasmid with mutation V32A in the wild-type pilin gene | This study |
| PK7 | E. coli BL21(DE3) containing pET23a plasmid with mutation F34A in the wild-type pilin gene | This study |
| PK8 | E. coli BL21(DE3) containing pET23a plasmid with mutation I38A in the wild-type pilin gene | This study |
| PK9 | E. coli BL21(DE3) containing pET23a plasmid with triple mutation (VFI → AAA) in the wild-type pilin gene | This study |
The upstream and signal sequences of the gene encoding the mature 17-kDa protein were determined as described by Reddy et al. (32). Briefly, genomic DNA was digested with BamHI, and the fragments were ligated to a double-stranded adapter sequence (Table 2) with a BamHI overhang. Following adapter ligation, PCR amplification was performed by using gene-specific biotinylated primer BP1 and adapter-specific primer WP1 (Table 2). The PCR product was purified by using paramagnetic beads coated with streptavidin. A second PCR with the purified DNA was performed by using another gene-specific primer, IP2 (internal to the biotinylated primer), and primer WP1 to verify the specificity of the amplified product. A 1-kb fragment obtained after PCR amplification was ligated in the pGEMT-easy vector, which produced plasmid pPK2, which was transformed into E. coli DH5α; this produced strain PK2. The plasmid from strain PK2 was sequenced to obtain the signal and upstream sequences. Oligonucleotide primers from the N terminus (including the leader sequence) and C terminus with restriction sites for BamHI and HindIII, respectively (Table 2), were used for PCR amplification of X. nematophila genomic DNA. A 537-bp fragment obtained by PCR amplification was ligated into the pGEMT-easy vector, and plasmid pPK3 was transformed into E. coli DH5α, producing strain PK3. The DNA fragment obtained after digestion of plasmid pPK3 with BamHI and HindIII was ligated into the pQE30 (QIAGEN) expression vector, and the resulting plasmid, pPK4, was transformed into E. coli M15, producing strain PK4. The BamHI/HindIII fragment was also ligated into pET23a, and the resulting plasmid, pPK5, was transformed into E. coli BL21(DE3), producing strain PK5.
TABLE 2.
Primers used in this study
| Primer | Sequence |
|---|---|
| Degenerate primers | |
| N terminal | 5′ GCNCCNACNCARGGNGAYGGNACN 3′ |
| C terminal | 5′ NARRTARTTNARNGTRAARTTNGC 3′ |
| Adapter sequence | 5′ GATCCTAATACCACTCACATAGGGCGGCCGCCCGGGC 3′, 3′ GATTATGGTGAGTGTATCCCGCCGGCGGGCCCG 5′ |
| WP1 | 5′ GATCCTAATACCACTCACATAGGGCGGCCGCCCGGGC 3′ |
| Biotinylated primer (BP1) | 5′ GTGTTCAGGTTTTGATTCACCACCATC 3′ |
| IP2 | 5′ GTTCTTGATTGAACAGGCTGC 3′ |
| Primers for amplifying 537-bp fragment | |
| Forward primer | 5′ GGATCCATGAAACTTAACACAATTGGC 3′ |
| Reverse primer | 5′ AAGCTTAAGGTAGTTGAGAGTGAAGTTG 3′ |
| Primers for point mutations | |
| Val32Ala (forward) | 5′ CTCAAGGTGACGGCGCAGCAAAATTCACCGGTTCTATTATTAATG 3′ |
| Phe34Ala (forward) | 5′ GGTGACGGCGCAGTTAAAGCAACCGGTTCTATTATTAATGC 3′ |
| Ile38Ala (forward) | 5′ GCAGTTAAATTCACCGGTTCTGCAATTAATGCAGCCTGTTCAATC 3′ |
| Triple mutation (forward) | 5′ CTCAAGGTGACGGCGCGCAAAGCAACCGGTTCGCAATTAATGCAGCCTGTTCAAT |
| (Val-Phe-Ile→Ala-Ala-Ala) | CAAG 3′ |
Phylogenetic analyses.
Phylogenic neighbors of the Xenorhabdus pilin protein were found by using the program PSI-BLAST (http://www.ncbi.nlm.nih.gov). Sequences were aligned by using the program CLUSTAL W (18). Phylogenetic analyses were carried out by using the PHYLIP (11) suite of programs, and the program SEQBOOT was used to carry out 1,000-fold bootstrapping, which involved generation of 1,000 independent data sets by random sampling. Pairwise distances between sequences were calculated for each of these data sets by using the program PROTDIST. Phylogenetic trees were generated by the neighbor-joining method by using the program NEIGHBOR. The branch lengths were generated by the Fitch-Margoliash method (13). To generate a majority rule consensus tree from the 1,000 trees generated from the bootstrapped data, the program CONSENSE was used. The majority rule consensus method selected for monophyletic groups which occurred the maximum number of times in the consensus trees.
Rationale for design of mutants.
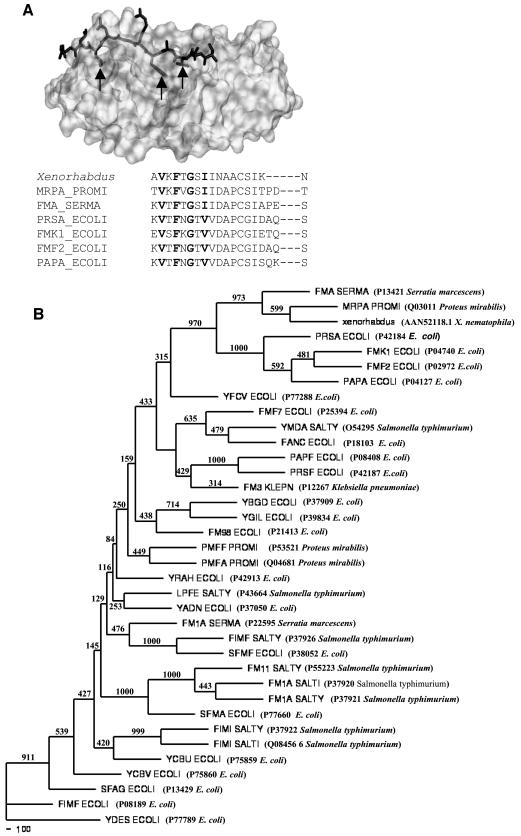
Pilin subunits of E. coli form multimers via head-to-tail interactions, in which the N-terminal segment (about 20 residues) of one subunit stabilizes the hydrophobic acceptor cleft in the carboxyl-terminal region of the preceding subunit by donor strand complementation (34). Sequence alignment of the N termini of several structural subunits known to participate in donor strand complementation showed a characteristic pattern of alternating hydrophobic residues (Fig. 1A), which are considered to be principal determinants of the specific interaction between two pilin subunits (33, 42). It is clear from Fig. 1A that the Val32, Phe34, and Ile38 residues in the Xenorhabdus sequence fulfill the conditions described above. Thus, it can be argued that if the Xenorhabdus sequence is a pilin subunit that undergoes polymerization by donor strand complementation, these three residues should be crucial for producing a structurally stable, oligomeric protein. Hence, we changed the three conserved hydrophobic residues, Val32, Phe34, and Ile38, in the N terminus of the protein to alanine separately and together (producing a triple mutant), and we tested their stability and polymerization behavior.
FIG. 1.
(A) X-ray crystallographic structure of PapE with donor strand complementing N-terminal peptide segment of PapK (indicated by sticks). Conserved deeply buried residues are indicated by arrows. An alignment of N-terminal sequences of various structural pilin proteins is also shown; conserved hydrophobic residues are indicated by boldface type. Sequences were aligned with the CLUSTALW program, followed by manual adjustments to minimize gaps within secondary structures. (B) Neighbor-joining tree showing the branching pattern of different pilin protein amino acid sequences and the phylogenic position of the Xenorhabdus pilin protein. Bootstrap resampling was done for 1,000 replicons. The numbers at the nodes in the consensus tree indicate the numbers of times that the subtree occurred in the 1,000 trees that were generated by NEIGHBOR. The designations of the proteins are followed by the accession numbers and names of the organisms in parentheses.
Site-directed mutagenesis was performed with a Quickchange mutagenesis kit (Stratagene). The colonies obtained after transformation in XL1-blue supercompetent cells were sequenced to confirm the altered residues. One plasmid of each type was digested with BamHI and HindIII and ligated to pET23a, and the resulting plasmids were transformed in E. coli BL21(DE3), producing strains PK6, PK7, PK8, and PK9.
Expression and purification.
For expression and purification of the recombinant 17-kDa protein from PK4, this strain was grown at 37°C in LB medium with 100 μg of ampicillin per ml and 25 μg of kanamycin per ml. After induction with 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG) for 4 h, cells were harvested by centrifugation at 6,000 × g for 10 min at 4°C. The cell pellet was resuspended in 10 ml of sonication buffer (50 mM sodium phosphate, 300 mM NaCl, 10 mM Tris-Cl; pH 8.0) and sonicated by using 15 30-s cycles at 50 W and 4°C. The lysate was centrifuged at 10,000 × g for 45 min at 4°C, and the pellet was collected. The pellet was washed three times with 1% deoxycholic acid and several times with 10 mM sodium phosphate buffer (pH 8.0). Then the pellet was suspended in 5 ml of denaturing buffer (8 M urea in 100 mM sodium phosphate buffer-10 mM Tris-Cl [pH 8.0]) and dialyzed against 10 mM sodium phosphate buffer (pH 8.0). For purification, the dialyzed protein was loaded onto a DEAE ion-exchange column and washed with several column volumes of wash buffer (10 mM sodium phosphate, pH 8.0), and the proteins were eluted with an NaCl gradient (0 to 250 mM) in wash buffer. Fractions containing the recombinant protein were pooled and dialyzed against the wash buffer or 10 mM Tris-Cl (pH 8.0).
All the other strains were grown at 37°C in LB medium with 100 μg of ampicillin per ml. Expression was induced with 1 mM IPTG for 4 h at 37°C, and the cells were harvested by centrifugation at 6,000 × g for 10 min at 4°C. Each cell pellet was resuspended in 5 ml of denaturing buffer and incubated at 37°C for 1 h on a rotary shaker. The lysate was centrifuged at 12,000 × g for 45 min at room temperature, and the supernatant was loaded onto an Ni-nitrilotriacetic acid (NTA) column previously equilibrated with the denaturing buffer. Recombinant protein bound to Ni-NTA was refolded by using a 8 to 0 M urea gradient in renaturation buffer (100 mM sodium phosphate, 10 mM Tris-Cl; pH 8.0). The proteins were eluted in the renaturation buffer with 250 mM imidazole. Fractions containing the recombinant protein were pooled and dialyzed against 10 mM Tris-Cl or 10 mM sodium phosphate (pH 8.0).
Identification of 17-kDa protein oligomers.
In E. coli, the pilin subunits in the native pilus rod resist disorganization by 1.5% SDS at room temperature (7, 37). To detect recombinant protein oligomers, the wild-type protein and the triple-mutant protein were incubated with 1.5% SDS in loading buffer at 25 or 95°C for 5 min and then resolved by SDS-PAGE and detected by Western blotting with anti-His antibodies (Clonetech) or antiserum against the native 17-kDa protein from X. nematophila. To check the presence of intermolecular disulfide bonds, the protein was heated in reducing and nonreducing buffers and resolved by SDS-PAGE.
Analytical gel chromatography.
A Sephadex G-200 column (35 by 1.2 cm; Pharmacia) was equilibrated with 20 mM sodium phosphate buffer (pH 8.0). The column was calibrated by using globular molecular weight marker proteins (Sigma). The void volume was determined with blue dextran 2000. Recombinant wild-type protein in the buffer described above was applied to the column and eluted with the same buffer at a flow rate of 0.1 ml/min. Protein concentrations in the samples were determined by measuring the absorbance at 220 nm (0.1 U of absorbance at 220 nm was equivalent to 3.35 μg of protein/ml as determined by using the Bradford reagent with bovine serum albumin [BSA] as the standard).
CD spectroscopy.
Circular dichroism (CD) spectra of the native and recombinant proteins were obtained with a JASCO J 715 spectropolarimeter at 20°C by using a 0.2-cm cell, wavelengths between 205 and 250 nm, and a scanning speed of 20 nm/min. All the proteins were dissolved in 10 mM sodium phosphate buffer (pH 8.0). The CD spectrum of the denatured protein was recorded after the protein was denatured in 8 M urea in 10 mM sodium phosphate buffer (pH 8.0). Twenty spectra were obtained in each case and were averaged; this was followed by baseline correction by subtraction of the buffer spectrum. The CD signals were converted to molar ellipticity by using the following relationship: θm = θo × 100/lc, where θm is molar ellipticity (in degrees per square centimeter per decimole), θo is the observed ellipticity (in degrees), l is the path length (in centimeters), and c is the molar concentration.
Hemagglutination and cytotoxicity assay of larval hemocytes.
The hemagglutination and cytotoxicity for larval hemocytes of the recombinant protein were determined as described previously (20). Inhibition of agglutination was determined with anti-pilin antiserum. To characterize the interaction between the 17-kDa protein and the hemocytes, various sugars and their derivatives were tested for the ability to inhibit the agglutination activity of the protein. The recombinant 17-kDa protein was incubated with various sugars for 1 h before it was added to larval hemocytes. Agglutination by wild-type 17-kDa protein was used as a positive control. Heat-inactivated wild-type 17-kDa protein, phosphate-buffered saline (PBS), and E. coli K-12 cells were used as controls.
Detection of binding of the 17-kDa protein to larval hemocytes by immunofluorescence.
The binding of the protein to H. armigera larval (fourth- or fifth-instar) hemocytes was determined by binding of a fluorescent substrate on the surface of the cells, as described previously (20).
Insect bioassay.
The larvicidal activity of the pilin protein was determined as described previously (19). The test protein preparations were diluted in 10 mM sodium phosphate buffer (pH 8.0) and mixed into the artificial diet. Each group contained 24 larvae that were placed individually on the surface of the diet. Mortality and larval weight were recorded periodically over the entire larval period. A dose of protein was the amount of protein mixed into the diet and was not always the actual amount consumed by the larvae. The bioassay was performed more than three times, and the data presented below are the data from one representative experiment. Heat-inactivated 17-kDa recombinant protein, BSA, E. coli K-12 cells, and buffer were used as controls. The 50% lethal dose (LD50) was determined by Probit analysis (12).
Histopathology.
Six-day-old insect larvae were fixed with 4% formaldehyde in PBS; several holes were punctured into the cuticle to allow penetration of the fixative. The larvae were embedded in paraffin wax, and 5-μm sections were cut. The sections were stained with eosin and hematoxylin and mounted with glycerol, and images were obtained.
Nucleotide sequence accession number.
The nucleotide sequence reported in this paper has been deposited in the GenBank database under accession number AY140909.
RESULTS
Isolation of 17-kDa protein from OMVs of X. nematophila.
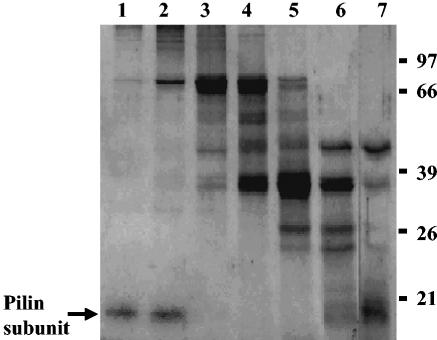
The OMVs were isolated from the culture filtrate of X. nematophila. When the OMV proteins were subjected to Sephacryl S-300 column chromatography, a 17-kDa polypeptide eluted in the void volume fractions (Fig. 2, lanes 1 and 2), indicating that the protein was likely present in an oligomeric form even after exposure to 1% SDS at 37°C. The N- and C-terminal sequences of the 17-kDa protein were found to be APTQGDGAVK and TGEFTAIANFTLNYL, respectively. The N-terminal sequence was same as that of the pilin subunit described previously (20).
FIG. 2.
Purification of 17-kDa pilin protein from OMVs of X. nematophila. OMV proteins were solubilized in TENS buffer and applied to a Sephacryl S-300 column. The proteins were eluted with TENS buffer. Peak fractions were pooled and examined. Lanes 1 and 2, void volume fractions; lanes 3 to 7, subsequent column fractions. The proteins were resolved on an SDS—12% PAGE gel and were visualized by staining with Coomassie brilliant blue. Numbers on the right are molecular weight markers (in thousands).
Cloning, expression, and purification of recombinant 17-kDa protein.
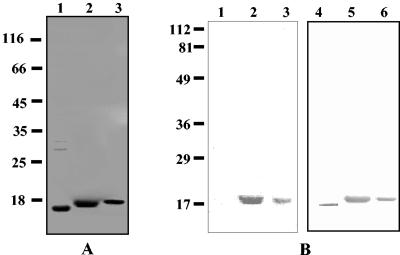
The 537-bp DNA fragment encoding the pilin subunit was amplified by PCR and cloned as described in Materials and Methods. Cells harboring the DNA produced a protein with an apparent molecular mass of 17 kDa as determined by SDS-PAGE (Fig. 3A). The gene showed 35% identity and 65% similarity at the amino acid level with the PapA subunit of the P pili of E. coli.
FIG. 3.
(A) SDS-PAGE profile of purified recombinant 17-kDa protein. The proteins were resolved on an SDS—12% PAGE gel and were visualized by staining with Coomassie brilliant blue or blotted with antiserum. Lane 1, purified recombinant 17-kDa protein without histidine tag after DEAE ion-exchange purification; lane 2, Ni-NTA affinity column-purified 17-kDa protein; lane 3, Ni-NTA affinity column-purified triple-mutant protein (Val-Phe-Ile → Ala-Ala-Ala). Numbers on the left are molecular weight markers (in thousands). (B) Western blot of recombinant 17-kDa pilin protein of X. nematophila. Lanes 1, 2, and 3 were developed with monoclonal antibodies against the six-His tag. Lanes 4, 5, and 6 were developed with polyclonal antibodies against native 17-kDa protein of X. nematophila. Lane 1, cell lysate of uninduced culture of strain PK5; lane 2, cell lysate of induced culture of strain PK5; lanes 3 and 5, Ni-NTA-purified 17-kDa protein; lane 4, DEAE column-purified 17-kDa protein (without tag); lane 6, Ni-NTA-purified triple-mutant protein (Val-Phe-Ile → Ala-Ala-Ala). Numbers on the left are prestained markers.
No recombinant protein was detected when a 468-bp DNA fragment (without the leader peptide sequence) was expressed in several commonly used expression vectors (data not shown). However, when the gene was expressed together with its signal sequence, the protein was recovered as inclusion bodies. The N-terminal sequence of the purified recombinant protein from both of the constructs used (PK4 and PK 5) showed that the signal sequence was processed in the E. coli host. Thus, the recombinant 17-kDa protein expressed by strain PK4 contained no histidine tag (cleaved with the signal sequence) in the N terminus and was obtained in substantially pure form after DEAE column chromatography (Fig. 3A, lane 1). Preliminary experiments were performed with this protein.
The recombinant 17-kDa protein with a six-His tag in the C terminus was obtained from strain PK5 (Fig. 3A, lane 2). The protein that eluted from the Ni-NTA column at the end of an 8 to 0 M urea gradient was found to be modestly stable in solution (it remained in solution at concentrations up to 0.5 mg/ml, but it slowly precipitated at concentrations above this concentration). A 25 μM solution of the protein showed no visible precipitation even after dialysis in a buffer without urea. The protein obtained from the column was more than 95% pure, as shown by SDS-PAGE (Fig. 3A, lane 2). The purified recombinant 17-kDa protein from PK5 reacted with both anti-His tag antibody and antiserum against the 17-kDa protein (Fig. 3B, lanes 2, 3, and 5). The recombinant 17-kDa protein with the six-His tag was used for the detailed structural and biological studies.
The yield of the purified wild-type protein was 4 mg per 100 ml of culture, and the yields of mutant proteins were 0.5, 2.5, 3.0, and 0.4 mg per 100 ml of culture for the Val32Ala, Phe34Ala, and Ile38Ala mutants and the triple mutant (Val-Phe-Ile → Ala-Ala-Ala), respectively.
Detection of protein oligomers.
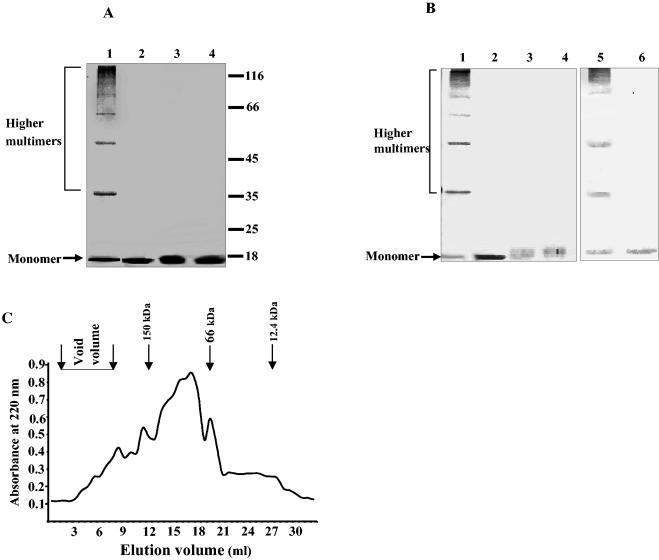
When the recombinant 17-kDa protein was incubated in the presence of 1.5% SDS at 25°C for 5 min and then subjected to SDS-PAGE and blotted with antibodies for the six-His tag, bands corresponding to higher oligomers were observed (Fig. 4A, lane 1). However, when the protein was heated at 95°C in 1.5% SDS, only the monomer was seen, as expected (Fig. 4A, lane 2). The recombinant protein without the histidine tag behaved just like the tagged protein, as shown by blotting with anti-pilin serum (Fig. 4B, lanes 5 and 6). Similar experiments performed with the triple mutant (Val-Phe-Ile → Ala-Ala-Ala) did not show any oligomers, and the protein migrated as monomers only (Fig. 4A, lane 3, and Fig. 4B, lane 3). The recombinant protein migrated as a monomer when the samples were boiled in the reducing or nonreducing loading dye before SDS-PAGE (data not shown), indicating that the oligomerization was not due to intermolecular disulfide bridges.
FIG. 4.
Detection of oligomers of recombinant 17-kDa pilin subunit by SDS-PAGE. (A) Ni-NTA-purified wild-type 17-kDa protein and triple-mutant 17-kDa protein (Val-Phe-Ile → Ala-Ala-Ala) were incubated at 25 or 95°C for 5 min in SDS loading buffer containing 1.5% SDS prior to loading onto the gel. The proteins were resolved on an SDS—10% PAGE gel and were visualized by staining with Coomassie brilliant blue. Lane 1, wild-type 17-kDa protein incubated at 25°C; lane 2, wild-type protein incubated at 95°C; lane 3, triple-mutant protein incubated at 25°C; lane 4, triple-mutant protein incubated at 95°C. Numbers on the right are molecular weight markers (in thousands). (B) Detection of protein oligomers by Western blotting. The protein samples were prepared and resolved as described above. Lanes 1 and 2, wild-type 17-kDa protein incubated at 25 and 95°C, respectively; lanes 3 and 4, triple-mutant protein incubated at 25 and 95°C, respectively; lanes 5 and 6, recombinant 17-kDa protein (without His tag) incubated at 25 and 95°C, respectively. Lanes 1, 2, 3, and 4 were developed with antibodies against the six-His tag, and lanes 5 and 6 were developed with polyclonal antibodies against native 17-kDa protein. (C) Recombinant 17-kDa pilin protein (His tagged) analyzed for oligomer formation by gel filtration on a Sephadex G-200 column (35 by 1.2 cm; Pharmacia). Protein fractions were eluted with 20 mM sodium phosphate buffer (pH 8.0). Protein concentrations in the fractions were determined by measuring the absorbance at 220 nm. Peaks of standard molecular mass markers are indicated by arrows.
Analytical gel chromatography.
Gel filtration was performed to determine the degree of oligomerization of the recombinant 17-kDa protein. The elution profile of the protein obtained with a Sephadex G-200 column is shown in Fig. 4C. The protein recovery estimates showed that about 22% of the recombinant protein was irreversibly adsorbed to the column, possibly due to the formation of large insoluble aggregates, and was not eluted, as has been shown previously (39). A small fraction of the protein eluted in the void volume as large multimers (whose molecular organization was not determined), while the major portion eluted as oligomers of different sizes; the molecular mass of the largest population was in the range from 50 to 160 kDa. In contrast, 65% of the triple-mutant protein was irreversibly adsorbed on the column, and the amount of the eluted protein was barely within detection limits (data not shown).
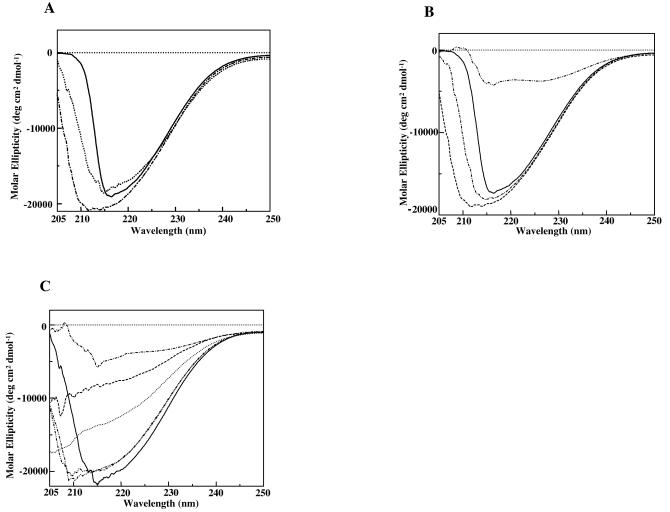
CD spectroscopy.
The CD spectra of the native pilin protein, the recombinant wild-type protein, and the mutant proteins are shown in Fig. 5A and C. The recombinant 17-kDa proteins (with and without the His tag), like the native pilin, displayed a deep negative Cotton effect in the far-UV CD spectrum, with a minimum around 210 to 215 nm, which was indicative of an ordered structure (Fig. 5A). When the proteins were denatured in 8 M urea, the CD signals were greatly reduced, as expected (Fig. 5B and C), reflecting the loss of structure. The proteins with the Phe34Ala and Ile38Ala individual mutations displayed CD spectra similar to that of the wild-type protein at higher concentrations (Fig. 5B), indicating that these mutations caused little change in the protein structure (Fig. 5C). However, a progressive decrease was observed in the molar ellipticity of the Val32Ala mutant and the triple mutant (Val-Phe-Ile → Ala-Ala-Ala). In each of these cases the negative peak between 210 and 215 nm was missing, and the spectrum more closely resembled that of the denatured wild-type protein. These results indicate that there was an order-disorder transition in the protein structure. Furthermore, the strongest destabilization observed for the triple mutant pointed towards the additive contributions of the three residues and thus their participatory role in donor strand complementation (Fig. 5C).
FIG. 5.
Comparision of far-UV CD spectra of the native and recombinant 17-kDa pilin proteins. Twenty spectra were averaged, and CD signals were converted to molar ellipticity. (A) CD spectra of both the native and recombinant 17-kDa pilin proteins at a concentration of 20 μM. Solid line, recombinant 17-kDa protein with six-His tag; dotted line, recombinant 17-kDa protein without tag; dotted and dashed line, purified native pilin protein. (B) CD spectra for different concentrations of recombinant 17-kDa protein. Solid line, 20 μM; dashed line with single dots, 30 μM; dashed line, 40 μM; dashed line with double dots, 20 μM recombinant 17-kDa pilin protein denatured with 8 M urea. (C) CD spectra at a protein concentration of 20 μM, showing relative CD signal intensities. Solid line, wild-type 17-kDa protein; dashed line with single dots, 17-kDa protein denatured with 8 M urea; dotted line, Val32Ala mutant; dashed lines with double dots, Phe34Ala and Ile38Ala mutants; dashed line, triple mutant (Val-Phe-Ile → Ala-Ala-Ala).
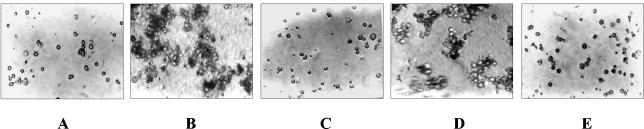
Hemocyte agglutination and cytotoxicity.
Similar to the native protein (20), the recombinant 17-kDa protein caused profuse blebbing and agglutination of H. armigera hemocytes (Fig. 6B and Table 3). No clumping was seen in the buffer control or in the presence of E. coli cells (Fig. 6A and E). Antiserum against the 17-kDa protein inhibited agglutination of hemocytes by the recombinant protein (Fig. 6C), while preimmune serum had no effect on hemocyte agglutination (Fig. 6D).
FIG. 6.
Agglutination of H. armigera fifth-instar larval hemocytes by recombinant 17-kDa protein (His tagged). Hemocytes obtained from H. armigera were suspended in Grace's insect medium. Protein samples were added, and the plate was incubated at 28°C for 2 h. Agglutination was observed by light microscopy. (A) Control (hemocytes in PBS); (B) 10 μg of recombinant 17-kDa protein; (C) recombinant 17-kDa protein preincubated with polyclonal antiserum (1:100) against native 17-kDa protein; (D) recombinant 17-kDa protein preincubated with preimmune serum; (E) 107 E. coli K-12 cells per ml.
TABLE 3.
Agglutination of rabbit erythrocytes and H. armigera fourth- or fifth-instar larval hemocytes by X. nematophila cells and recombinant 17-kDa proteina
| Sample | Agglutination titerb
|
|
|---|---|---|
| Erythrocytes | Hemocytes | |
| Buffer control | NA | NA |
| X. nematophila cells (107 cells/ml) | 64 | 64 |
| E. coli cells (107 cells/ml) | ND | <2 |
| Wild-type 17-kDa protein with six-His tag | NA | 32 |
| Wild-type 17-kDa protein without tag | NA | 32 |
| Heat-inactivated 17-kDa protein with six-His tag | NA | <2 |
| Triple mutant with six-His tag | NA | 8 |
| E. coli K-12 purified pilin protein | ND | <2 |
| Preimmune serum (undiluted) | NA | <2 |
| Wild-type 17-kDa protein + preimmune serum | NA | 32 |
| Wild-type 17-kDa protein + anti-17-kDa serum | ND | <2 |
The initial amount of protein was 100 μg.
NA, no activity; ND, not determined.
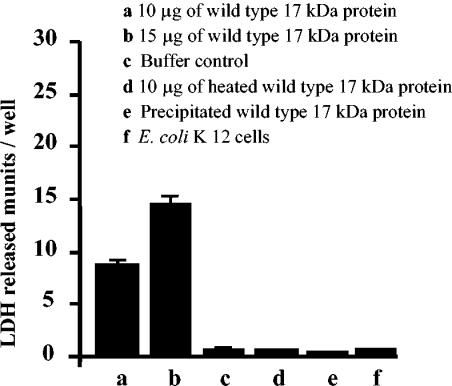
A cytoplasmic enzyme, lactate dehydrogenase (LDH), was released into the medium as a consequence of an interaction of the recombinant 17-kDa protein with the target cells (Fig. 7). The total LDH content of the hemocytes was 27.6 ± 1.4 mU/well, while 0.71 ± 0.12 mU of LDH per well was released in the buffer control. About 8.5 ± 0.32 and 14.2 ± 1.2 mU of the intracellular LDH per well were released when the cells were incubated in the presence of 10 and 15 μg of the recombinant 17-kDa protein, respectively (Fig. 7). Only 1.01 ± 0.12 mU of LDH per well was detected in the presence of E. coli K-12 cells. Heat-inactivated recombinant wild-type protein resulted in the release of 0.61 ± 0.22 mU of LDH per well. The protein that precipitated at a higher concentration was not toxic to cells.
FIG. 7.
Cytotoxicity for larval hemocytes of recombinant 17-kDa proteins (His tagged). The cells were incubated with the test proteins at 28°C for 4 to 5 h. The supernatants were separated, and LDH activity was determined.
The sugar galactose and the sugar derivatives xylitol, glucaric acid, glucouronic acid, and gluconic acid inhibited hemocyte agglutination by the protein, as reflected by values for inhibition of LDH release ranging from 50 to 80% (Table 4). Interestingly, when whole cells of X. nematophila were used as a source of agglutinating protein, in addition to all the compounds mentioned above, N-acetyl-lactosamine and arabinose also inhibited hemocyte agglutination (Table 4). These results suggest that there are two distinct binding specificities present on the cells of X. nematophila.
TABLE 4.
Inhibition of Xenorhabdus cell-mediated and wild-type 17-kDa protein-mediated hemocyte agglutination by various sugars and their derivatives
| Sugar or derivativea | Concn (mM) | Inhibition of LDH releasea
|
|
|---|---|---|---|
| Wild-type 17-kDa protein (7.5 μg/well) | Xenorhabdus (107 cells/ml) | ||
| Lactose | 500 | − | − |
| Xylitol | 250 | + | + |
| Mannose | 500 | − | − |
| Sorbitol | 500 | − | − |
| Trehalose | 500 | − | − |
| d-Glucouronic acid | 100 | + | + |
| N-Acetyl-lactosamine | 25 | − | + |
| Glucose | 500 | − | − |
| Glucaric acid | 50 | + | + |
| Fructose | 500 | − | − |
| d-Gluconic acid | 100 | + | + |
| Mannitol | 500 | − | − |
| Galactose | 100 | + | + |
| Arabinose | 500 | − | + |
+, more than 50% inhibition; −, no inhibition.
Binding of recombinant 17-kDa protein to larval hemocytes.
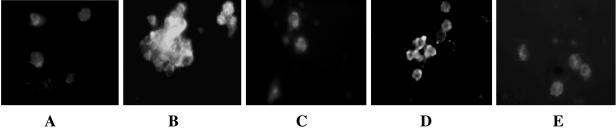
Binding of the recombinant 17-kDa protein to the larval hemocytes was reflected by strong fluorescence on the cell surface (Fig. 8B). Preincubation of the protein with antiserum against the 17-kDa protein inhibited binding of the recombinant 17-kDa protein to the hemocytes (Fig. 8C), while incubation with preimmune serum had no effect on binding of the protein as the intensity of emission from the cells remained unchanged (Fig. 8D). The binding of proteins to the hemocyte surface was consistent with cell clumping activity.
FIG. 8.
Immunofluorescence, showing binding of recombinant 17-kDa protein (His tagged) on H. armigera fourth- or fifth-instar larval hemocytes. Hemocyte monolayers were incubated with protein samples for 30 min and fixed with 0.4% formaldehyde in PBS. Binding of the protein was detected by using polyclonal antibodies against native 17-kDa protein of X. nematophila, followed by fluorescein-labeled conjugate. (A) Control hemocytes with buffer; (B) recombinant 17-kDa protein; (C) recombinant 17-kDa proteins preincubated with polyclonal antiserum against native 17-kDa protein; (D) recombinant 17-kDa protein preincubated with preimmune serum before binding to the cells; (E) E. coli K-12 cells.
Insecticidal activity of the toxin.
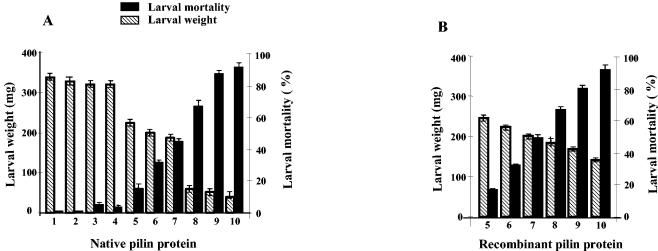
The native pilin and the purified recombinant 17-kDa pilin subunit of X. nematophila showed oral larvicidal activity when they were incorporated into the diet of neonatal H. armigera (Fig. 9). A dose-dependent effect on larval mortality was observed throughout the larval period (Fig. 9). The LD50s were 3.3 and 3.6 μg/cm3 for the native and recombinant wild-type 17-kDa proteins, respectively. A corresponding decrease in the larval weight was observed at all of the concentrations of the proteins (Fig. 9). Heating the protein at 80°C reduced the larvicidal activity substantially; this treatment resulted in 5.1% ± 2% larval death at a protein dose of 10 μg/cm3 of the diet. Furthermore, E. coli whole cells, BSA at a concentration of 20 μg/cm3 of the diet, and the buffer used to resuspend the proteins showed no larval toxicity (Fig. 9A).
FIG. 9.
Bioassay of purified native (A) and wild-type (B) 17-kDa proteins of X. nematophila, showing larval mortality and growth inhibition. Each group contained 24 larvae, and mortality was recorded for the entire larval period, which varied from 12 and 15 days in the controls and the test groups, respectively. Bar 1, control; bar 2, 20 μg of BSA/cm3 of diet; bar 3, 20 μg of heat-inactivated recombinant 17-kDa protein/cm3 of diet; bar 4, E. coli K-12 cells; bars 5 to 10, 0.4, 2, 4, 6, 8, and 12 μg of 17-kDa protein/cm3 of diet, respectively.
Histopathology.
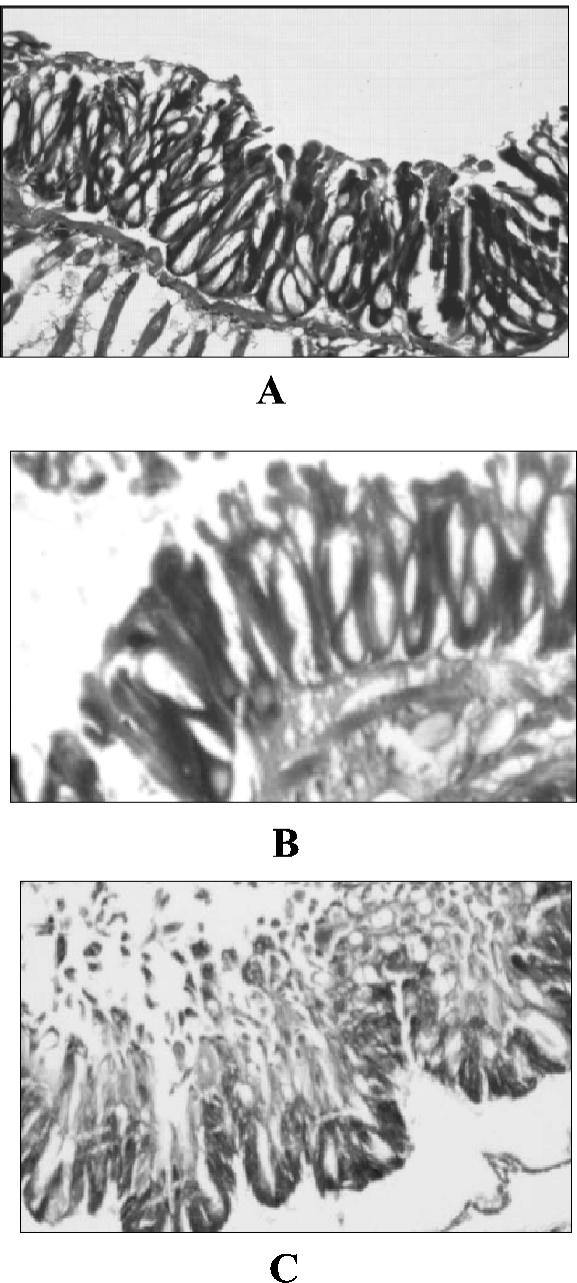
The midgut membrane of a 6-day-old H. armigera larva showed typical cellular morphology of the epithelial lining, limited by a basement membrane (Fig. 10A). The membrane integrity was not affected in an E. coli-fed larva (Fig. 10B). However, when a larva was fed the 17-kDa pilin protein, breakdown of the epithelial lining was observed. Both the basement membrane and the cellular lining were disorganized, and extensive damage to the integrity of the gut lining was observed at later stages, resulting in sloughing of cell debris in the lumen (Fig. 10C).
FIG. 10.
Histopathological sections of gut epithelial membrane of H. armigera larvae treated with the 17-kDa protein. (A) Control; (B) larva fed E. coli K-12; (C) purified recombinant 17-kDa protein. (A and C) Magnification, ×10; (B) magnification, ×20.
DISCUSSION
In this study we investigated the insecticidal potential of a 17-kDa protein present in the large protein complex secreted by X. nematophila as OMVs (19). The purified protein was cytotoxic to the larval hemocytes of H. armigera (20). To gain insight into the basis of the toxicity of the 17-kDa protein in the host, we cloned and expressed the gene encoding the pilin subunit of X. nematophila. Phylogenetic comparison of the deduced amino acid sequence of the 17-kDa protein with other fimbrial sequences identified by PSI-BLAST demonstrated that the Xenorhabdus protein forms a distinct tight cluster with the structural subunits of mannose-resistant fimbrial subunits, including MrpA of P. mirabilis and SfmA of Serratia marcescens (Fig. 1B). Several E. coli fimbrial proteins, like PrsA, KS71A, F7-2, and PapA, the major structural proteins of the P pilus, also belong to this cluster. The bootstrap value based on 1,000 random trials at the node separating the cluster from the remaining fimbrial proteins was as high as 970, showing that the cluster can be reliably considered to be distinct.
The high level of sequence similarity of the Xenorhabdus fimbrial gene with the papA gene, which encodes the structural subunit of the P pilus of E. coli, provided the opportunity to include the wealth of structural information available for the PapA protein and on this basis verify structural characteristics of the recombinant Xenorhabdus protein. It was necessary to ensure that the protein had attained a native-like structure, as the recombinant protein was refolded under in vitro conditions in the absence of the cellular machinery thought to be essential for pilin assembly.
The recombinant protein was expressed in E. coli in an insoluble form. Analysis of the recombinant 17-kDa protein showed that the protein was expressed as it is in native Xenorhabdus, without its leader sequence or a tag at the N terminus. Upon refolding, the recombinant 17-kDa protein (no tag) had an ordered conformation (as demonstrated by its CD spectrum [Fig. 5A]) and was organized into small, native-like, SDS-resistant oligomers showing biological activity in the in vitro assay. However, since the yields of the purified protein were low (1 to 2 mg/liter), we expressed the protein in the pET23a vector with a C-terminal His tag. When the structural and functional properties of the histidine-tagged protein were compared with those of the recombinant (no histidine tag) and native proteins, it was evident that a significant portion of the recombinant protein had refolded and had a structure similar to that of the native protein; hence, we performed all further investigations with the protein containing a six-His tag at the C terminus.
Elution of the recombinant 17-kDa pilin subunit as a broad peak at molecular masses ranging from 50 to 160 kDa from the gel filtration column and the CD spectra together indicated that the oligomeric protein consisting of monomers with an ordered structure was formed during in vitro refolding. The oligomers were similar to those formed by the interaction of monomers in the mature, native pilin, as 1.5% SDS at 25°C could not destabilize them (Fig. 4A, lane 1). Although the degree of oligomerization of the native protein was substantially higher than that of the recombinant wild-type proteins, the fact that even small oligomers were produced in the absence of the chaperone is significant. Like the Xenorhabdus protein, the recombinant PapA protein of E. coli was also shown to produce only small oligomers, even when it was expressed in the presence of the cognate chaperone (7, 37). Furthermore, the oligomers were produced by the interaction between neighboring subunits through the conserved N-terminal donor strand, as point mutations in the conserved N-terminal region of the protein substantially reduced this interaction, causing structural defects in the protein. Taken together, the results described above reinforce the role of the N-terminal region in the assembly of a stable, biologically active pilin protein of X. nematophila.
It is important that refolding and oligomerization of the Xenorhabdus protein occurred in the absence of the cognate chaperone and produced stable oligomers. This is rather surprising, because the presence of the chaperone is considered indispensable during pilin assembly (34), as the chaperone binds to the newly synthesized pilin monomers and protects them from premature aggregation and degradation by proteases (24). So far, there has been only one report demonstrating expression and oligomerization of pilin subunits in vitro in the absence of the chaperone. Vetch et al. (39) showed that refolding of the FimH adhesin protein of E. coli occurred in vitro in the absence of its cognate chaperone, FimC; however, the stability of the refolded pilin domain was low. In the case of the Xenorhabdus protein, the role of the chaperone is presumably partially taken over by the N-terminal overhang of the monomers, which act as a chaperone for a neighboring subunit, albeit with reduced efficiency. In the process of in vitro refolding in the presence of a relatively high concentration of the recombinant protein, there is ample opportunity for the molecules to visit various conformational states. At least some of these states can be involved in productive interactions with the N-terminal extension of another molecule, forming stable multimeric structures.
The inhibition of cytotoxicity for cultured hemocytes by the sugar derivatives points to a specific recognition event between the pilin protein and the insect cells. At this stage we do not know if the pilin protein has any role in the recognition of the nematode host. Interestingly, the two compounds which inhibited the interaction of whole bacterial cells with the hemocytes, N-acetyl-lactosamine and arabinose, showed no inhibition when they were added to the hemocytes in the presence of the 17-kDa protein, suggesting that there is an additional molecule with a distinct binding specificity on the bacterial surface. This could be the adhesin subunit, which is normally responsible for host cell recognition in other gram-negative bacteria. Recently, the mrx fimbrial operon of X. nematophila has been reported, and this operon contains an open reading frame corresponding to the adhesin subunit (17); however, no information concerning the characteristics of the adhesin protein of X. nematophila is available. Moureaux et al. (27) also described inhibition of agglutination of sheep erythrocytes by arabinose and N-acetyl-lactosamine in the presence of X. nematophila cells. So far, there is no direct evidence showing biological activity of the structural subunit of pilin. Our results demonstrate for the first time sugar-specific recognition of host cells by a structural subunit of a pilin.
The most significant finding of this study is the association of oral larvicidal activity and concomitant damage of the gut membrane with the structural subunit of the pilin protein. Several protein toxins of Xenorhabdus and the related genus Photorhabdus have been described previously. The LD50s of the individual Xpt toxins isolated from X. nematophila were shown to range from 2 to 5 μg for different species of Pieris and Heliothis (36); these values are comparable to the values for the pilin protein studied here. The high-molecular-weight TcA protein isolated from Photorhabdus had a lower LD50 (870 ng) for Galleria larvae (5). At this stage we cannot comment on the contribution of hemocyte cytotoxicity to the overall insecticidal activity of the protein.
X. nematophila is a successful endosymbiont of the soil nematode S. carpocapsae, but it switches to an aggressive mode inside an insect larva. Recent sequencing of several endosymbiotic bacterial genomes has revealed that obligate host-associated bacteria have some of the smallest genomes known in nature (25). Although the whole genome of Xenorhabdus is not available yet, it should be mentioned that the recently published mrx operon of X. nematophila is considered to be the smallest fimbrial operon studied to date (17). In this context it is tempting to speculate that in X. nematophila, synthesis of proteins with dual functions may be a mechanism to minimize the genome size in order to get a competitive edge in the natural habitat. Thus, the presence of abundant pilin protein on the surface of the cells, which is primarily used for host recognition, also serves as a convenient tool to intoxicate the insect larva rapidly. Furthermore, OMVs, which have been recognized as a means of effector protein secretion in several gram-negative pathogens (40), provide a ready vehicle for transportation of the protein outside the cell if necessary.
In conclusion, this is a first report demonstrating the larvicidal nature of a structural subunit of X. nematophila pilin. Our results also show that the recombinant protein can be refolded and oligomerized in vitro without the cognate chaperone, which has not been reported previously. It can acquire an oligomeric organization similar to that of the native protein through donor strand complementation of the N-terminal extension, between successive monomers. The finding that the protein can assume a structure competent for biological activity without the assistance of any other factor demonstrates its potential as a biological control agent, particularly in the context of an urgent need for discovering novel biopesticides.
Acknowledgments
This work was funded by ICGEB, New Delhi and Ministry of Environment and Forest, Government of India. P.K. is a recipient of a senior research fellowship from CSIR, Government of India.
We acknowledge Faizan Ahmed, Jamia Millia Islamia, and R. P. Roy, National Institute of Immunology, for use of their CD spectrophotometer. The immunofluorescence experiments were carried out with the support of C. Chitnis, ICGEB. We are also thankful to Shiv Kumar for help with the larval hemocyte studies.
REFERENCES
- 1.Akhurst, R. J. 1982. Antibiotic activity of Xenorhabdus spp. bacteria symbiotically associated with insect pathogenic nematodes of the families Heterorhabditae and Steinernematidae. J. Gen. Microbiol. 128:3061-3065. [DOI] [PubMed] [Google Scholar]
- 2.Akhurst, R. J., and G. B. Dunphy. 1993. Symbiotically associated entomopathogenic bacteria, nematodes and their insect hosts, p. 1-23. In N. Beckage, S. Thompson, and B. Federici (ed.), Parasites and pathogens of insects, vol. 2. Academic Press, Inc., New York, N.Y. [Google Scholar]
- 3.Beveridge, T. J. 1999. Structures of Gram-negative cell walls and their derived membrane vesicles. J. Bacteriol. 181:4725-4733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boemare, N. E., and R. J. Akhrust. 1988. Biochemical and physiological characterization of colony form variants in Xenorhabdus ssp. ( Enteriobacteriaceae). J. Gen. Microbiol. 134:751-761. [Google Scholar]
- 5.Bowen, D., T. A. Rocheleau, M. Blackburn, O. Andreev, E. Golubeva, R. Bhartia, and R. H. Ffrench-Constant. 1998. Insecticidal toxins from the bacterium Photorhabdus luminescens. Science 280:2129-2132. [DOI] [PubMed] [Google Scholar]
- 6.Brown, S. E., A. T. Cao, E. R. Hines, and R. J. Akhurst. 2004. A novel secreted protein toxin from the insect-pathogenic bacterium Xenorhabdus nematophila. J. Biol. Chem. 279:14595-14601. [DOI] [PubMed] [Google Scholar]
- 7.Bullitt, E., C. H. Jones, R. Striker, G. Soto, F. Jacob-Dubuisson, J. Pinkner, M. J. Wick, L. Makowski, and S. J. Hultgren. 1996. Development of pilus organelle subassemblies in vitro depends on chaperone uncapping of a beta zipper. Proc. Natl. Acad. Sci. USA 93:12890-12895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Choudhury, D., A. Thompson, V. Stojanoff, S. Langermann, J. Pinkner, S. J. Hultgren, and S. D. Knight. 1999. X-ray structure of the FimC-FimH chaperone-adhesion complex from uropathogenic E. coli. Science 285:1061-1066. [DOI] [PubMed] [Google Scholar]
- 9.Converse, V., and P. S. Grewal. 1998. Virulence of entomopathogenic nematodes to the western masked chafer Cyclocephala hirta. (Coleoptera: Scarabaeidae) J. Econ. Entomol. 91:428-432. [DOI] [PubMed] [Google Scholar]
- 10.Converse, V., and R. W. Miller. 1999. Development of the one-on-one quality assessment assay for entomopathogenic nematodes. J. Invertebr. Pathol. 74:143-148. [DOI] [PubMed] [Google Scholar]
- 11.Felsenstein, J. 1989. PHYLIP—phylogeny inference package (version 3.2). Cadistics 5:164-166. [Google Scholar]
- 12.Finney, D. J. 1971. Probit analysis. Cambridge University Press, Cambridge, United Kingdom.
- 13.Fitch, W. M., and E. Margoliash. 1967. Construction of phylogenetic trees. Science 155:279-284. [DOI] [PubMed] [Google Scholar]
- 14.Forst, S., and K. Nealson. 1996. Molecular biology of the symbiotic-pathogenic bacteria Xenorhabdus spp. and Photorhabdus spp. Microbiol. Rev. 60:21-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gaastra, W., and E. K. De Graaf. 1992. Host-specific fimbrial adhesin of noninvasive enterotoxigenic Escherichia coli strains. Microbiol. Rev. 46:129-161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Givaudan, A., and A. Lanois. 2000. flhDC, the flagellar master operon of Xenorhabdus nematophilus: requirement for motility, lipolysis, extracellular hemolysis, and full virulence in insects. J. Bacteriol. 182:107-115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.He, H., H. A. Snyder, and S. Frost. 2004. Unique organization and regulation of the MRX fimbrial operon in Xenorhabdus nematophila. Microbiology 150:1439-1446. [DOI] [PubMed] [Google Scholar]
- 18.Higgins, D. G., J. D. Thompson, and T. J. Gibson. 1996. Using CLUSTAL for multiple sequence alignments. Methods Enzymol. 266:383-402. [DOI] [PubMed] [Google Scholar]
- 19.Khandelwal, P., and N. B. Bhatnagar. 2003. Insecticidal activity associated with the outer membrane vesicles of Xenorhabdus nematophilus. Appl. Environ. Microbiol. 69:2032-2037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Khandelwal, P., D. Choudhury, R. Bhatnagar, and N. Banerjee. 2004. Characterization of a cytotoxic pilin subunit of Xenorhabdus nematophila. Biochem. Biophys. Res. Commun. 314:943-949. [DOI] [PubMed] [Google Scholar]
- 21.Klemm, P., M. Schembri, and D. L. Hasty. 1996. The FimH protein of type 1 fimbriae. An adaptable adhesin. Adv. Exp. Med. Biol. 408:193-195. [DOI] [PubMed] [Google Scholar]
- 22.Kolling, G. L., and K. R. Matthews. 1999. Export of virulence genes and Shiga toxin by membrane vesicles of Escherichia coli O157:H7. Appl. Environ. Microbiol. 65:1843-1848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Korhonen, T. K., E. L. Nurmaha, Rantah, and C. S. Eden. 1980. New methods for isolation of immunologically pure pili from Escherichia coli. Infect. Immun. 27:569-575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kuehn, M. J., S. Normark, and S. J. Hultgren. 1991. Immunoglobulin-like papD chaperone caps and uncaps interactive surfaces of nascently translocated pilus subunits. Proc. Natl. Acad. Sci. USA 88:10586-10590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Moran, N. A. 2002. Microbial minimalism: genome reduction in bacterial pathogens. Cell 108:583-586. [DOI] [PubMed] [Google Scholar]
- 26.Morgan, J. A., M. Sergeant, D. Ellis, M. Ousley, and P. Jarrett. 2001. Sequence analysis of insecticidal genes from Xenorhabdus nematophilus PMFI296. Appl. Environ. Microbiol. 67:2062-2069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Moureaux, N., T. Karjalainen, A. Givaudan, P. Bourlioux, and N. Boemare. 1995. Biochemical characterization and agglutinating properties of Xenorhabdus nematophilus F1 fimbriae. Appl. Environ. Microbiol. 61:2707-2712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nishimatsu, T., and J. J. Jackson. 1998. Interaction of insecticides, entomopathogenic nematodes and larvae of the western corn rootworm (Coleoptera: Chrysomelidae). J. Econ. Entomol. 91:410-418. [DOI] [PubMed] [Google Scholar]
- 29.Pearce, W. A., and T. M. Buchanan. 1978. Attachment role of gonococcal pili. Optimum conditions and quantification of adherence of isolated pili to human cells in vitro. J. Clin. Investig. 61:931-943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Poinar, G. O., Jr. 1996. The presence of. Achromobacter nematophilus in the infective stage of a Neoplacenta sp. (Steinernematidae: Nematoda). Nematologica 12:105-108. [Google Scholar]
- 31.Pugsley, A. P. 1993. The complete general secretary pathway in gram-negative bacteria. Microbiol. Rev. 57:50-108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Reddy, M. K., S. Nair, and S. K. Sopory. 2002. A new approach for efficient directional genome walking using polymerase chain reaction. Anal. Biochem. 306:154-158. [DOI] [PubMed] [Google Scholar]
- 33.Sauer, F. G., S. J. Pinker, G. Waksman, and S. J. Hultgren. 2002. Chaperone priming of pilus subunits facilitates a topological transition that drives fiber formation. Cell 111:543-551. [DOI] [PubMed] [Google Scholar]
- 34.Sauer, F. G., K. Fiitter, J. S. Pinkner, K. W. Dodson, S. J. Hultgren, and G. Waksman. 1999. Structural basis of chaperone function and pilus biogenesis. Science 285:1058-1061. [DOI] [PubMed] [Google Scholar]
- 35.Schroeder, P. C., C. S. Ferguson, A. M. Shelton, W. T. Wilsey, M. P. Hoffmann, and C. Petzoldt. 1996. Greenhouse and field evaluations of entomopathogenic nematodes (Nematoda: Heterorhabditidae and Steinernematidae) for control of cabbage maggot (Diptera: Anthomyiidae) on cabbage. J. Econ. Entomol. 89:1109-1115. [DOI] [PubMed] [Google Scholar]
- 36.Sergeant, M., P. Jarrett, M. Ousley, and A. W. Morgan. 2003. Interactions of insecticidal toxin gene products from Xenorhabdus nematophilus PMF 1296. Appl. Environ. Microbiol. 69:3344-3349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Striker, R., F. Jacob-Dubuisson, C. Friden, and S. J. Hultgren. 1994. Stable fiber-forming and nonfiber-forming chaperone-subunit complexes in pilus biogenesis. J. Biol. Chem. 269:12233-12239. [PubMed] [Google Scholar]
- 38.Tinker, J. K., L. S. Hancox, and S. Clegg. 2001. FimW is a negative regulator affecting type I fimbrial expression in Salmonella enterica serovar Typhimurium. J. Bacteriol. 183:435-442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vetch, M., B. Sebbel, and R. Glockshuber. 2002. Chaperone-independent folding of type 1 pilus domains. J. Mol. Biol. 322:827-840. [DOI] [PubMed] [Google Scholar]
- 40.Wai, S. N., B. Lindmark, T. Soderblom, A. Takade, M. Westermark, J. Oscarsson, J. Jass, A. Richter-Dahlfors, Y. Mizunoe, and B. E. Uhlin. 2003. Vesicle-mediated export and assembly of pore-forming oligomers of the enterobacterial ClyA cytotoxin. Cell 115:25-35. [DOI] [PubMed] [Google Scholar]
- 41.Zavialov, A. V., J. Berglund, A. F. Pudney, L. J. Fooks, T. M. Ibrhahim, S. MacIntyre, and S. D. Knight. 2003. Structure and biogenesisof the capsular F1 antigen from Yersinia pestis: preserved folding energy drives fiber formation. Cell 113:587-596. [DOI] [PubMed] [Google Scholar]
- 42.Zavialov, A. V., J. Kersley, T. Korpela, V. P. Zaviyalov, S. MacIntyre, and S. D. Knight. 2002. Donor strand complementation mechanism in the biogenesis of nonpilus systems. Mol. Microbiol. 45:983-995. [DOI] [PubMed] [Google Scholar]
- 43.Zhao, H., X. Li, D. E. Johnson, I. Blomfield, and H. I. Mobley. 1997. In vivo phase variation of MR/P fimbrial gene expression in Proteus mirabilis infecting the urinary tract. Mol. Microbiol. 23:1009-1019. [DOI] [PubMed] [Google Scholar]