Abstract
Accurate DNA replication involves polymerases with high nucleotide selectivity and proofreading activity. We show here why both fidelity mechanisms fail when normally accurate T7 DNA polymerase bypasses the common oxidative lesion 8-oxo-7, 8-dihydro-2′-deoxyguanosine (8oG). The crystal structure of the polymerase with 8oG templating dC insertion shows that the O8 oxygen is tolerated by strong kinking of the DNA template. A model of a corresponding structure with dATP predicts steric and electrostatic clashes that would reduce but not eliminate insertion of dA. The structure of a postinsertional complex shows 8oG(syn)·dA (anti) in a Hoogsteen-like base pair at the 3′ terminus, and polymerase interactions with the minor groove surface of the mismatch that mimic those with undamaged, matched base pairs. This explains why translesion synthesis is permitted without proofreading of an 8oG·dA mismatch, thus providing insight into the high mutagenic potential of 8oG.
Keywords: DNA damage, DNA polymerase, protein crystallography, replication fidelity
Introduction
The high fidelity of DNA synthesis that typifies replicative DNA polymerases (Kornberg and Baker, 1992) is challenged by ongoing oxidative damage to DNA caused by normal metabolism and exposure to toxicants (Cooke et al, 2003). Although a number of repair enzymes cleanse the genome of oxidatively damaged nucleosides, it is inevitable that some of the ongoing damage remains unrepaired and it must be negotiated by DNA polymerases during replication. The highly mutagenic lesion 8-oxo-7, 8-dihydro-2′-deoxyguanosine (8oG) (Kuchino et al, 1987) is one of the most abundant oxidation products, approaching levels of one 8oG lesion per 104–105 guanines (Fraga et al, 1990; Beckman and Ames, 1997; Marnett, 2000). The presence of 8oG in DNA has been implicated in a variety a diseases including cancers, neurodegenerative diseases, cystic fibrosis and aging (Cooke et al, 2003). 8oG is a particularly insidious type of DNA damage because it efficiently templates the insertion of both C and A during DNA synthesis by many different polymerases (Shibutani et al, 1991).
At physiological pH 8oG is predominately in the 6, 8-diketo form and protonated at N7 (Uesugi and Ikehara, 1977; Cho et al, 1990). The 8oG nucleoside can adopt both syn and anti conformations seen in standard nucleosides by rotating about the glycosidic bond (Figure 1A; Uesugi and Ikehara, 1977); however, the syn conformation of 8oG may actually be favored because of the interference of the bulky O8 group with the sugar phosphate backbone in the anti conformation of 8oG (Figure 1A; Uesugi and Ikehara, 1977). The syn conformation of 8oG's glycosidic bond exposes the Hoogsteen edge of 8oG for interaction with dA in the anti conformation (Figure 1B). In the anti conformation, 8oG can pair with dC (anti) in a normal Watson–Crick base pair. NMR (Kouchakdjian et al, 1991; Oda et al, 1991) and X-ray crystallographic studies (McAuley-Hecht et al, 1994; Lipscomb et al, 1995) of DNA containing an 8oG (syn)·dA (anti) base pair or an 8oG (anti)·dC (anti) base pair show that both pairings are readily accommodated in a B-form DNA helix without a significant distortion. These altered physical and chemical properties of 8oG have been proposed to underlie its dual coding characteristics, with both syn and anti conformations templating DNA synthesis (Shibutani et al, 1991; Furge and Guengerich, 1997, 1998; Einolf et al, 1998; Einolf and Guengerich, 2001; Freisinger et al, 2004). However, there is little physical evidence showing how the 8oG lesion is accommodated in the active site of a DNA polymerase during lesion bypass.
Figure 1.
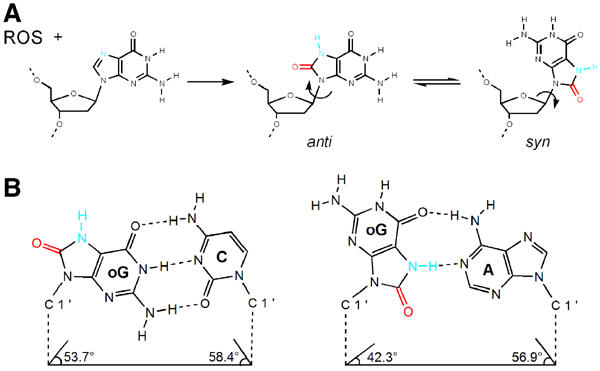
The dual coding potential of 8oG is explained by its unique chemical characteristics and physical properties. (A) Oxidation of guanosine by reactive oxygen species (ROS) leads to the formation of 8oG, which is distinguished from guanosine by the O8 group (colored red) and the protonation of nitrogen N7 (colored blue). Nucleosides have free rotation at their glycosidic bond (arrow); thus, they exist in equilibrium between syn and anti conformations. Whereas guanosine predominates in an anti conformation, the syn conformation is favored in 8oG, presumably due to the unfavorable steric interaction between the O8 group and the ribose O4 oxygen. (B) 8oG forms both Watson–Crick and Hoogsteen base pairs (Lipscomb et al, 1995; McAuley-Hecht et al, 1994). An 8oG (anti)·dC base pair (left) is structurally identical to a canonical dG·dC base pair, in that the anti conformation of the 8oG allows for hydrogen bonding between the Watson–Crick faces of both bases, and that the C1′–C1′ distance and the minor groove hydrogen bond donors and acceptors are conserved. The protonated N7 and O8 groups project into the major groove. The 8oG (syn)·dA base pair (right) is formed from the Hoogsteen face of 8oG in syn with the Watson–Crick face of adenine in anti. In this mismatch, the N1, N2 and O6 atoms of the 8oG (syn) project into the major grove and the O8 groups project into the minor groove of the base pair.
Most DNA polymerases exhibit an error rate of about 10−3–10−5 per nucleotide incorporated in the absence of proofreading activity (Echols and Goodman, 1991; Kool, 2002; Kunkel, 2004). This accuracy is much greater than can be accounted for by the different base-pairing energies of matched and mismatched base pairs, leading to the proposal that the shape of the polymerase active site further exaggerates the penalty for mispairing during DNA synthesis (Goodman, 1997). Crystal structures of several DNA polymerases complexed to DNA and nucleotide substrates support the proposal that ‘geometric selection' of the correct nucleotide for incorporation results from a conformational change of the fingers subdomain of the polymerase that constrains base pairing in the active site (reviewed in Doublie and Ellenberger, 1998; Steitz, 1999). The open conformation of the fingers allows nucleotide substrates to sample the polymerase active site, and the closed conformation brings catalytically important residues into contact with the nucleotide substrate. This is a type of induced fit mechanism in which the fingers close, or perhaps some other conformational change occurs (Yang et al, 2002), at different rates around matched versus mismatched nucleotide pairs.
Occasional mistakes during DNA synthesis are subject to editing by the 3′–5′ proofreading exonuclease activity of T7 DNA polymerase and other accurate polymerases. The rapid rate of DNA synthesis catalyzed by T7 polymerase (300 nt s−1) slows dramatically (to 0.01 s−1) following misincorporation. The exonucleolytic reaction (2.3 s−1) is then favored, and the DNA shifts from the polymerase site to the 3′–5′ exonuclease site without dissociating (Wong et al, 1991). How does the polymerase detect a misincorporated base and initiate the transfer of DNA to the exonuclease site? In crystal structures of T7 DNA polymerase and other polymerases complexed to DNA, the DNA is in the A-form with a widened minor groove near the polymerase active site (Brautigam and Steitz, 1998) where conserved residues interact with the universal hydrogen bond acceptors (Seeman et al, 1976) in the minor groove of the bound DNA. We have previously proposed that these interactions could detect misincorporated nucleotides because a mispair at the 3′ end of the primer would prevent an intimate interaction with the polymerase, resulting in dissociation of the DNA and rebinding to the proofreading exonuclease active site.
Crystal structures of two different DNA polymerases in complex with an 8oG·dCTP base pair bound were recently reported (Krahn et al, 2003; Freisinger et al, 2004). However, the structural consequences of an 8oG (syn)·dA (anti) mispair and its elongation remain to be determined. Below we describe crystal structures of a highly accurate replicative DNA polymerase from bacteriophage T7 in complexes with DNA containing 8oG either during nucleotide insertion or in postinsertional complexes with 8oG base paired with dC or dA at the 3′ end of the primer. We show that the efficiency of bypass and the mutational spectrum of 8oG are unaffected by the presence of proofreading activity, and the crystal structures explain how the 8oG·dA base pair defeats the normal error detection mechanisms that would otherwise suppress mutations during DNA synthesis.
Results
Translesion synthesis past 8oG
Kinetic analysis of single nucleotide incorporation with an exonuclease-deficient T7 DNA polymerase has shown that the rates of dCTP insertion opposite 8oG (Furge and Guengerich, 1997) and incorporation of the next correct nucleotide into 8oG-containing template–primer (Furge and Guengerich, 1998) are reduced in comparison to rates with undamaged dG. Here we have examined bypass parameters using a different experimental approach (Kokoska et al, 2003), one that measures the efficiency of each successive incorporation event during a complete 8oG bypass reaction with all four dNTPs present. Under these conditions, exonuclease-deficient T7 DNA polymerase bypasses 8oG with 7% efficiency as compared to bypass of undamaged template G in the same sequence context (Figure 2). As anticipated by the steady-state kinetic data using single nucleotides (Furge and Guengerich, 1997, 1998), this reflects less efficient dNTP insertion opposite 8oG (16%, lane 4) and less efficient extension from the 8oG-containing template–primer (34%). Interestingly, after adding only one nucleotide beyond the lesion, subsequent nucleotides are incorporated with efficiencies similar to those seen with the undamaged template. For comparison, we also measured bypass parameters for wild-type T7 DNA polymerase. The results (Figure 2, lane 8) indicate that the efficiencies of insertion opposite 8oG (30%), extension from the damaged substrate (45%) and overall bypass of template 8oG (15%) are all slightly higher for the wild-type enzyme than for the exonuclease-deficient polymerase (Table I). Thus, the intrinsic 3′ exonuclease activity of T7 DNA polymerase does not reduce the 8oG bypass efficiency of the polymerase.
Figure 2.
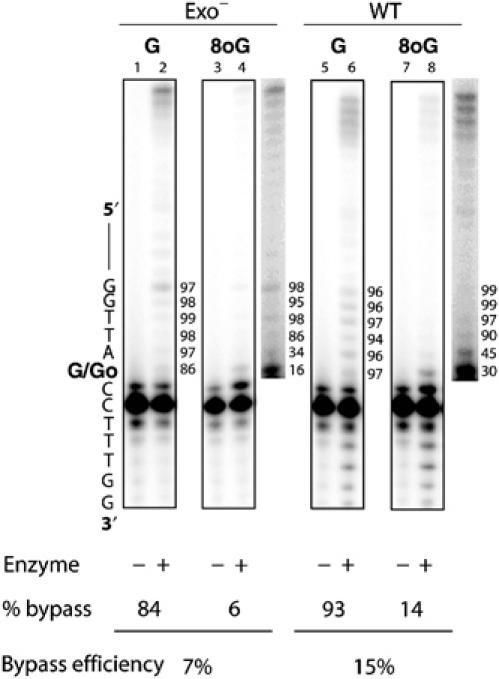
T7 DNA polymerase bypass of an 8oG lesion. Primer extension reactions were performed with exo− (left) and wild-type (right) T7 DNA polymerase with undamaged guanine (G) and 8-oxoguanine (8oG) in comparison to controls containing no enzyme. The images shown are for 3 min incubations of reaction mixtures containing 200- to 400-fold excess of DNA over polymerase. The most intense band in each lane is unreacted primer, at least 80% of which remains unextended for all efficiency reactions performed in this study. The location of 8oG within the template strand is as indicated and enhanced images of products using 8oG are shown to the right of the boxed images. The probability of insertion at each template site, listed in percent to the right of each lane, is an average of 7–16 determinations and is calculated as described previously (Kokoska et al, 2003).
Table 1.
T7 DNA polymerase fidelity during copying of a template 8oG lesion
| Polymerase | Template | Plaques |
Dark blue frequency | dNMP incorporated |
||
|---|---|---|---|---|---|---|
| Dark blue | Totala | Dark blue | Light blue | |||
| Exo− | 8oG | 187 | 876 | 0.21 | 31/31 dAMP | 32/32 dCMP |
| 477 | 2059 | 0.24 | 24/24 dAMP | 24/24 dCMP | ||
| G | 3 | 2137 | 0.0014 | |||
| 2 | 4071 | 0.00049 | ||||
| WT | 8oG | 127 | 662 | 0.19 | 32/32 dAMP | 29/29 dCMP |
| 469 | 1755 | 0.27 | 23/23 dAMP | 24/24 dCMP | ||
| G | 0 | 5052 | <0.00020 | |||
| aFor all variables, a few colorless plaques are included in the totals. These were observed at frequencies between 0.5 and 2.1%, with no significant differences in frequencies among the variables listed. WT: wild type. | ||||||
To determine if the 3′ exonucleolytic proofreading activity of this replicative DNA polymerase influences the fidelity of 8oG bypass, we measured error rates using a recently developed lesion bypass fidelity assay (Kokoska et al, 2003). Undamaged and 8oG-containing templates were copied by exonuclease-deficient and wild-type T7 DNA polymerases to generate primarily full-length DNA products. These products were gel purified and hybridized to gapped M13lac DNA molecules, which were then introduced into Escherichia coli cells and plated to obtain M13 plaques (Kokoska et al, 2003). Base substitution errors were scored as dark blue plaque revertants of a TAG amber codon in this LacZ α-complementation gene sequence, which encodes a plaque phenotype that is light blue due to slight read through of the stop codon. The identity of the base incorporated opposite 8oG was determined by sequence analysis of DNA isolated from independent M13 plaques. As shown in Table I, synthesis past 8oG by exonuclease-deficient T7 polymerase generated products with a dark blue plaque phenotype at a frequency of 21%, a value much higher than for copying the undamaged G (Table I). Among 31 blue plaques examined by sequence analysis, all resulted from misincorporation of dA opposite 8oG. In this assay system, 60% of polymerization errors are scored upon introduction into E. coli cells (Kokoska et al, 2003). This allows us to calculate that 32% of all bypass events by exonuclease-deficient T7 polymerase result from stable misincorporation of dA opposite 8oG, which requires both misinsertion of dAMP opposite 8oG and multiple subsequent incorporations of the damaged, mismatched template–primer. The light blue plaque frequency and sequencing data (Table I) demonstrate that the vast majority of other bypass events involve stable misincorporation of dC opposite 8oG. This ambiguous coding potential of 8oG is consistent with previous kinetic studies of exonuclease-deficient T7 polymerase (Furge and Guengerich, 1997, 1998). More interestingly, synthesis past 8oG by wild-type T7 polymerase produced misincorporation frequency and specificity results that were indistinguishable from the results with the exonuclease-deficient polymerase (Table I). The observed error rate of about 1 in 3 for this normally highly accurate replicative DNA polymerase demonstrates that the 3′ to 5′ exonuclease activity, which can very efficiently proofread undamaged base-base mismatches, does not excise dAMP inserted opposite 8oG. This agrees with earlier studies showing that the proofreading activity of two other polymerases has little effect on the outcome of synthesis past 8oG (Shibutani et al, 1991).
In order to investigate the structural basis for the dual coding potential of an 8oG lesion, we determined four crystal structures of T7 DNA polymerase in complexes with DNA and nucleotide substrates (Table II). The structures correspond to two different steps of lesion bypass—the insertion of a nucleotide opposite 8oG (insertion complexes) and the elongation of DNA synthesis from an 8oG-containing base pair (postinsertion complexes). Three of the structures reveal a catalytically active, closed conformation of the polymerase during dCTP incorporation (8oG·dCTP insertion complex) or during elongation from 8oG paired with dC (8oG·dC postinsertion complex) or dA (8oG·dA postinsertion complex). An attempt to crystallize the polymerase with an 8oG·dATP base pair in the polymerase active site resulted in an open conformation of the polymerase lacking bound dATP (8oG open complex; Table II)
Table 2.
Data collection, phasing and refinement statistics
| Template guanosine | Template 8-oxoguanosine | ||||
|---|---|---|---|---|---|
| Complex | Native G·dCTP insertion | 8oG·dCTP insertion | 8oG open | 8oG·dC postinsertion | 8oG·dA postinsertion |
| Position | Active site | Active site | Active site | Extension | Extension |
| Active site ddNTP | ddCTP | ddCTP | None | ddTTP | ddTTP |
| Conformation | Closed | Closed | Open | Closed | Closed |
| Data collection | |||||
| Beamline | Laboratory source | NSLS X26-C | APS 19-ID | NSLS X26-C | NSLS X26-C |
| Space group | P21212 | P21212 | P21212 | P21212 | P21212 |
| Unit cell parameters | |||||
| a | 106.3 | 106.2 | 105.3 | 105.5 | 106.2 |
| b | 218.1 | 215.1 | 213.3 | 215.1 | 215.7 |
| c | 52.1 | 52.1 | 52.2 | 52.0 | 52.2 |
| Resolution (Å) | 2.54 | 2.3 | 2.0 | 2.48 | 2.3 |
| Total reflections | 325 241 | 1 006 949 | 1 681 015 | 773 206 | 779 077 |
| Unique reflections | 39 002 | 51 127 | 79 532 | 40 910 | 52 299 |
| Completeness (%) | 94.3 (86.0) | 93.3 (66.1) | 99.6 (99.9) | 96.4 (86.0) | 96.5 (77.0) |
| 〈I〉/〈σ(I)〉 | 13.8 (2.9) | 23.8 (5.1) | 17.9 (5.0) | 24.6 (8.4) | 23.6 (12.7) |
| Rsym b | 0.069 (0.360) | 0.075 (0.224) | 0.097 (0.423) | 0.071 (0.253) | 0.063 (0.083) |
| Refinement | |||||
| Resolution | 40–2.54 | 50–2.3 | 50–2.2 | 50–2.48 | 50–2.5 |
| No. of reflections | 37 036 | 48 466 | 57 565 | 38 896 | 40 258 |
| Rcryst c | 0.220 | 0.217 | 0.214 | 0.202 | 0.206 |
| Rfree c | 0.255 | 0.263 | 0.246 | 0.264 | 0.261 |
| Number of atoms | |||||
| Protein/DNA | 7328 | 6856 | 6672 | 7081 | 7085 |
| Solvent | 414 | 574 | 811 | 762 | 574 |
| Ave. B-factor (Å2) | 39.2 | 38.0 | 34.6 | 31.1 | 28.5 |
| R.m.s.d. bonds (Å) | 0.006 | 0.006 | 0.005 | 0.005 | 0.005 |
| R.m.s.d. angles (deg) | 1.2 | 1.3 | 1.1 | 1.2 | 1.2 |
| aValues in parentheses refer to data in the highest resolution shell. | |||||
| bRsym=∑hkl∑j∣Ij−〈I〉∣/∑hkl∑jIj, where 〈I〉 is the mean intensity of j observations of reflection hkl and its symmetry equivalents. | |||||
| cRcryst=∑hkl∣Fobs−kFcalc∣/∑hkl∣Fobs∣. Rfree=Rcryst for 5% of reflections that were not used in refinement. | |||||
A structural rationale for deoxycytidine insertion opposite 8oG
As a reference structure, a ternary complex was crystallized in which dCTP is paired with dG in the polymerase active site (native dG·dCTP insertion complex). Aside from the identity of the base pair in the polymerase active site, the native complexes of T7 DNA polymerase with dG·dCTP and dC·dGTP (Doublie and Ellenberger, 1998) are virtually identical. However, a subtle shift in the crystal packing arrangement of the native dG·dCTP complex stabilized two loops (residues 294–320 and residues 576–588) that were disordered and the duplex DNA exiting the polymerase also became better ordered. The dG in the templating position forms a Watson–Crick base pair with the incoming ddCTP, and two Mg2+ ions that are ligated by the nucleotide are well positioned to participate in catalysis (Figure 3A).
Figure 3.

Structural rationale for deoxycytidine insertion opposite 8oG. The σA-weighted Fo–Fc annealed omit electron density maps, contoured at 3σ, are shown superimposed onto the refined models of incoming ddCTP across from template guanosine (A) and template 8oG (B). In the 8oG·dCTP insertion complex, Lys536 (gold bonds) moves 3 Å relative to its position in the native dG·dCTP complex and forms a hydrogen bond with the O8 group. The positions of the Mg2+ ions (yellow spheres) are similar to previously determined T7 DNA polymerase closed complexes. (C) Superposition of ternary complexes of T7 DNA polymerase and Pol β with G and 8oG at the templating position. The conformations of an 8oG·dCTP insertion complex (orange) and native G·dCTP insertion complex (yellow) are almost identical. A superposition between both complexes reveals the conservation of a DNA kink of the 5′ phosphate backbone at the templating base. A superposition of a dG·dCTP insertion of pol β (green) shows that the kink is not as pronounced and a steric clash between the 5′ phosphate and the O8 group is prone to occur. However, the structure of pol β in an 8oG·dCTP insertion complex (blue) reveals that pol β is able to incorporate dCTP across from the lesion because of a local conformational change of the DNA backbone.
The 8oG·dCTP insertion complex crystallized under the same conditions as the native ternary complexes (Doublie et al, 1998), producing crystals in the same space group and similar unit cell parameters (Table II). However, in contrast to the native complexes in which the nucleotide remains bound without adding ddNTP to the harvest buffer, crystals of the 8oG·dCTP complex required the addition of 5 mM ddCTP in the crystal harvest buffer in order to obtain high-quality diffracting crystals of the complex. Based on our experiences with the crystallization of T7 DNA polymerase in complex with other lesioned DNAs (Li et al, 2004), we suspect that base-pairing interactions between the 8oG template and the nucleotide bound in the polymerase active site are less stable, and the lesion interferes with the closure of the fingers.
The conformation of T7 DNA polymerase in complex with 8oG·dCTP is nearly identical to the native dG·dCTP complex (root mean square deviation of 0.2731 Å). The nascent base pair between 8oG (anti) and incoming ddCTP (anti) (Figure 3B) fits snugly within the active site and the fingers are in a fully closed conformation. However, there is one notable change in comparison to the native dG·dCTP complex; the side chain of residue Lys536 moves about 3 Å and its ɛ amino group is in position to donate a hydrogen bond to the aberrant O8 group of 8oG (Figure 3B). This favorable interaction might stabilize 8oG in the anti conformation for base pairing with dCTP. Although we anticipated a clash between the 5′ phosphate of 8oG (anti) and the O8 oxygen, a sharp kink in the DNA template draws the phosphate out of the way, more than 3.5 Å away from the O8 oxygen. A sharp kinking of the DNA template at its point of entry into the polymerase active site is a conserved feature of many different DNA polymerases (Doublie et al, 1998; Johnson et al, 2003) including human DNA polymerase β (Pol β) (Sawaya et al, 1997) and bacteriophage RB69 DNA polymerase (Franklin et al, 2001). A recent structure of a Pol B family DNA polymerase from bacteriophage RB69 in complex with 8oG·dCTP (Freisinger et al, 2004) also shows that the 8oG template is accommodated with no rearrangement of residues in the active site compared with a native dA·dTTP complex (Franklin et al, 2001).
In the light of the crystallographic information available for Pol β in catalytic complexes with 8oG (Krahn et al, 2003) and dG (Sawaya et al, 1997) templating bases, we were curious to compare these structures with the analogous structures of T7 DNA polymerase. The structures were superimposed by aligning the nascent base pair bound to the active site of each structure (Figure 3C). It is evident that the kinking of the DNA template in the native Pol β complexes is less pronounced than that seen in the T7 DNA polymerase complexes. Without the strong kinking of the DNA, a local shift in the position of the 5′ phosphate is required to accommodate 8oG (anti) in the templating position of the Pol β active site, thereby avoiding a clash with the O8 group in the complex. Thus, Pol β negotiates the 8oG (anti) nucleotide by a local conformational change in the 5′ phosphate to permit dCTP incorporation opposite the lesion (Krahn et al, 2003).
dATP does not stably pair with 8oG in the polymerase active site
Attempts to crystallize T7 DNA polymerase in the process of inserting dATP across from 8oG resulted in the crystallization of a binary complex (Figure 4). Although the DNA binds to the polymerase in the same register as for native catalytic complexes, the fingers adopt a catalytically inactive, open conformation, and there is no convincing electron density for dATP or the 5′ end of the template strand including the 8oG nucleotide. Residues 531–536 are disordered in a segment spanning the C-terminal end of the O helix and its connection with helix O1. The side chain of Tyr530 (not shown) has rotated upwards and it occupies the binding site normally occupied by the templating base in a closed complex (Figure 4).
Figure 4.
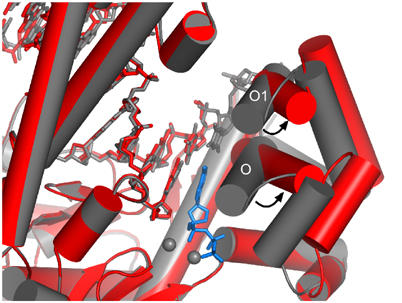
Comparison of an open 8oG complex and a closed T·ddATP insertion complex. The open 8oG complex (red) and a dT·dATP insertion complex (gray) were superimposed using Cα atoms. The proteins are depicted as cylinders and the DNA as sticks. Both structures are largely similar but they specifically differ in the orientation adopted by their fingers subdomains. In the closed structure, α-helices O and O1 pack against the incoming ddATP (blue) and the template thymine, respectively. In the open structure, the fingers move outwards from the palm subdomain, as shown by the ∼45° rotation of the O and O1 helices relative to the closed conformation. Residue Tyr530, which moves to the position that would correspond to the templating base of the closed complex, has been omitted for clarity. The templating 8oG, the 5′ template strand, and residues 532–536 located at the junction between α helices O and O1 are disordered in the open complex. No interpretable electron density is observed for the metal ions or incoming nucleotide.
The failure of the 8oG template to bind in the polymerase active site is notable because the crystal structure of a different binary polymerase–DNA complex shows a well-ordered templating base bound in the polymerase active site, in the absence of dNTP and with the fingers in an open conformation (S Doublie and T Ellenberger, unpublished results). The crystallographic evidence suggests that 8oG binds less tightly to the polymerase active site than an undamaged base and that the interaction with dATP is not sufficient to stabilize a closed, active conformation of the polymerase during crystallization. The O8 oxygen of 8oG imposes local structural changes in the DNA backbone (Malins et al, 2000) that evidently are overcome by base-pairing interactions with dCTP in the polymerase active site (Figure 3B). These observations are in agreement with evidence that T7 DNA polymerase inserts dA less favorably than dC opposite 8oG (Furge and Guengerich, 1997), and the suggestion that chemistry is rate limiting for insertion of dA opposite 8oG (Furge and Guengerich, 1997). To better understand why pairing of 8oG with dATP is less favorable in the polymerase active site, we have modeled this base pair in a catalytically active, closed conformation.
Modeling a catalytic complex of dATP paired with 8oG
A docking model for the 8oG·dATP base pair bound to the closed, catalytically active conformation of T7 DNA polymerase was created based on a native dT·ddATP complex (Li et al, 2004). The 8oG (syn)·dA (anti) Hoogsteen-type base pair from one of the postinsertion complexes (Table II) was superimposed onto the Watson–Crick T·dATP base pair bound to the active site of T7 DNA polymerase by aligning the sugar phosphate backbones of the nucleotides. The 10.71 Å distance between the C1′–C1′ atoms of the 8oG (syn)·dATP (anti) base pair is within the normal range for Watson–Crick base pairs, and this Hoogsteen base pair fits snugly in the polymerase active site (Figure 5). Several steric and electrostatic clashes (red patches) are evident between the 8oG (syn) base and the closed fingers. The O8 oxygen impinges upon the edge of the Tyr530 side chain, creating a steric clash that is not readily relieved because of van der Waals interactions that constrain the position of the Tyr530 side chain. The N2 nitrogen of 8oG brushes up against the aliphatic carbons of side chains from Ile540 and Lys536, creating an unfavorable interaction that might prevent the fingers from closing around the 8oG (syn) nucleotide. In general terms, the 8oG (syn) base protrudes into the major groove (see also Figure 6D) where it would presumably interfere with closure of the fingers and formation of the catalytically active complex.
Figure 5.
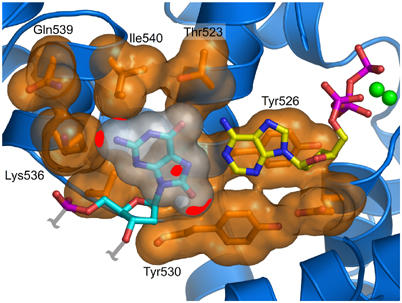
Model of a catalytic complex of dATP paired with 8oG. Modeling an 8oG(syn)·dATP pair from the 8oG·dA postinsertion complex into the polymerase active site shows that the amino acids from the O and O1 helices hinder the incorporation of dATP (yellow carbons) across from an 8oG lesion (cyan carbons). van der Waals surfaces are shown for protein side chains (orange) and 8oG (white). Unfavorable interactions between the 8oG and protein atoms are represented as red patches on the van der Waals surfaces. The N1 and N2 atoms of 8oG make detrimental electrostatic interactions with the ɛ and δ amino groups of Lys536 and the γ carbon of Ile540. The O8 group also makes steric clashes with residue Tyr530.
Figure 6.

Structural rationale for elongation from an 8oG lesion. The 8oG (anti)·dC (anti) postinsertion complexes are shown in panels A, C and E. 8oG (syn)·dA (anti) postinsertion complexes are shown in panels B, D and F. (A, B) The σA-weighted Fo–Fc annealed omit electron density maps, contoured at 3σ, for the elongated 8oG·dCMP (A) and 8oG·dAMP (B) structures are shown against the refined models. The topology of the major groove is highlighted with dashed arcs. In the 8oG (syn)·dA base pair, the adenine N1, N2 and O6 atoms protrude into the major groove and the O8 group is located at the minor groove. (C, D) Molecular surface representations of the DNA in the elongation complexes. The major groove surfaces corresponding to the 8oG atoms are colored red and the Mg2+ ions are represented as green spheres. The major groove surface of the 8oG·dC postinsertion complex (C) is similar to that of a canonical dG·dC base pair, whereas 8oG (syn) paired with dA forms a bump in the major groove (D). However, no steric clashes are observed between the polymerase and the bulky 8oG·dA pair. (E, F) Superpositions between 8oG postinsertion complexes and normal DNA. (E) A superposition between the 8oG·dC postinsertion complex (blue) and a normal postinsertion dA·dT complex (yellow) shows that the minor groove groups are positioned in an identical manner between the lesioned and the unmodified purine·pyrimidine base pairs. The acceptors are recognized by residues Gln615 and Arg429, which hydrogen bond with N7 of the 8oG base and O2 of the incorporated dC. The phosphate backbone of the 8oG rotates by 90° about its γ torsion angle in comparison to an unlesioned base, in order to avoid a steric clash with the O8 group. (F) A superposition between an 8oG·dA postinsertion complex (green) and a postinsertion dT·dA complex (orange) shows that the minor groove is identical between the lesioned and the unmodified pyrimidine·purine base pairs. Residues Gln615 and Arg429 form hydrogen bonds with the O8 group of 8oG and with the N3 group of the incorporated dA. The O8 group mimics the O2 group of a thymidine.
Extension of DNA synthesis past an 8oG lesion
In order to investigate the structural rationale for the extension of DNA synthesis after insertion of dC or dA opposite 8oG (Figure 2), we determined two crystal structures of postinsertional complexes. In these complexes, 8oG is paired with dC or dA at the 3′ end of the primer strand and dA is paired with dTTP in the polymerase active site (Table II). In both complexes, the fingers subdomain is in a fully closed conformation and two Mg2+ ions are bound in the polymerase active site.
The 8oG nucleotide in the 8oG (anti)·dC (anti) postinsertional complex (Table II and Figure 6A) does not grossly affect the structure of DNA bound to the polymerase. A normal Watson–Crick base pair is formed between 8oG (anti) and dC (anti) (Figure 6A). As in other polymerase–DNA complexes (Brautigam and Steitz, 1998; Doublie et al, 1999), the bound DNA has a widened minor groove and it resembles A-form DNA in the vicinity of the polymerase active site. Although the 8oG·dC base pair has no global effect, there is a local perturbation of the DNA backbone that accommodates O8 of 8oG in the anti conformation (Figure 7). The 5′ phosphate of 8oG is rotated out of the way by a 90° rotation around the bond connecting C4′ and C5′ of the DNA backbone (the γ torsion angle), relative to the backbone conformation of an unmodified purine (Figure 7). The orientation of the His607 side chain also adjusts so that it can participate in hydrogen bonding interactions with the phosphate of 8oG (anti) (Figure 7). The local adjustment of the DNA backbone that accommodates the 8oG (anti) nucleotide in the postinsertional complex is reminiscent of the changed phosphate position of an 8oG-containing template bound to Pol β (Krahn et al, 2003).
Figure 7.
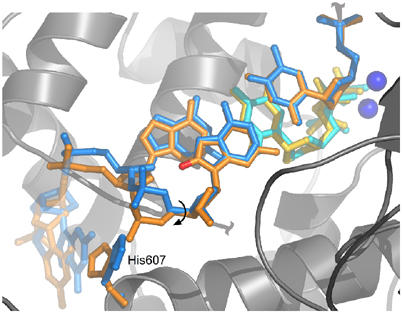
Bias against elongation from 8oG·dC. A superposition between the 8oG (anti)·dC (anti) postinsertion complex (blue) and a dG·dCTP insertion complex (orange) shows that a rotation around the C5′–C4′ bond of the 8oG phosphate backbone is necessary to avoid a steric clash with oxygen O8. This movement is translated to the adjacent nucleotide and it shifts the position of the sugar moiety of the templating strand by ∼1.5 Å in comparison to an unmodified template. As the C5′–C4′ bond of an 8oG (syn)·dA (anti) postinsertion complex is similar to a native dG·dCTP insertion complex, the subtle shift in the sugar moiety of an 8oG (anti)·dC (anti) pair may decrease the catalytic efficiency of nucleotide insertion, explaining the preference for elongation of a mismatch versus a correct pair. The imidazole ring of His607 tracks along with the 5′ phosphate for both elongation complexes as is observed for the original ternary complex, supporting its role in positioning the template.
In the 8oG (syn)·dA (anti) postinsertion complex, a Hoogsteen-like base pair is present at the 3′ end of the primer and its conformation is defined by the omit electron density (Figure 6B). The glycosidic torsion angle (χ) of 8oG (syn) is 75° and that of its dA (anti) partner is −148°. The 8oG (syn)·dA (anti) base pair is highly propeller twisted (ω=122°) and buckled (κ=115°). The geometry of the 8oG·dA base pair suggests two hydrogen bonding interactions between the bases. The protonated N7 of 8oG's Hoogsteen edge can donate a hydrogen bond to N1 of dA, and N6 of dA can donate a hydrogen bond to O6 of 8oG (Figure 6B). In contrast to the 8oG (anti)·dC (anti) base pair that does not alter the DNA major groove (Figure 6C), the Watson–Crick base-pairing edge of 8oG (syn) protrudes into the major groove, but there are no interactions of the polymerase with this region of the DNA and the Hoogsteen base pair is well tolerated (Figure 6D).
Minor groove readout fails to detect 8oG·A mispairs
Why is the 8oG·dA mispair not subject to proofreading by T7 DNA polymerase (Table I) The interactions between residues on the palm and the widened minor groove are thought to function as a sensor for misincorporated bases (Doublie et al, 1998). In catalytic complexes of T7 DNA polymerase, the strictly conserved residues Arg429 and Gln615 contact the universal hydrogen bond acceptors (Seeman et al, 1976) located on the minor groove surface of the base pair at the 3′ end of the primer. It is remarkable that 8oG-containing base pairs at the primer 3′ terminus faithfully engage in the same minor groove interactions with DNA polymerase as a normal base pair (Figure 6E and F). The 8oG (anti)·dC base pair from the postinsertion complex superimposes well onto the corresponding native dG·dC base pair from another complex (Figure 6E). The side chains from Gln615 and Arg429 contact N3 of 8oG and O2 of dC, respectively. Similarly, in the 8oG (syn)·dA (anti) postinsertion complex, the O8 of 8oG (syn) stands in to accept a hydrogen bond from Gln615 and resembles a pyrimidine O2 in the minor groove (Figure 6F). Thus, the minor groove surface of the 8oG·dA Hoogsteen pair impersonates a normal dT·dA base pair (Kouchakdjian et al, 1991; McAuley-Hecht et al, 1994; Lipscomb et al, 1995) and thereby defeats the polymerase's ability to detect the misincorporated base as being aberrant. Although the Hoogsteen base pair protrudes into the major groove near the polymerase active site (Figure 6B and D), the lesion does not alter the overall structure of the DNA or interfere with contacts made by the polymerase. Thus, there is no means for the polymerase to detect a mispaired adenine once it is inserted opposite 8oG.
Discussion
The crystal structures of T7 DNA polymerase reveal different consequences of the 8oG nucleoside at two different steps of lesion bypass. With 8oG in the templating position, the crystal structures of the insertional complexes (Table II) suggest that the oxidized base binds less tightly than a normal purine, destabilizing the closed, active conformation of the polymerase. Specifically, the 8oG (anti)·dCTP insertion complex required high concentrations of dCTP to stabilize the closed conformation of the fingers during crystal growth and harvest. The 8oG (syn)·dATP pairing was even less stable than an 8oG (anti)·dCTP, as we failed to trap the polymerase in a closed complex with this promutagenic base pair bound in the active site. Instead, the fingers remained open and there is no electron density for dATP or the 8oG templating base in the open conformation of the polymerase.
A molecular model of an 8oG (syn)·dATP pair in the closed conformation of the polymerase suggests clashes with 8oG that might interfere with closure of the fingers (Figure 5). An unfavorable contact between Tyr530 and the O8 group of 8oG may weaken binding of the lesioned base, even to the open conformation of the polymerase. Tyr530 is a conserved residue of Pol A family (Braithwaite and Ito, 1993) polymerases that contributes strongly to the fidelity of DNA synthesis (Minnick et al, 1999). The syn–anti equilibrium of the 8oG nucleotide that results from an unhindered rotation about the glycosidic bond may also increase the entropic cost of binding. The crystallographic data agree well with functional studies of T7 DNA polymerase showing the decreased catalytic efficiency of nucleotide insertion opposite 8oG (Figure 2; Furge and Guengerich, 1997) and a preference for the insertion of dC over dA (Table I).
After a nucleotide is inserted opposite the lesion, the 8oG-containing base pair has a modest effect on insertion of the next nucleotide and no effect on insertion of subsequent nucleotides, allowing bypass of the lesion to occur (Figure 2). This is also evinced by the relatively normal base-pairing geometry within the polymerase active site seen in crystal structures of postinsertion complexes in which 8oG is paired with dC or dA at the 3′ end of the primer. Thus, the structure of the bound DNA and its interactions with the polymerase are little affected by the 8oG lesion (Figure 6A and B). Of particular note is the preservation of minor groove interactions between Arg429 and Gln615 and the 8oG-containing base pairs, especially 8oG·dA (Figure 6E and F). The observation that the polymerase interacts with this damaged and mismatched primer terminus in a normal manner implies that dA will not efficiently be partitioned to the exonuclease active site, thus explaining as to why the proofreading activity of T7 DNA polymerase has no apparent effect on the rate of stable misincorporation of dA opposite 8oG (Table I).
The reduced 8oG bypass synthesis efficiency we see here (Figure 2) is consistent with the catalytic efficiencies of exonuclease-deficient T7 DNA polymerase during elongation from 8oG (syn)·dA (anti) and 8oG (anti)·dC (anti) pairs, which are reduced ∼50- and 300-fold, respectively, compared to elongation from a normal dG·dC pair (Furge and Guengerich, 1998). It is counterintuitive that elongation from a mismatched, damaged base pair (8oG·dA) is more efficient than elongation from a matched, damaged base pair (8oG·dC). In this regard, the crystal structure of the 8oG (anti)·dC (anti) postinsertion complex is informative in revealing a 1.3 Å shift in the sugar moiety of the templating nucleotide coupled with the 90° rotation around the C5′–C4′ bond (γ torsion angle) of the neighboring 8oG (anti) nucleotide (Figure 7). This change in the conformation of the phosphosugar backbone of the templating nucleotide might destabilize base pairing in the active site and explain the lower efficiency of elongation from an 8oG·dC base pair. Additionally, the change in the templating nucleotide might misalign the primer 3′ OH, but we would not detect this change in our studies because we have used a 2′, 3′-dideoxy nucleotide to halt DNA synthesis. In the other postinsertion complex that was crystallized, the 8oG (syn)·dA base pair at the 3′ end of the primer has no significant effect on the geometry of base pairing in the active site.
The lesion bypass activities of polymerases from the Y superfamily have been ascribed to a loose active site that accommodates lesioned bases, at the price of lower fidelity when copying undamaged DNA (Ling et al, 2001; Trincao et al, 2001). Paradoxically, the Y family polymerase η (Pol η) bypasses 8oG in a predominately error-free manner, with a 19-fold preference for insertion of dCMP over insertion of dAMP opposite the lesion (Haracska et al, 2000). This nearly error-free synthesis by Pol η is difficult to reconcile with the mutagenic bypass of 8oG by the otherwise faithful T7 DNA polymerase (Table I). A unique C-terminal domain of Y family polymerases, termed the ‘little finger' (Ling et al, 2001), is an important determinant of lesion bypass activity (Boudsocq et al, 2004). The little finger domain binds to DNA near the template base where it could influence the syn–anti conformational equilibrium of the 8oG template to favor the nonmutagenic pairing of 8oG (anti) with dCTP in the polymerase active site. Structures of Y family polymerases bypassing the normally mutagenic 8oG lesion should be informative in this regard.
Conclusions
The relatively high efficiency and low fidelity of DNA synthesis templated by 8oG result from the good fit of the lesion with the polymerase during insertion and elongation. The strong kinking of the DNA template facilitates the insertion of dC opposite 8oG and thereby moderates the mutagenic potential of the lesion. A mismatched 8oG (syn)·dA base pair in the polymerase active site may interfere somewhat with closure of the fingers and thereby attenuate misincorporation. For reactions templated by 8oG, the catalytic advantage of the closed conformation of the polymerase might not be fully realized. Nonetheless, dA is readily incorporated and the mutagenic potential of this mistake is fully achieved during elongation because an 8oG (syn)·dA mispair is accommodated as a native Watson–Crick pair. Remarkably, the proofreading activity of the polymerase is completely ineffective at correcting this mutagenic outcome.
Materials and methods
8oG bypass efficiency and fidelity assays
For reactions to determine bypass efficiency, the primer strand oligonucleotide 5′AATTTCTGCAGGTCGACTCCAAAG3′ was 5′ end labeled with 32P using T4 polynucleotide kinase and hybridized to a damaged or undamaged template strand 5′CCAGCTCGGTACCGGGTTAGoCCTTTGGAGTCGAC CTGCAGAAATT3′, where Go indicates the location of 8-oxoguanine on the damaged template or guanine on the undamaged template. Reaction mixtures contained 40 mM Tris–HCl (pH 7.5), 5 mM MgCl2, 50 mM NaCl, 5 mM dithiothreitol (DTT), 25 μM each of the four dNTPs, 130 nM substrate and either 0.325 or 0.65 nM wild-type or exonuclease-deficient T7 DNA polymerase. Reactions were incubated at 37° for 3–9 min and terminated by adding an equal volume of formamide stop solution (95% formamide, 25 mM EDTA, 0.1% xylene cynanol and 0.01% bromophenol blue). DNA products were separated on 12% denaturing polyacrylamide gels and analyzed by phosphoimagery. Reactions for assaying bypass fidelity were similar but used unlabeled substrates and [α-32P]dCTP, either 6.5 nM wild-type T7 DNA polymerase or 130 nM exo− T7 DNA polymerase, and incubation times of 5 or 30 min, respectively. DNA products were recovered and annealed to gapped M13lacDNA, which was then introduced into E. coli for determining mutant frequencies as described (Kokoska et al, 2003).
DNA synthesis and DNA substrate assembly
Oligonucleotides were synthesized on an Applied Biosystems DNA synthesizer. β-Mercaptoethanol (0.25 M) was added during deprotection to avoid oxidation of the 8oG base. Oligonucleotides were purified by PAGE, desalted using a C18 SEP-PAK cartridge (Waters Corp.) and concentrated to 2 mM in H2O. Equimolar amounts of DNA primer and template strands were annealed in a low-salt buffer prior to crystallization.
Protein expression and purification
T7 DNA polymerase (Δ6 exo−) and thioredoxin were overexpressed as described (Doublie et al, 1998). Lysates were mixed and a 1:1 complex was purified to homogeneity by phosphocellulose, anion exchange and hydroxyapatite chromatography. The complex was concentrated to 50 mg ml−1 in 50 mM ACES pH 7.5, 50% glycerol, 2 mM EDTA and 5 mM DTT.
Crystallization of polymerase–DNA complexes
A complex of T7 DNA polymerase (1 × 10−4 M) with thioredoxin was assembled using equimolar amounts of primer–template DNA in 50 mM HEPES pH 7.5, 10 mM MgCl2, 2 mM DTT, 0.5 mM terminal 2′, 3′-dideoxynucleotide and 10 mM ddNTP. To avoid misincorporation during the formation of the elongation complexes, a chain-terminating ddNTP (ddATP or ddCTP) was added and the reaction was incubated for 30 min on ice before the addition of the next incoming nucleotide.
Seed crystals were grown by hanging drop vapor diffusion by mixing 1 μl each of protein–DNA solution and a reservoir solution containing 14–17% PEG 8000, 100 mM ACES pH 7.5, 120 mM ammonium sulfate, 30 mM MgCl2 and 5 mM DTT. These crystals were used for streak-seeding in the presence of lower concentrations of PEG 8000. Rod-shaped crystals appear overnight and grow to ∼70 × 200 × 70 μm3 after 2–3 days. Crystals were soaked overnight in mother liquor containing 10% PEG 400 and 5 mM incoming ddNTP, mounted in nylon loops and flash frozen in liquid nitrogen.
X-ray data collection and structure determination
Data were collected at beamlines X26-C at the National Synchrotron Light Source, and 19-ID at the Advanced Photon Source, or with a rotating anode source. X-ray data were processed with the HKL2000 (Otwinowski and Minor, 1996) program. Molecular replacement solutions were obtained with the program EPMR (Kissinger et al, 1999) or CNS (Brunger et al, 1998) using only the protein atoms from the original structure of T7 DNA polymerase (Doublie et al, 1998) as a search model. The fingers subdomain (residues 500–560) was omitted from the search model used to determine the structure of the 8oG open complex.
Model building and refinement
Model building was performed by visual inspection of electron density maps using the programs O (Jones et al, 1991) and XtalView (McRee, 1991). DNA and protein atoms missing from the original model of T7 DNA polymerase were built into 2Fo–Fc and Fo–Fc electron density maps, followed by successive rounds of conjugate gradient minimization and individual B-factor refinement (Brunger et al, 1998) using CNS. Nucleotides in the bypass and active site positions were positioned using annealed Fo–Fc omit maps. After refinement of protein and DNA atoms, solvent molecules were positioned manually into the refined 2Fo–Fc and Fo–Fc electron density maps and refined. Figures were constructed using PyMOL (DeLano, 2002) and Ribbons (Carson, 1997)
Model coordinates
The atomic coordinates have been deposited in the Protein Data Bank under the entry codes 1T8E for the native dG·dCTP insertion complex, 1TK0 for the 8oG·dCTP insertion complex, 1TKD for the 8oG·dC postinsertion complex, 1TK8 for the 8oG·dA postinsertion complex and 1TK5 for the 8oG open complex.
Acknowledgments
We are indebted to John Pascal, Ying Li, Tapan Biswas, Hideki Aihara and Oleg Tsodikov for their generous help. We thank Caroline Kisker for communicating results previous to their publication, and the staff of beamlines X26-C (National Synchrotron Light Source, Brookhaven National Laboratory) and 19-ID (Advanced Photon Source, Argonne National Laboratory) for their assistance. This work was supported by a grant from the National Institutes of Health (GM55390) awarded to TE, and the resources of the Harvard-Armenise Structural Biology Center. LGB is supported by the Pew Latinoamerican Postdoctoral Fellowship Program. TE is the Hsien Wu and Daisy Yen Wu Professor of Biological Chemistry and Molecular Pharmacology.
References
- Beckman KB, Ames BN (1997) Oxidative decay of DNA. J Biol Chem 272: 19633–19636 [DOI] [PubMed] [Google Scholar]
- Boudsocq F, Kokoska RJ, Plosky BS, Vaisman A, Ling H, Kunkel TA, Yang W, Woodgate R (2004) Investigating the role of the little finger domain of Y-family DNA polymerases in low-fidelity synthesis and translesion replication. J Biol Chem, (in press) [DOI] [PubMed] [Google Scholar]
- Braithwaite DK, Ito J (1993) Compilation, alignment, and phylogenetic relationships of DNA polymerases. Nucleic Acids Res 21: 787–802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brautigam CA, Steitz TA (1998) Structural and functional insights provided by crystal structures of DNA polymerases and their substrate complexes. Curr Opin Struct Biol 8: 54–63 [DOI] [PubMed] [Google Scholar]
- Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, Read RJ, Rice LM, Simonson T, Warren GL (1998) Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr D 54: 905–921 [DOI] [PubMed] [Google Scholar]
- Carson M (1997) Ribbons. Methods Enzymol 277: 493–505 [PubMed] [Google Scholar]
- Cho BP, Kadlubar FF, Culp SJ, Evans FE (1990) 15N nuclear magnetic resonance studies on the tautomerism of 8-hydroxy-2′-deoxyguanosine, 8-hydroxyguanosine, and other C8-substituted guanine nucleosides. Chem Res Toxicol 3: 445–452 [DOI] [PubMed] [Google Scholar]
- Cooke MS, Evans MD, Dizdaroglu M, Lunec J (2003) Oxidative DNA damage: mechanisms, mutation, and disease. FASEB J 17: 1195–1214 [DOI] [PubMed] [Google Scholar]
- DeLano WL (2002) The PyMOL Molecular Graphics System on World Wide Web http://www.pymol.org
- Doublie S, Ellenberger T (1998) The mechanism of action of T7 DNA polymerase. Curr Opin Struct Biol 8: 704–712 [DOI] [PubMed] [Google Scholar]
- Doublie S, Sawaya MR, Ellenberger T (1999) An open and closed case for all polymerases. Struct Fold Des 7: R31–R35 [DOI] [PubMed] [Google Scholar]
- Doublie S, Tabor S, Long AM, Richardson CC, Ellenberger T (1998) Crystal structure of a bacteriophage T7 DNA replication complex at 2.2 Å resolution. Nature 391: 251–258 [DOI] [PubMed] [Google Scholar]
- Echols H, Goodman MF (1991) Fidelity mechanisms in DNA replication. Annu Rev Biochem 60: 477–511 [DOI] [PubMed] [Google Scholar]
- Einolf HJ, Guengerich FP (2001) Fidelity of nucleotide insertion at 8-oxo-7,8-dihydroguanine by mammalian DNA polymerase delta. Steady-state and pre-steady-state kinetic analysis. J Biol Chem 276: 3764–3771 [DOI] [PubMed] [Google Scholar]
- Einolf HJ, Schnetz-Boutaud N, Guengerich FP (1998) Steady-state and pre-steady-state kinetic analysis of 8-oxo-7,8-dihydroguanosine triphosphate incorporation and extension by replicative and repair DNA polymerases. Biochemistry 37: 13300–13312 [DOI] [PubMed] [Google Scholar]
- Fraga CG, Shigenaga MK, Park JW, Degan P, Ames BN (1990) Oxidative damage to DNA during aging: 8-hydroxy-2′-deoxyguanosine in rat organ DNA and urine. Proc Natl Acad Sci USA 87: 4533–4537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franklin MC, Wang J, Steitz TA (2001) Structure of the replicating complex of a pol alpha family DNA polymerase. Cell 105: 657–667 [DOI] [PubMed] [Google Scholar]
- Freisinger E, Grollman AP, Miller H, Kisker C (2004) Lesion (in) tolerance reveals insights into DNA replication fidelity. EMBO J 23: 1494–1505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furge LL, Guengerich FP (1997) Analysis of nucleotide insertion and extension at 8-oxo-7,8-dihydroguanine by replicative T7 polymerase exo- and human immunodeficiency virus-1 reverse transcriptase using steady-state and pre-steady-state kinetics. Biochemistry 36: 6475–6487 [DOI] [PubMed] [Google Scholar]
- Furge LL, Guengerich FP (1998) Pre-steady-state kinetics of nucleotide insertion following 8-oxo-7,8-dihydroguanine base pair mismatches by bacteriophage T7 DNA polymerase exo. Biochemistry 37: 3567–3574 [DOI] [PubMed] [Google Scholar]
- Goodman MF (1997) Hydrogen bonding revisited: geometric selection as a principal determinant of DNA replication fidelity. Proc Natl Acad Sci USA 94: 10493–10495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haracska L, Yu SL, Johnson RE, Prakash L, Prakash S (2000) Efficient and accurate replication in the presence of 7,8-dihydro-8-oxoguanine by DNA polymerase eta. Nat Genet 25: 458–461 [DOI] [PubMed] [Google Scholar]
- Johnson SJ, Taylor JS, Beese LS (2003) Processive DNA synthesis observed in a polymerase crystal suggests a mechanism for the prevention of frameshift mutations. Proc Natl Acad Sci USA 100: 3895–3900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones TA, Zou JY, Cowan SW, Kjeldgaard M (1991) Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr A 47: 110–119 [DOI] [PubMed] [Google Scholar]
- Kissinger CR, Gehlhaar DK, Fogel DB (1999) Rapid automated molecular replacement by evolutionary search. Acta Crystallogr D 55: 484–491 [DOI] [PubMed] [Google Scholar]
- Kokoska RJ, McCulloch SD, Kunkel TA (2003) The efficiency and specificity of apurinic/apyrimidinic site bypass by human DNA polymerase eta and Sulfolobus solfataricus Dpo4. J Biol Chem 278: 50537–50545 [DOI] [PubMed] [Google Scholar]
- Kool ET (2002) Active site tightness and substrate fit in DNA replication. Annu Rev Biochem 71: 191–219 [DOI] [PubMed] [Google Scholar]
- Kornberg A, Baker T (1992) DNA Replication. New York, NY: WH Freeman [Google Scholar]
- Kouchakdjian M, Bodepudi V, Shibutani S, Eisenberg M, Johnson F, Grollman AP, Patel DJ (1991) NMR structural studies of the ionizing radiation adduct 7-hydro-8-oxodeoxyguanosine (8-oxo-7H-dG) opposite deoxyadenosine in a DNA duplex. 8-Oxo-7H-dG(syn).dA(anti) alignment at lesion site. Biochemistry 30: 1403–1412 [DOI] [PubMed] [Google Scholar]
- Krahn JM, Beard WA, Miller H, Grollman AP, Wilson SH (2003) Structure of DNA polymerase beta with the mutagenic DNA lesion 8-oxodeoxyguanine reveals structural insights into its coding potential. Structure 11: 121–127 [DOI] [PubMed] [Google Scholar]
- Kuchino Y, Mori F, Kasai H, Inoue H, Iwai S, Miura K, Ohtsuka E, Nishimura S (1987) Misreading of DNA templates containing 8-hydroxydeoxyguanosine at the modified base and at adjacent residues. Nature 327: 77–79 [DOI] [PubMed] [Google Scholar]
- Kunkel TA (2004) DNA replication fidelity. J Biol Chem 279: 16895–16898 [DOI] [PubMed] [Google Scholar]
- Li Y, Dutta S, Doublie S, Moh'd Bdour H, Taylor JS, Ellenberger T (2004) Nucleotide insertion opposite a cis–syn thymine dimer by a replicative DNA polymerase from bacteriophage T7. Nature Struct Mol Biol, (in press) [DOI] [PubMed] [Google Scholar]
- Ling H, Boudsocq F, Woodgate R, Yang W (2001) Crystal structure of a Y-family DNA polymerase in action: a mechanism for error-prone and lesion-bypass replication. Cell 107: 91–102 [DOI] [PubMed] [Google Scholar]
- Lipscomb LA, Peek ME, Morningstar ML, Verghis SM, Miller EM, Rich A, Essigmann JM, Williams LD (1995) X-ray structure of a DNA decamer containing 7,8-dihydro-8-oxoguanine. Proc Natl Acad Sci USA 92: 719–723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malins DC, Polissar NL, Ostrander GK, Vinson MA (2000) Single 8-oxo-guanine and 8-oxo-adenine lesions induce marked changes in the backbone structure of a 25-base DNA strand. Proc Natl Acad Sci USA 97: 12442–12445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marnett LJ (2000) Oxyradicals and DNA damage. Carcinogenesis 21: 361–370 [DOI] [PubMed] [Google Scholar]
- McAuley-Hecht KE, Leonard GA, Gibson NJ, Thomson JB, Watson WP, Hunter WN, Brown T (1994) Crystal structure of a DNA duplex containing 8-hydroxydeoxyguanine-adenine base pairs. Biochemistry 33: 10266–10270 [DOI] [PubMed] [Google Scholar]
- McRee DE (1991) Xtal View/Xfit—a versatile program for manipulating atomic coordinates and electron density. J Struct Biol 125: 156–165 [DOI] [PubMed] [Google Scholar]
- Minnick DT, Bebenek K, Osheroff WP, Turner RM Jr, Astatke M, Liu L, Kunkel TA, Joyce CM (1999) Side chains that influence fidelity at the polymerase active site of Escherichia coli DNA polymerase I (Klenow fragment). J Biol Chem 274: 3067–3075 [DOI] [PubMed] [Google Scholar]
- Oda Y, Uesugi S, Ikehara M, Nishimura S, Kawase Y, Ishikawa H, Inoue H, Ohtsuka E (1991) NMR studies of a DNA containing 8-hydroxydeoxyguanosine. Nucleic Acids Res 19: 1407–1412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otwinowski Z, Minor W (1996) Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol 276: 307–326 [DOI] [PubMed] [Google Scholar]
- Sawaya MR, Prasad R, Wilson SH, Kraut J, Pelletier H (1997) Crystal structures of human DNA polymerase beta complexed with gapped and nicked DNA: evidence for an induced fit mechanism. Biochemistry 36: 11205–11215 [DOI] [PubMed] [Google Scholar]
- Seeman NC, Rosenberg JM, Rich A (1976) Sequence-specific recognition of double helical nucleic acids by proteins. Proc Natl Acad Sci USA 73: 804–808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibutani S, Takeshita M, Grollman AP (1991) Insertion of specific bases during DNA synthesis past the oxidation-damaged base 8-oxodG. Nature 349: 431–434 [DOI] [PubMed] [Google Scholar]
- Steitz TA (1999) DNA polymerases: structural diversity and common mechanisms. J Biol Chem 274: 17395–17398 [DOI] [PubMed] [Google Scholar]
- Trincao J, Johnson RE, Escalante CR, Prakash S, Prakash L, Aggarwal AK (2001) Structure of the catalytic core of S. cerevisiae DNA polymerase eta: implications for translesion DNA synthesis. Mol Cell 8: 417–426 [DOI] [PubMed] [Google Scholar]
- Uesugi S, Ikehara M (1977) Carbon-13 magnetic resonance spectra of 8-substituted purine nucleosides. Characteristic shifts for the syn conformation. J Am Chem Soc 99: 3250–3253 [DOI] [PubMed] [Google Scholar]
- Wong I, Patel SS, Johnson KA (1991) An induced-fit kinetic mechanism for DNA replication fidelity: direct measurement by single-turnover kinetics. Biochemistry 30: 526–537 [DOI] [PubMed] [Google Scholar]
- Yang L, Beard WA, Wilson SH, Broyde S, Schlick T (2002) Polymerase beta simulations suggest that Arg258 rotation is a slow step rather than large subdomain motions per se. J Mol Biol 317: 651–671 [DOI] [PubMed] [Google Scholar]


