Abstract
The active RNA-dependent RNA polymerase of poliovirus, 3Dpol, is generated by cleavage of the 3CDpro precursor protein, a protease that has no polymerase activity despite containing the entire polymerase domain. By intentionally disrupting a known and persistent crystal packing interaction, we have crystallized the poliovirus polymerase in a new space group and solved the complete structure of the protein at 2.0 Å resolution. It shows that the N-terminus of fully processed 3Dpol is buried in a surface pocket where it makes hydrogen bonds that act to position Asp238 in the active site. Asp238 is an essential residue that selects for the 2′ OH group of substrate rNTPs, as shown by a 2.35 Å structure of a 3Dpol–GTP complex. Mutational, biochemical, and structural data further demonstrate that 3Dpol activity is exquisitely sensitive to mutations at the N-terminus. This sensitivity is the result of allosteric effects where the structure around the buried N-terminus directly affects the positioning of Asp238 in the active site.
Keywords: allostery, poliovirus, polymerase, proteolysis, replication
Introduction
Poliovirus is a member of Picornaviridae, a family of viruses that includes the heart disease causing coxsackie virus, hepatitis A virus, foot and mouth disease viruses, and the rhinoviruses that are a major cause of the common cold (Semler and Wimmer, 2002). These highly homologous viruses contain ∼7500 nt positive sense single-stranded RNA genomes that are translated in a cap-independent manner via an internal ribosome entry site. This produces a single large polyprotein that is subsequently cleaved by cis-acting viral proteases into about a dozen different proteins required for viral propagation. RNA replication and virion assembly are coupled and occur at large viral replication center structures that are found on the surfaces of small vesicles, and all aspects of the viral life cycle, including RNA replication, take place in the cytoplasm.
The last protein in the viral polyprotein is 3Dpol, an RNA-dependent RNA polymerase (RdRp). 3Dpol is responsible for both replicating the infecting positive sense genome into minus sense complements and then using these as templates for the synthesis of positive sense genomes that are packaged into new virions. Poliovirus 3Dpol has an unusual priming reaction in which every RNA is covalently attached to the hydroxyl group of Tyr3 on the 22-residue viral 3B protein. This priming reaction is stimulated by the viral 3CDpro protein that is a fusion of the 3Dpol polymerase and the 3Cpro protease (Paul et al, 2000; Rieder et al, 2000). 3CDpro functions as an RNA-binding protein with a major role in controlling translation and replication of the viral genome (Andino et al, 1993; Murray and Barton, 2003). It binds predicted stem–loop structures in the 5′ and 3′ noncoding regions of the poliovirus genome as well as 2C(cre), an important regulatory RNA sequence in the middle of the genome. 3CDpro retains protease activity and cleaves capsid proteins up to 1000-fold more efficiently than 3Cpro alone (Ypma-Wong et al, 1988; Parsley et al, 1999). Proper proteolytic processing of the 3Dpol N-terminus is required to activate the polymerase as the 3CDpro precursor has absolutely no RNA polymerase activity despite the fact that it contains the entire polymerase domain (Flanegan and Van Dyke, 1979; Harris et al, 1992). The addition of 11 residues from the C-terminal end of 3Cpro to the N-terminus of 3Dpol results in a total loss of polymerase activity (Rothstein et al, 1988), as does the deletion of the first six residues (Hobson et al, 2001) or of only Trp5 (Plotch et al, 1989). The proteolytic processing-dependent activation of 3Dpol is likely important for viral replication because it prevents precursor proteins that have other functions in viral replication from acting as polymerases.
A partial crystal structure of poliovirus 3Dpol showed that RdRps shared the fingers–palm–thumb domain structure of DNA polymerases (Hansen et al, 1997). This original 3Dpol structure (PDB code 1RDR) and three other crystal forms (Hobson, 2000) crystallized in similar space groups whose lattices were dominated by a persistent crystal contact called Interface I. Interface I results in a head-to-tail oligomerization of the polymerase, which potentially reflects biologically relevant interactions in the membrane-bound replication complexes (Hobson et al, 2001; Lyle et al, 2002). Unfortunately, the electron density for the fingers domain was missing from all these structures, precluding a structural explanation of many aspects of polymerase function such as the critical importance of having a proper N-terminus in order for the polymerase to function. The original structure does show a small segment of the fingers domain where residues 25–37 interact with the top of the thumb and residues 12–24 descend toward the active site in the palm domain. Although the first 11 residues were not observed, the structure suggested that the N-terminus could be an integral component of the catalytic site. Such a structure would be very different from those of other RdRps, such as the rabbit hemorrhagic disease virus (Ng et al, 2002) and hepatitis C virus polymerases (Ago et al, 1999; Bressanelli et al, 1999; Lesburg et al, 1999), whose N-terminal are located on the back of the fingers domain.
To determine the complete structure of the poliovirus polymerase and ascertain the structural and functional role of the very N-terminus, we intentionally disrupted the persistent Interface I observed in the previous structures. This allowed the protein to crystallize in a new packing arrangement where the entire structure could be solved at 2.0 Å resolution. The complete 3Dpol structure reveals a novel polymerase activation motif where the very N-terminus of the protein is buried in a pocket on the back of the fingers domain. The buried terminus stabilizes a structure that directly positions aspartate 238 for binding to the 2′ OH group of the incoming nucleoside in the active site. We confirm this interaction by also presenting the 2.35 Å structure of a 3Dpol–GTP complex and the structure of a G1A mutant that alters the positioning of Asp238 and has partial activity.
Results
Crystallization
Interface I is an interaction between the thumb domain of one polymerase and the back of the palm domain of a second that buries ∼2000 Å2 of solvent-accessible surface area (Hansen et al, 1997; Hobson et al, 2001). A major molecular interaction along this interface is the insertion of Leu446 from the thumb into a hydrophobic pocket on the bottom of the palm of another polymerase molecule. The interface is further stabilized by intermolecular salt bridges linking Arg455–Asp349, Arg456–Asp339, and Asp459–Lys384. Mutations at these residues have been shown to disrupt the formation of 3Dpol sheet structures observed in electron microscopy studies (Lyle et al, 2002) and affect viral viability (Hobson et al, 2001). In addition, it is also possible that residues 12–36, observed to bind the top of the thumb domain in the original structure, were contributed in trans from another molecule through an interaction called Interface II (Hansen et al, 1997). Based on these observations, four different 3Dpol mutants were made in an effort to eliminate the native crystal contacts and force the formation of a new crystal lattice. First, Interface I was disrupted by mutating Leu446 to either alanine or aspartic acid and Arg455 to aspartic acid, that is, L446A/R455D and L446D/R455D. Second, these mutations were combined with deletion of the first 68 residues of 3Dpol, that is, Δ68/L446A/R455D and Δ68/L446D/R455D, to eliminate the proposed Interface II by initiating protein synthesis at the first ordered residue of the original structure. All four proteins expressed well and were purified at significantly higher yield than the wild-type protein. This is probably due to the higher solubility of the mutants, which increases from ∼3 mg/ml for the wild-type protein to ∼20 mg/ml for Δ68/L446A/R455D and greater than 40 mg/ml for L446D/R455D in 200 mM NaCl.
New crystallization conditions were found for two of the mutants, and their structures were solved by molecular replacement using the partial 3Dpol structure. Δ68/L446A/R455D crystallized in the space group P3121 and provided a 2.5 Å resolution structure (Table I). Despite the increased solubility of the mutant due to destabilization of Interface I, this protein crystallized with Interface I intact. The fingers domain continued to be disordered and the structure is essentially identical to that of the wild-type 3Dpol with an r.m.s. difference in Cα positions of 0.6 Å. The full-length L446D/R455D mutant, on the other hand, crystallized in a new P65 lattice (Table I) where 3Dpol molecules did not interact across Interface I. Electron density for the missing fingers domain was clearly visible in the maps after molecular replacement and the entire structure was solved at 2.0 Å resolution.
Table 1.
Data collection and refinement statistics
| 3D-Δ68 | 3D full length | 3D with GTP | 3D G1A mutant | |
|---|---|---|---|---|
| PDB code | 1RAJ | 1RA6 | 1RA7 | 1TQL |
| Space group | P3(1)21 | P6(5) | P6(5) | P6(5) |
| Unit cell | a=b=87.7, c=107.3 | a=b=127.6, c=113.0 | a=b=128.0, c=112.9 | a=b=127.8, c=113.3 |
| X-ray source | Cu-Kα | NSLS X25—1.1 Å | Cu-Kα | ALS 4.2.2—1.0 Å |
| Resolution limits | 30–2.5 (2.59–2.50) | 30–2.0 (2.07–2.00) | 30–2.35 (2.43–2.35) | 30–2.30 (2.38–2.30) |
| Reflections | ||||
| Total collected | 169 949 | 236 338 | 215 638 | 279 462 |
| Unique | 16 973 | 67 129 | 43 126 | 46 290 |
| Redundancy | 10 (8) | 3.5 (3.1) | 5 (3.9) | 6.0 (4.4) |
| I/σa | 35 (3.3) | 16.5 (2.5) | 16.2 (2.1) | 13.8 (2.3) |
| Completeness (%) | 99.9 (99.6) | 96.3 (93.7) | 98.6 (96.2) | 99.2 (96.1) |
| Rmerge (%) | 6.2 (56.9) | 5.4 (45.6) | 8.2 (55.7) | 7.4 (50.4) |
| Refinement | ||||
| Resolution range | 30–2.5 | 30–2.0 | 30–2.35 | 30–2.3 |
| R | 24.2 | 24.3 | 22.4 | 23.7 |
| Rfree (10% of data) | 26.2 | 25.6 | 26.2 | 26.2 |
| Model statistics | ||||
| Number of atoms (waters) | 2138 (67) | 4091 (298) | 4030 (263) | 3869 (137) |
| Average B-factor | 56.9 | 51.4 | 45.9, 59.1 for GTP | 62.1 |
| R.m.s.d. bond length (Å) | 0.008 | 0.007 | 0.007 | 0.007 |
| R.m.s.d. bond angle (deg) | 1.4 | 1.4 | 1.3 | 1.3 |
| Ramachandran statisticsb | ||||
| Favored | 209 | 380 | 379 | 373 |
| Allowed | 16 | 28 | 30 | 36 |
| Generous | 0 | 2 | 1 | 1 |
| Disfavored |
0 |
1 |
1 |
1 |
| Data in parentheses are for the highest resolution shell. | ||||
| Statistics do not include the 28 glycine and 21 proline residues in the protein. | ||||
Structural overview
The domain structure of poliovirus 3Dpol follows the usual right hand analogy of a thumb, palm, and fingers domain that was first used to describe the structure of the DNA polymerase I Klenow fragment (Ollis et al, 1985). Like the HCV and RHDV polymerase structures, 3Dpol adopts a ‘closed' conformation where extensive interactions between the thumb and fingers domains completely encircle the active site and create an NTP entry tunnel at the back of the polymerase (Figure 1). The structures of the palm and thumb domains are essentially identical to those seen in the initial partial structure (Figure 1A and B). There is a minor ∼2.5° change in the relative orientations of these domains that may be due to the removal of crystal packing constraints along Interface I. The new aspect of our complete 3Dpol structure is the fingers domain, which is composed of four separate stretches of amino-acid sequence. These form four tightly intertwined finger structures that we describe by analogy to primate anatomy where there are index, middle, ring, and pinky fingers in addition to a thumb (Figure 1C).
Figure 1.
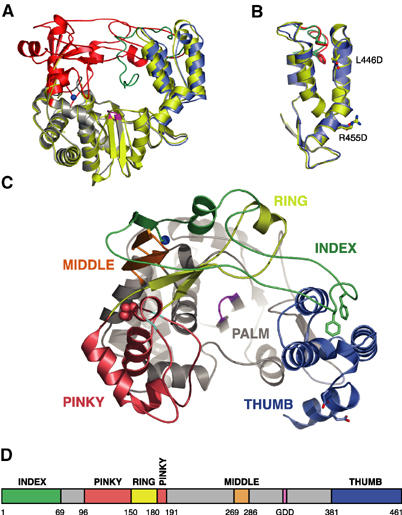
Overview of poliovirus 3Dpol RdRp structure. (A) Comparison of the original partial structure (yellow) with the complete structure shown with the fingers domain in red, the palm in gray, the thumb in blue, and the active site colored magenta. The N-terminal strand (residues 12–36) of the original structure that descended toward the active site is shown in green. The two structures were superimposed using the backbone atoms of the active site GDD motif and three residues on either side of it (i.e. residues 324–332). (B) Superimposition of the thumb domains from the original structure (yellow) and new complete structure (blue) showing that the thumb structure is largely unchanged by the two mutations (L446D and R455D) used to break Interface I and crystallize 3Dpol in a new lattice. The side chains of Phe30 and Phe34 are shown in green for the original structure and red for the new complete structure. (C) Top view of the complete 3Dpol structure highlighting the individual fingers of the fingers domain. The index finger is shown in green, the middle finger in orange, the ring finger in yellow, and the pinky finger in pink. As in (A), the palm is shown in gray, the thumb is in blue, and the active site is colored magenta. Phe30 and Phe34 are shown as sticks, Pro119 on the pinky finger is indicated with spheres, and glycines 117 and 124 are colored in cyan. (D) Bar representation of the 3Dpol sequence colored according to the structural elements shown in (C). Sections of the sequence in the palm are in gray and the numbers correspond to the first residue in a given structural motif.
The fingers begin with a buried N-terminus at the back of the palm, which plays a critical role in structuring the active site (see below). The index finger (residues 1–68, green in Figure 1C) then rises from the palm domain for eight residues before folding into a loop and reaching across the palm to interact with the thumb. This conformation is anchored by the insertion of Phe30 and Phe34 into the hydrophobic core at the top of the thumb. Following this, residues 35–68 fold back toward the palm domain in a generally extended structure that completes the index finger. The middle finger (residues 269–285, orange) consists of an antiparallel β-sheet with a β-turn at its fingertip. This sheet also includes the first eight residues of the index finger, and the tip of the middle finger is inserted into the loop formed by residues 9–17 of the index finger. Lys276, the only Ramachandran plot outlier in the structure, is located at the tip of the middle finger. There is not an obvious structural reason for the distorted geometry, but there may be a functional requirement for a basic residue at this position because a K276L mutation reverts in vivo to an arginine residue (Richards and Ehrenfeld, 1997). Next, the ring finger (residues 150–179, yellow) forms an elongated β-sheet structure that reaches across the active site and protrudes from the polymerase by traversing under the index finger to present a short α-helix on the surface of the protein. The ring finger forms the roof of the NTP entry tunnel and contains conserved basic residues (Arg163, Lys167, Arg174) that are poised for interactions with the incoming NTP. The pinky finger (residues 96–149 and 180–190, pink) is a fairly large structure that is separated from the other fingers by a groove at the top of the fingers domain (Figures 1C and 2A). The top of the pinky finger appears to be quite flexible because it has relatively weak electron density. The folding of the pinky is likely dependent on the ring finger adopting its proper conformation as the roof of the NTP entry tunnel because the ring finger is effectively an insertion in the middle of the pinky sequence (Figures 1D and 2A).
Figure 2.
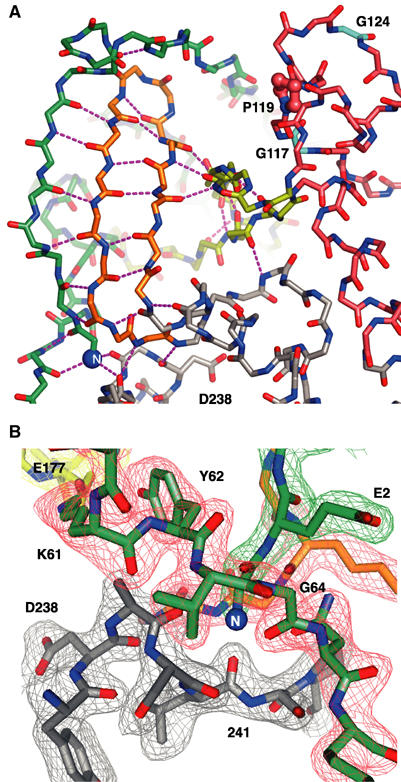
Structural details of the 3Dpol fingers domain and buried N-terminus. (A) Structure of the fingers domain highlighting the extensive network of hydrogen bonds linking the N-terminus (blue sphere at lower left) and N-terminal strand of the index finger to the middle and ring fingers. Note the putative template entry channel separating pinky finger (pink carbon atoms) from the rest of the fingers domain and how the ring finger (yellow) is an insertion in the pinky finger structure. Proline 119 and glycines 117 and 124 that may play a role in template binding are highlighted (see Discussion). The view is from the left side of Figure 1A and C. (B) Electron density map of the region surrounding the buried N-terminus. The map is a 2.0 Å resolution simulated annealing (1500 K) composite omit 2Fo–Fc map contoured at 1.6σ. The view is from the left side as compared to (A), the carbon atoms of the various structural motifs are colored as in Figure 1C and D, and corresponding sections of the density map are colored differently for clarity.
The intertwined structures of the four fingers are stabilized by a multitude of interactions where one set of interactions will form the platform upon which another set of interactions is built. For example, there is an extensive hydrogen bonding network in which the first eight residues of the index finger and the entire β-turn middle finger together form a three-stranded antiparallel β-sheet (Figure 2A). The edge of this sheet is then hydrogen bonded to the backbone of the ring finger, stabilizing the ring finger as it crosses the top of the active site to form the roof of the NTP entry tunnel. The finger structure is further anchored by a well-ordered salt bridge between Lys61 on the index finger and Asp177 on the ring finger that lines the side of the NTP entry tunnel. A mutation of Lys61 to leucine abolishes polymerase activity (Richards et al, 1996). At the top of the NTP entry tunnel, there are a number of hydrogen bonding and van der Waals packing interactions between the index finger and the ring finger that are dependent on a kink in the index finger structure. This kink is stabilized by hydrogen bonding interactions involving Pro40, Glu47, and Arg49, and these residues are highly conserved across picornaviruses, suggesting that the kink is common to all these viral polymerases.
Buried N-terminus
The most remarkable and unique feature of the poliovirus 3Dpol structure is an elegant proteolytic processing-dependent allosteric switch for polymerase activation that involves burying the N-terminal glycine residue in a pocket at the base of the fingers domain (Figure 2B). A comparison of the complete structure with the original wild-type and the Δ68/L446A/R445D structures shows that the buried N-terminus is involved in positioning Asp238 in the active site. When the three 3Dpol structures are superimposed using the protein backbone of the active site Gly-Asp-Asp motif and three residues on either side of it (residues 324–332), there is a clear 1.4 Å movement of Asp238 toward the active site, which is apparent only in the complete structure (Figure 3A). Importantly, this movement is relative to the other active site residues, which are highly superimposable among the three structures. In RNA polymerases, this aspartate hydrogen bonds to the 2′ OH of the incoming NTP in an interaction that is important for the selection of rNTPs over dNTPs (Huang et al, 1997; Gohara et al, 2000). The residue is essential in poliovirus 3Dpol, as a mutation of Asp238 to alanine abolishes poliovirus polymerase activity and viral viability (Gohara et al, 2000). The source of the Asp238 movement can be traced to a pair of hydrogen bonds between the buried N-terminus and the backbone carbonyls of residues 239 and 241. These act to pull the polypeptide backbone of residues 239–242 up toward the buried N-terminus, pushing Asp238 into the active site. This conformation is further stabilized by pairs of backbone hydrogen bonds linking residues 285–241 and 286–238 (Figure 3A).
Figure 3.
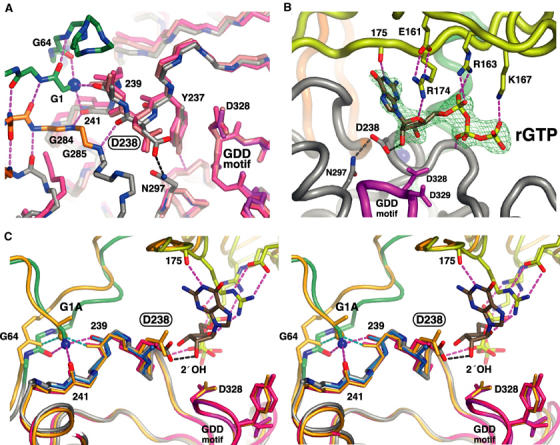
Molecular details of the 3Dpol nucleotide-binding site illustrating how the buried N-terminus positions Asp238 for interactions with the 2′ OH group of the bound NTP. (A) Superposition of three 3Dpol structures showing the selective ∼1.4 Å movement of Asp238 toward the active site when the N-terminus is properly positioned. The original partial wild-type structure is in pink, the 3Dpol Δ68/L446A/R455D structure is in salmon, and the complete structure is colored by atom type with carbons colored according to structural motifs as in Figure 1C. Most side chains have been omitted for clarity and residues 324–332 of the active site (magenta) were used for the superimpositions. (B) Electron density map and model of the GTP molecule bound to 3Dpol with the 2′ OH group making a 2.8 Å long hydrogen bond with Asp238. The GTP makes bridging interactions between the fingers and palm domains. The base is staked on Arg174 from the ring finger, the ribose interacts with Arg174 from the ring finger and Asp238 in the palm, and the triphosphate interacts with Arg163 and Lys167 from the ring finger and the backbone of the palm domain. The map is a 2.35 Å resolution 2Fo–Fc simulated annealing (1500 K) composite omit map contoured at 1.6σ around the rGTP molecule bound after soaking crystals in 10 mM GTP. (C) Stereo view showing how the buried N-terminus of 3Dpol positions Asp238 for rNTP interactions. The N-terminus forms three hydrogen bonds with the carbonyl oxygens of residues 64, 239, and 241 (magenta bonds) that act to position Asp238 for interaction with the 2′ OH of rNTPs. The structures of the G1A mutant (orange), D238A mutant (teal, only residues 238–241 are shown), and original partial structure without a buried N-terminus (red) are superimposed using the active site.
The binding pocket for the N-terminal glycine residue is itself almost entirely composed of glycines and the structure makes use of both the small size and backbone torsional flexibility of this amino acid. There is no formal charge counter-ion for the buried N-terminus. Instead, the terminal amino group makes hydrogen bonds with the backbone carbonyls of residues 64, 239, and 241 in almost perfect tetrahedral geometry. One side of the binding pocket for the N-terminus is made up of glycines 284 and 285 from the base of the middle finger. These glycines are 100% conserved among picornaviral polymerase sequences and their backbone conformations are in the disallowed region of the Ramachandran plot. The outside of the pocket consists of Gly64 from the end of the index finger sequence that is also in the disallowed region of the Ramachandran plot. Gly64 makes two reciprocal amide–carbonyl backbone hydrogen bonds with Gly1 that link together the two ends of the index finger sequence. Overall, the structure around the N-terminus relies on backbone contacts and glycine flexibility to make interactions that leave little room to accommodate mutations in the protein or improper proteolytic processing of the polymerase.
3Dpol–GTP complex
The role of Asp238 in rNTP binding was confirmed by the co-crystal structures of 3Dpol in complex with rGTP, which is bound with its 2′ OH group making the anticipated (2.8 Å long) hydrogen bond with Asp238 (Figure 3B). The nucleoside is bound with the base moiety interacting with the guanidinium group of Arg174 from the ring finger in a cation-π stacking interaction similar to that observed in single-stranded nucleic acid-binding proteins (Theobald and Schultz, 2003). The Arg174 guanidinium group is further locked in place by a hydrogen bond with the ribose sugar ring oxygen and a charge interaction with Glu161 from the other end of the ring finger. The base-pairing face of the rGTP is facing the protein and its N1 atom is hydrogen bonded to the backbone carbonyl group of residue 175 in the ring finger. Thus, the nucleotide is not yet positioned to form a base pair with the template strand. Similarly, the phosphate groups are not positioned above the active site aspartate residues. They are instead trailing out through the rNTP entry tunnel where they interact with conserved basic residues in a structure that appear to select for a complete triphosphate. There are ionic interactions with Arg163 and Lys167 that drop down from the ring finger in the roof of the rNTP entry tunnel and the β-phosphate is hydrogen bonded to the backbone amide of residue 236 in the palm domain.
The structure of the 3Dpol–GTP complex could be obtained either by co-crystallization or by soaking of 3Dpol crystals, indicating that the binding of rGTP is not accompanied by large conformational changes in the protein that would break the crystal lattice. We also generated and crystallized a D238A mutant that results in an inactive polymerase (Table II). In this structure, the N-terminus was buried normally and the backbone structure around the active site is the same as with the native Asp238 (Figure 3C). However, we did not see any nucleoside density upon soaking crystals of the D238A mutant in 10 mM GTP. This is in stark contrast to what was observed with the native Asp238 residue (Figure 3B) and highlights the importance of Asp238 for rNTP binding.
Table 2.
Polymerase extension activities
| Protein | % Activity |
|---|---|
| 3Dpol WT | 215±7 |
| 3Dpol L446D/R455D | 100±4 |
| 3CDpro WT | 0.1±0.2 |
| Mutants below are in addition to L446D/R455D | |
| 3Dpol Δ68 | 0.0±0.2 |
| 3Dpol ΔG1 | −0.1±0.2 |
| 3Dpol +Q | 0.1±0.2 |
| 3Dpol +SQ | −0.4±0.2 |
| 3Dpol G1A | 54±2 |
| 3Dpol G1S | 1.6±0.3 |
| 3Dpol P119A | −0.1±0.2 |
| 3Dpol P119G | −0.1±0.2 |
| 3Dpol D238A |
−0.3±0.2 |
| Errors are based on propagation of 2σ scintillation counting confidence limits from three activity measurements per protein. | |
Mutation of the N-terminus
To investigate the importance of the N-terminus for the structure and function of 3Dpol, we made a series of mutations that added or removed residues from the protein or altered Gly1. The elongation activities of these mutants were tested using a poly(A)/oligo(dT) extension assay to measure [32P]UMP incorporation into a product RNA strand. We generated two mutants that added a single glutamine residue or a serine–glutamine dipeptide to the N-terminus. These correspond to the last two residues of 3Cpro and thus mimic the natural precursor junction sequence of 3CDpro. Both mutants had less than 0.2% the activity of the full-length protein (Table II), demonstrating that the addition of even a single residue completely abolishes enzymatic activity. Likewise, deletion of the first residue, Gly1, also inactivates the polymerase. We then made two more subtle mutations of Gly1 to alanine and serine. These mutations retained partial polymerase activity, with G1A having ∼54% activity and G1S having only 1.6% activity.
With the exception of G1A, none of the N-terminus mutants could be crystallized under the conditions used for the full-length protein and they failed to crystallize in more extensive screens for different conditions. This could be because small changes at the terminus affect the stability of the finger structure or the ability of Trp5 to participate in a crystal contact (see Materials and methods). The G1A mutant did crystallize, and the resulting structure further demonstrates the importance of positioning Asp238 in the active site for proper enzymatic activity. In this structure, the side-chain methyl group of the G1A mutation is packed into the area normally occupied by the Gly1 alpha carbon. As a result, the N-terminus is pushed out of its binding pocket by ∼0.9 Å and the hydrogen bond to the backbone carbonyl of residue 241 is lost. This has a direct effect on the positioning of Asp238, which is now only pushed about halfway into the active site as compared to the native Gly1 residue (Figure 3C). Thus, there is a direct correlation between enzymatic activity and the positioning of Asp238 for the interaction with the 2′ OH of the incoming NTP.
Discussion
The poliovirus 3Dpol structure provides the first structural insight into the molecular mechanism responsible for the proteolysis-dependent activation of a polymerase. It shows that the newly created 3Dpol N-terminus is buried in a pocket at the junction of the fingers and palm domains. This in turn positions the essential Asp238 residue in the active site for interactions with the 2′ OH group of the incoming rNTP. Asp238 is perfectly pre-positioned by the buried N-terminus and the location of the side chain does not change significantly upon rGTP binding. Moreover, the nucleotide is bound in a precatalytic site that appears to test for a ribose triphosphate with little or no base discrimination. In the case of rGTP, the nucleoside is bound in the syn conformation with its hydrogen bonding face interacting with the protein. For the base to be incorporated into the product strand, it must both flip into the anti conformation and be moved into the catalytic site of the polymerase.
The overall fold of poliovirus 3Dpol is similar to that of other viral RdRps in adopting a conformation where the active site is fully enclosed as a result of extensive interactions between the fingers and thumb domains. This conformation has now been observed in hepatitis C virus, rabbit hemorrhagic disease virus, bacteriophage φ6, bovine viral diarrhea virus, Norwalk virus, and poliovirus polymerase structures (Ago et al, 1999; Bressanelli et al, 1999; Lesburg et al, 1999; Butcher et al, 2001; Ng et al, 2002, 2004; Choi et al, 2004). A number of important interactions in the poliovirus 3Dpol structure are dependent on residues that are highly conserved among the picornaviruses, suggesting that many aspects of the structure are also retained among these viral polymerases. These interactions include the Lys61–Glu177 salt bridge between index and ring fingers that lines the NTP entry tunnel, the hydrophobic residues on the index finger that are inserted into the top of the thumb (Figure 1C), the kink in the index finger structure where it interacts with the ring finger to form the top of the NTP entry tunnel (Figure 1C), and glycines 284 and 285 in the binding pocket for the buried N-terminus (Figure 3A).
The 3Dpol fingers domain has its own hydrophobic core that includes residues from the index, middle, and ring fingers and extends from the base of the fingers, across the top of the NTP entry tunnel, and over to the thumb. The pinky finger also has a small self-contained hydrophobic core that is separate from that of the other fingers. As noted previously, the ring finger is effectively an insertion in the pinky finger sequence (Figure 1D) and as a result the folding of the two hydrophobic cores within the fingers domain is probably linked. Importantly, there are relatively few interactions between the palm and fingers domains. A number of observations suggest that the poliovirus polymerase palm and thumb domains may fold independently of its fingers domain. The lattice of the original partial structure showed that the fingers can be selectively disordered while leaving the palm and thumb intact. Our structure of the 68-residue N-terminal deletion mutant further showed that the entire index finger can be deleted without affecting the folding of the palm and thumb. Finally, our observations that we could not crystallize many of the N-terminus mutant proteins may indicate that the structure of the fingers domain is appreciably perturbed by small changes to the N-terminus. The buried N-terminus sits at the heart of an extensive array of hydrogen bonds and may play a significant role in nucleating the structure of the entire fingers domain (Figure 2A). If this is the case, then the structure of the fingers domain may be quite different in the context of 3CDpro where the proper 3Dpol N-terminus does not exist. In other words, the structure of 3CDpro may not simply be a fusion of the 3Cpro structure (Mosimann et al, 1997) and the 3Dpol structure presented here. Notably, it is the palm and thumb domains that create the surfaces involved in forming head-to-tail fibers along Interface I. These domains could therefore provide a platform for the oligomerization of the polymerase that is independent of the fingers structure. Such a platform may also allow for the self-association of polymerase precursor proteins such as 3CDpro or 3ABCD (i.e. the entire P3 region) at various stages of the viral life cycle. Consistent with this, both the L446A and L446D mutations also significantly increase the solubility of 3CDpro (data not shown).
The complete structure of 3Dpol also provides an explanation as to why the fingers domain was disordered in the original partial wild-type structure solved by Hansen et al (1997). When our complete structure is superimposed into the P3221 crystal lattice of the original structure, there are two significant steric clashes involving the fingers domain: one is at the loop of the index finger as it wraps around the middle finger and the other involves the pinky finger. Neither of these crystal contacts is a direct result of interactions across Interface I, the interface that was intentionally disrupted to obtain the complete structure. Rather, the clashes arise from secondary crystal packing interactions between parallel Interface I fibers. Based on this, we find it unlikely that the proposed Interface II of the original structure exists in solution. This interface, where residues 12–35 interacting with the top of the thumb were proposed to come in trans from another polymerase molecule, involves the burial of two large hydrophobic residues into the top of the thumb (Phe30 and Phe34; see Figure 1B and C). We feel that this interaction is likely too strong to have been disrupted by the unfolding of the fingers domain and therefore remained intact in the crystal lattice of the original structure. However, we cannot rule out that 3Dpol precursors such as 3CDpro have alternate folding schemes where this interface is used to interconnect fibers of molecules oligomerized along Interface I.
A number of mechanisms have been proposed for translocation of the template–primer during nucleotide polymerization by polymerases (Patel et al, 1995; Ding et al, 1998). In DNA polymerases and DNA-templated RNA polymerases, the lack of a direct connection between the fingers and thumb domains may allow for large ratchet-like conformational changes of the thumb during nucleotide polymerization. However, the enclosed active sites of RdRps make it unlikely that such mechanisms are used and instead nucleic acid binding to the fingers domain may play a role in translocation of nucleic acid during synthesis (Najmudin et al, 2000). There are several structures of the DNA-dependent RNA polymerase from bacteriophage T7 in complex with a number of nucleic acid templates and template–product pairs available that provide insights into the potential function of the 3Dpol fingers domain. First, at the top of the 3Dpol pinky, there is a helix followed by a large loop structure (residues 124–149) that appears to be quite flexible by virtue of weak electron density. This loop is most likely involved in RNA binding based on a structural alignment of the 3Dpol active site with that of bacteriophage T7 RNA polymerase (Figure 4A). In this superimposition, the pinky loop roughly superimposes on the T7 polymerase specificity loop, which makes sequence-specific contacts with the T7 promoter in the initiation complex (Cheetham et al, 1999) and nonspecific contacts in the T7 elongation complex (Temiakov et al, 2000; Yin and Steitz, 2002). Second, in poliovirus 3Dpol, we observe a groove between the index and pinky fingers that may serve as a template entry channel (Figure 2A). A superposition of the 3Dpol active site with the T7 polymerase active site in that enzyme's elongation complex structure (Yin and Steitz, 2002) shows that the template strand of the T7 complex roughly threads into this groove in 3Dpol (Figure 4B).
Figure 4.
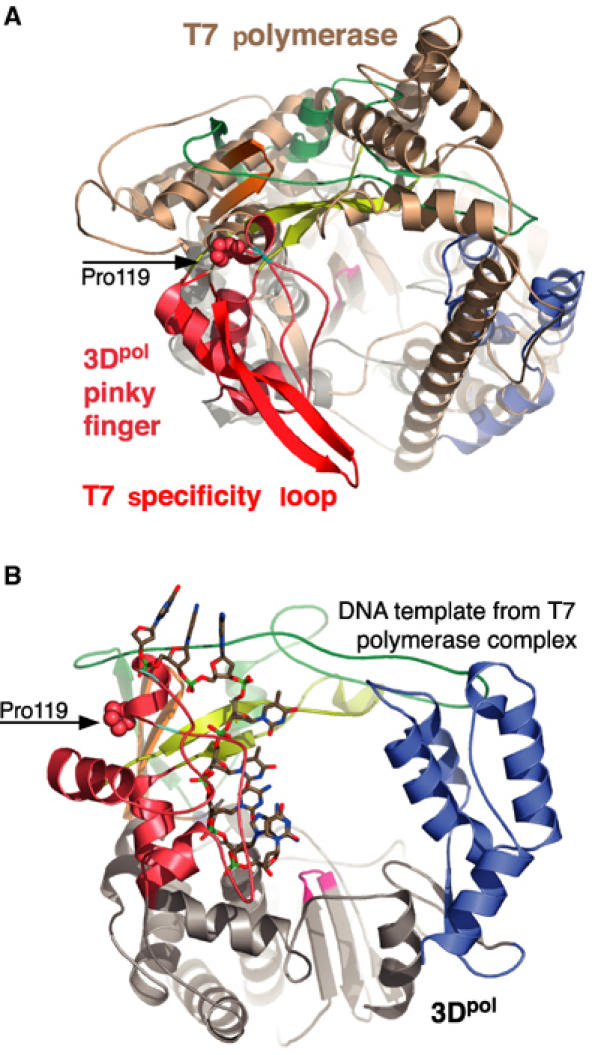
Comparison of the poliovirus 3Dpol and bacteriophage T7 polymerases based on superimposing the Cα atoms of their ‘motif C' structures. This motif contains the pair of β-strands in the core of the palm that present the β-turn GDD motif in the active site. (A) Superposition with the T7 protein structure (brown) from the initiation complex (1CEZ) showing the structural alignment of the 3Dpol pinky finger with the T7 specificity loop (red). (B) Position of the DNA template strand from the T7 elongation complex (1MSW) after the motif C superimposition showing how this predicted template strand path in 3Dpol would collide with the helix/loop containing the cis Pro119 residue (spheres) flanked by glycines 117 and 124 (cyan).
Interestingly, there is a 100% conserved cis proline (Pro119) lining this groove that is bracketed by 100% conserved glycines 117 and 124 (Figures 1C and 2A). This sequence includes the conserved RdRp G motif (Gorbalenya et al, 2002) and is consistent with an emerging structural motif used to control the conformation of small loops via proline cis–trans isomerization (Andreotti, 2003). To ascertain the biochemical importance of Pro119 in 3Dpol, we mutated the residue to alanine and glycine and found that both mutations totally abolish elongation of a poly(A)/oligo(dT) substrate (Table II). Thus, Pro119 is essential for activity and the total loss of activity suggests that the cis peptide bond conformation may be a required structural feature. It has been shown that upon incubation of 3Dpol with primer–template there is a slow isomerization (t1/2∼12 s) to an elongation-competent complex whose half-life is ∼2 h and this increases to ∼8 h after incorporation of the first nucleotide (Arnold and Cameron, 2000). The timescale for formation of this stable 3Dpol–substrate complex is consistent with the 0.1–5 min timescales observed for proline cis–trans isomerization in protein folding studies (Brandts et al, 1975; Reimer et al, 1998). We therefore hypothesize that cis–trans isomerization of Pro119 is a key to conformational change in the pinky finger that locks the enzyme–substrate complex into the stable elongation-competent mode.
The structure of the G1A mutant further highlights the correlation between the burial of the native N-terminus, the positioning of Asp238 in the active site, and the enzymatic activity of 3Dpol. It is interesting to note that the native Gly1 residue makes a pair of reciprocal amide–carbonyl hydrogen bonds with Gly64 located on the exterior side of the binding pocket. This residue is the site of a serine mutation found in virus resistant to the antiviral drug ribavirin (Pfeiffer and Kirkegaard, 2003). Interestingly, the G64S mutation leads to a polymerase with increased fidelity, allowing it to discriminate against ribavirin, a nucleoside analog (Crotty et al, 2001). We do not yet know the structure of the G64S 3Dpol, but based on the structure of 3Dpol with the native Gly1 and mutant Ala1 N-termini we believe that one effect of the G64S mutation is to alter the structure of the backbone at residue 64 and thus slightly alter the positioning of Asp238 in the active site. The Cameron laboratory has identified a conformational change step during 3Dpol elongation that occurs after NTP binding but before the phosphoryl transfer catalytic step and they correlated changes in the rate of this step with the position of Asp238 (Gohara et al, 2004). Based on our structure of the 3Dpol–GTP complex we hypothesize that this conformational step is the movement of the NTP from the position observed in our structure into the catalytic site where it would be base paired with the template and poised for the phosphoryl transfer reaction. The hydrogen bond between the 2′ OH and Asp238 may remain intact during this conformational step and the positioning of Asp238 could then have a direct effect on the rate at which the NTP moves into the catalytic site. In the case of the G64S mutant, an altered positioning of Asp238 may increase the time required for this step and thus allow more time for the dissociation of incorrectly base-paired complexes.
In conclusion, specific proteolytic processing is a common strategy employed by many classes of proteins to regulate proper activation of proteins at the desired time and location. The classic examples are digestive enzymes such as trypsin and chymotrypsin that are initially released as inactive proenzymes whose cleavage results in a slight refolding of the active site to yield the active enzyme. Such allosteric effects are also common among RNA viruses that have evolved mechanisms to take advantage of variable proteolytic processing to regulate many aspects of the viral life cycle. In poliovirus, the 3CDpro protein plays an integral role in orchestrating viral RNA replication by binding to both ends of the viral RNA genome in the context of large membrane-associated replication centers (Murray and Barton, 2003). This has the effect of localizing an inactive polymerase domain to the sites of negative and positive strand RNA synthesis, where it is poised to initiate RNA replication after cleavage to the mature 3Dpol protein. Our structure of poliovirus 3Dpol shows the molecular details of the switch responsible for the complete allosteric activation of an RNA polymerase and provides insights into the structural conservation of picornaviral polymerases.
Materials and methods
Protein purification and crystallization
All polymerases were expressed in Escherichia coli BL-21 (DE3) pLysS using inducible T7-based expression plasmids. These cells were lysed by sonication and the polymerase was precipitated with ammonium sulfate at 40% of saturation. The protein was dialyzed, loaded onto an S-Sepharose column, and eluted with a linear gradient from 50 mM to 1 M NaCl in 25 mM HEPES (pH 8.5), 15% glycerol, 0.1 mM EDTA, 0.02% NaN3, and 2 mM DTT. Fractions containing the polymerase were pooled and diluted to reduce the NaCl concentration to ∼0.15 M. This was then loaded onto a Q-Sepharose column and eluted with a linear gradient to 1 M NaCl with 25 mM Tris (pH 8.5), 15% glycerol, 0.1 mM EDTA, 0.02% (w/v) NaN3, and 2 mM DTT. The polymerase was concentrated and run over a Superdex 200 column equilibrated in 200 mM NaCl, 5 mM Tris (pH 7.5), 0.1 mM EDTA, 0.02% (w/v) sodium azide, and 2 mM DTT. Upon elution from this column, the proteins were >99% pure as estimated from SDS–PAGE. Crystals were grown by hanging drop vapor diffusion at 16°C using 12 mg/ml protein. 3Dpol Δ68/L446A/R455D crystals grew in 8 days with a precipitant/well solution containing 1.5 M ammonium formate, 0.1 M sodium chloride, 50 mM HEPES (pH 7.0), 2 mM DTT, and 0.02% (w/v) sodium azide. 3Dpol L446D/R455D crystals grew in 4 days with a precipitant/well solution containing 2 M sodium acetate, 0.1 M cacodylic acid (pH 7.1), 2 mM DTT, and 0.02% (w/v) sodium azide. All crystals were transferred into corresponding precipitant solutions containing 30% (v/v) glycerol prior to freezing.
Structure determination
Diffraction data for L446D/R455D were collected at National Synchrotron Light Source (Brookhaven National Laboratory) beamline X-25 and data from the G1A mutant were collected at the MBC beamline 4.2.2 at the Advanced Light Source (Berkeley, CA). Data for the Δ68/L446A/R455D and 3Dpol–GTP complex crystals were collected using R-AXIS IV imaging plate detector with Cu-Kα radiation. Reflections were integrated, merged, and scaled using Denzo/Scalepack (Otwinowski and Minor, 1997) and d*TREK (Pflugrath, 1999). The initial structure solutions were obtained by using molecular replacement with the program CNS (Brunger et al, 1998) with the partial polymerase structure as the search model. Manual model rebuilding was performed using O (Jones et al, 1991) and refined with the CNS package using the MLI target. The figures were generated with Pymol Molecular Graphics System (DeLano, 2002). Coordinates have been deposited at the PDB with access codes as listed in Table I.
We would like to note that the observed conformation of Trp5 is likely a crystallization artifact. One face of the side chain is packed against a dimethyl arsenic adduct on Cys281 of the middle finger and the other face is packed against a hydrophobic patch on a neighboring 3Dpol molecule in the crystal lattice. The crystallization required cacodylic acid and DTT, consistent with a proposed mechanism (Tsao and Maki, 1991) for the formation of the dimethyl arsenic adduct that has previously been observed in several structures (Maignan et al, 1998; Tete-Favier et al, 2000; Raman et al, 2001). We have not been able to crystallize the protein using other buffers and/or precipitants and therefore feel that the observed conformation of Trp5 is the result of crystal packing forces where the hydrophobic dimethyl arsenic adduct likely stabilizes a flipped out conformation of the side chain enough to create a crystal packing interface. Cystine residues modified with the dimethyl arsenic adducts are listed as CAS residues in the PDB files and the structure was refined in CNS using CAS residues and a parameter set from the HIC-up database (Kleywegt and Jones, 1998).
Polymerase activity assays
A 20 μl volume of poly(A)/oligo(dT) polymerase extension reaction contained 0.01 μg/μl poly(A) template (average length 300 nt), 0.005 μg/μl oligo(dT15), 50 mM HEPES (pH 8.0), 25 μM UTP, 0.5 mM each GTP, CTP, ATP, 4 mM DTT, 0.1 mM MgAc2 60 μM ZnCl2 0.1% NP-40, 0.1 Ci/μl [α-32P]UTP, and 300 nM 3Dpol. Reactions were preincubated on ice for at least 5 min and then incubated for 30 min at 30°C. A 15 μl portion of each reaction was then transferred to DE81 Whatmann filters and washed three times with 5% (w/v) Na2HPO4, twice with ddH2O, once with 95% ethanol, and once with 100% ethyl ether. The activity was then assessed by scintillation counting.
Acknowledgments
We thank Ollie Richards, Cathy Radebaugh, and John Anderson for assistance with the activity assays, Vandy Johnson for help with protein purification, and Michael Becker and Howard Robinson of NSLS for help with data collection. This work was supported by grant R01-AI059130 from the NIH.
References
- Ago H, Adachi T, Yoshida A, Yamamoto M, Habuka N, Yatsunami K, Miyano M (1999) Crystal structure of the RNA-dependent RNA polymerase of hepatitis C virus. Struct Fold Des 7: 1417–1426 [DOI] [PubMed] [Google Scholar]
- Andino R, Rieckhof GE, Achacoso PL, Baltimore D (1993) Poliovirus RNA synthesis utilizes an RNP complex formed around the 5′-end of viral RNA. EMBO J 12: 3587–3598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreotti AH (2003) Native state proline isomerization: an intrinsic molecular switch. Biochemistry 42: 9515–9524 [DOI] [PubMed] [Google Scholar]
- Arnold JJ, Cameron CE (2000) Poliovirus RNA-dependent RNA polymerase 3D(pol). Assembly of stable, elongation-competent complexes by using a symmetrical primer–template substrate (sym/sub). J Biol Chem 275: 5329–5336 [DOI] [PubMed] [Google Scholar]
- Brandts JF, Halvorson HR, Brennan M (1975) Consideration of the possibility that the slow step in protein denaturation reactions is due to cis–trans isomerism of proline residues. Biochemistry 14: 4953–4963 [DOI] [PubMed] [Google Scholar]
- Bressanelli S, Tomei L, Roussel A, Incitti I, Vitale RL, Mathieu M, De Francesco R, Rey FA (1999) Crystal structure of the RNA-dependent RNA polymerase of hepatitis C virus. Proc Natl Acad Sci USA 96: 13034–13039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, Read RJ, Rice LM, Simonson T, Warren GL (1998) Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr D 54 (Part 5): 905–921 [DOI] [PubMed] [Google Scholar]
- Butcher SJ, Grimes JM, Makeyev EV, Bamford DH, Stuart DI (2001) A mechanism for initiating RNA-dependent RNA polymerization. Nature 410: 235–240 [DOI] [PubMed] [Google Scholar]
- Cheetham GM, Jeruzalmi D, Steitz TA (1999) Structural basis for initiation of transcription from an RNA polymerase–promoter complex. Nature 399: 80–83 [DOI] [PubMed] [Google Scholar]
- Choi KH, Groarke JM, Young DC, Kuhn RJ, Smith JL, Pevear DC, Rossmann MG (2004) The structure of the RNA-dependent RNA polymerase from bovine viral diarrhea virus establishes the role of GTP in de novo initiation. Proc Natl Acad Sci USA 101: 4425–4430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crotty S, Cameron CE, Andino R (2001) RNA virus error catastrophe: direct molecular test by using ribavirin. Proc Natl Acad Sci USA 98: 6895–6900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLano WL. (2002) The PYMOL Molecular Graphics System, DeLano Scientific (www.pymol.org), San Carlos, CA, USA [Google Scholar]
- Ding J, Das K, Hsiou Y, Sarafianos SG, Clark AD Jr, Jacobo-Molina A, Tantillo C, Hughes SH, Arnold E (1998) Structure and functional implications of the polymerase active site region in a complex of HIV-1 RT with a double-stranded DNA template–primer and an antibody Fab fragment at 2.8 Å resolution. J Mol Biol 284: 1095–1111 [DOI] [PubMed] [Google Scholar]
- Flanegan JB, Van Dyke TA (1979) Isolation of a soluble and template-dependent poliovirus RNA polymerase that copies virion RNA in vitro. J Virol 32: 155–161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gohara DW, Arnold JJ, Cameron CE (2004) Poliovirus RNA-dependent RNA polymerase (3Dpol): kinetic, thermodynamic, and structural analysis of ribonucleotide selection. Biochemistry 43: 5149–5158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gohara DW, Crotty S, Arnold JJ, Yoder JD, Andino R, Cameron CE (2000) Poliovirus RNA-dependent RNA polymerase (3Dpol): structural, biochemical, and biological analysis of conserved structural motifs A and B. J Biol Chem 275: 25523–25532 [DOI] [PubMed] [Google Scholar]
- Gorbalenya AE, Pringle FM, Zeddam JL, Luke BT, Cameron CE, Kalmakoff J, Hanzlik TN, Gordon KH, Ward VK (2002) The palm subdomain-based active site is internally permuted in viral RNA-dependent RNA polymerases of an ancient lineage. J Mol Biol 324: 47–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen JL, Long AM, Schultz SC (1997) Structure of the RNA-dependent RNA polymerase of poliovirus. Structure 5: 1109–1122 [DOI] [PubMed] [Google Scholar]
- Harris KS, Reddigari SR, Nicklin MJ, Hammerle T, Wimmer E (1992) Purification and characterization of poliovirus polypeptide 3CD, a proteinase and a precursor for RNA polymerase. J Virol 66: 7481–7489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hobson S (2000) Crystallographic and Biochemical Studies of Higher Order Poliovirus Polymerase Structures. Boulder, CO, USA: Department of Chemistry and Biochemistry, University of Colorado [Google Scholar]
- Hobson SD, Rosenblum ES, Richards OC, Richmond K, Kirkegaard K, Schultz SC (2001) Oligomeric structures of poliovirus polymerase are important for function. EMBO J 20: 1153–1163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y, Eckstein F, Padilla R, Sousa R (1997) Mechanism of ribose 2′-group discrimination by an RNA polymerase. Biochemistry 36: 8231–8242 [DOI] [PubMed] [Google Scholar]
- Jones TA, Zou JY, Cowan SW, Kjeldgaard M (1991) Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr A 47 (Part 2): 110–119 [DOI] [PubMed] [Google Scholar]
- Kleywegt GJ, Jones TA (1998) Databases in protein crystallography. Acta Crystallogr D 54: 1119–1131 [DOI] [PubMed] [Google Scholar]
- Lesburg CA, Cable MB, Ferrari E, Hong Z, Mannarino AF, Weber PC (1999) Crystal structure of the RNA-dependent RNA polymerase from hepatitis C virus reveals a fully encircled active site. Nat Struct Biol 6: 937–943 [DOI] [PubMed] [Google Scholar]
- Lyle JM, Bullitt E, Bienz K, Kirkegaard K (2002) Visualization and functional analysis of RNA-dependent RNA polymerase lattices. Science 296: 2218–2222 [DOI] [PubMed] [Google Scholar]
- Maignan S, Guilloteau JP, Zhou-Liu Q, Clement-Mella C, Mikol V (1998) Crystal structures of the catalytic domain of HIV-1 integrase free and complexed with its metal cofactor: high level of similarity of the active site with other viral integrases. J Mol Biol 282: 359–368 [DOI] [PubMed] [Google Scholar]
- Mosimann SC, Cherney MM, Sia S, Plotch S, James MN (1997) Refined X-ray crystallographic structure of the poliovirus 3C gene product. J Mol Biol 273: 1032–1047 [DOI] [PubMed] [Google Scholar]
- Murray KE, Barton DJ (2003) Poliovirus CRE-dependent VPg uridylylation is required for positive-strand RNA synthesis but not for negative-strand RNA synthesis. J Virol 77: 4739–4750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Najmudin S, Cote ML, Sun D, Yohannan S, Montano SP, Gu J, Georgiadis MM (2000) Crystal structures of an N-terminal fragment from Moloney murine leukemia virus reverse transcriptase complexed with nucleic acid: functional implications for template–primer binding to the fingers domain. J Mol Biol 296: 613–632 [DOI] [PubMed] [Google Scholar]
- Ng KK, Cherney MM, Vazquez AL, Machin A, Alonso JM, Parra F, James MN (2002) Crystal structures of active and inactive conformations of a caliciviral RNA-dependent RNA polymerase. J Biol Chem 277: 1381–1387 [DOI] [PubMed] [Google Scholar]
- Ng KK, Pendas-Franco N, Rojo J, Boga JA, Machin A, Alonso JM, Parra F (2004) Crystal structure of norwalk virus polymerase reveals the carboxyl terminus in the active site cleft. J Biol Chem 279: 16638–16645 [DOI] [PubMed] [Google Scholar]
- Ollis DL, Brick P, Hamlin R, Xuong NG, Steitz TA (1985) Structure of large fragment of Escherichia coli DNA polymerase I complexed with dTMP. Nature 313: 762–766 [DOI] [PubMed] [Google Scholar]
- Otwinowski Z, Minor W (1997) Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol 276: 307–326 [DOI] [PubMed] [Google Scholar]
- Parsley TB, Cornell CT, Semler BL (1999) Modulation of the RNA binding and protein processing activities of poliovirus polypeptide 3CD by the viral RNA polymerase domain. J Biol Chem 274: 12867–12876 [DOI] [PubMed] [Google Scholar]
- Patel PH, Jacobo-Molina A, Ding J, Tantillo C, Clark AD Jr, Raag R, Nanni RG, Hughes SH, Arnold E (1995) Insights into DNA polymerization mechanisms from structure and function analysis of HIV-1 reverse transcriptase. Biochemistry 34: 5351–5363 [DOI] [PubMed] [Google Scholar]
- Paul AV, Rieder E, Kim DW, van Boom JH, Wimmer E (2000) Identification of an RNA hairpin in poliovirus RNA that serves as the primary template in the in vitro uridylylation of VPg. J Virol 74: 10359–10370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeiffer JK, Kirkegaard K (2003) A single mutation in poliovirus RNA-dependent RNA polymerase confers resistance to mutagenic nucleotide analogs via increased fidelity. Proc Natl Acad Sci USA [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pflugrath JW (1999) The finer things in X-ray diffraction data collection. Acta Crystallogr D 55 (Part 10): 1718–1725 [DOI] [PubMed] [Google Scholar]
- Plotch SJ, Palant O, Gluzman Y (1989) Purification and properties of poliovirus RNA polymerase expressed in Escherichia coli. J Virol 63: 216–225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raman CS, Li H, Martasek P, Babu BR, Griffith OW, Masters BS, Poulos TL (2001) Implications for isoform-selective inhibitor design derived from the binding mode of bulky isothioureas to the heme domain of endothelial nitric-oxide synthase. J Biol Chem 276: 26486–26491 [DOI] [PubMed] [Google Scholar]
- Reimer U, Scherer G, Drewello M, Kruber S, Schutkowski M, Fischer G (1998) Side-chain effects on peptidyl-prolyl cis/trans isomerisation. J Mol Biol 279: 449–460 [DOI] [PubMed] [Google Scholar]
- Richards OC, Baker S, Ehrenfeld E (1996) Mutation of lysine residues in the nucleotide binding segments of the poliovirus RNA-dependent RNA polymerase. J Virol 70: 8564–8570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richards OC, Ehrenfeld E (1997) One of two NTP binding sites in poliovirus RNA polymerase required for RNA replication. J Biol Chem 272: 23261–23264 [DOI] [PubMed] [Google Scholar]
- Rieder E, Paul AV, Kim DW, van Boom JH, Wimmer E (2000) Genetic and biochemical studies of poliovirus cis-acting replication element cre in relation to VPg uridylylation. J Virol 74: 10371–10380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothstein MA, Richards OC, Amin C, Ehrenfeld E (1988) Enzymatic activity of poliovirus RNA polymerase synthesized in Escherichia coli from viral cDNA. Virology 164: 301–308 [DOI] [PubMed] [Google Scholar]
- Semler BL, Wimmer E (eds) (2002) Molecular Biology of Picornaviruses. Washington, DC: ASM Press [Google Scholar]
- Temiakov D, Mentesana PE, Ma K, Mustaev A, Borukhov S, McAllister WT (2000) The specificity loop of T7 RNA polymerase interacts first with the promoter and then with the elongating transcript, suggesting a mechanism for promoter clearance. Proc Natl Acad Sci USA 97: 14109–14114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tete-Favier F, Cobessi D, Boschi-Muller S, Azza S, Branlant G, Aubry A (2000) Crystal structure of the Escherichia coli peptide methionine sulphoxide reductase at 1.9 Å resolution. Struct Fold Des 8: 1167–1178 [DOI] [PubMed] [Google Scholar]
- Theobald DL, Schultz SC (2003) Nucleotide shuffling and ssDNA recognition in Oxytricha nova telomere end-binding protein complexes. EMBO J 22: 4314–4324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsao DH, Maki AH (1991) Optically detected magnetic resonance study of the interaction of an arsenic(III) derivative of cacodylic acid with EcoRI methyl transferase. Biochemistry 30: 4565–4572 [DOI] [PubMed] [Google Scholar]
- Yin YW, Steitz TA (2002) Structural basis for the transition from initiation to elongation transcription in T7 RNA polymerase. Science 298: 1387–1395 [DOI] [PubMed] [Google Scholar]
- Ypma-Wong MF, Dewalt PG, Johnson VH, Lamb JG, Semler BL (1988) Protein 3CD is the major poliovirus proteinase responsible for cleavage of the P1 capsid precursor. Virology 166: 265–270 [DOI] [PubMed] [Google Scholar]


