Current autophagy models suggest that Atg8 family LC3/GABARAP proteins are essential mediators of autophagosome biogenesis. Nguyen et al. exploit CRISPR/Cas9-generated knockouts of the LC3 or GABARAP subfamilies, or both subfamilies, to show that Atg8s are dispensable for autophagosome biogenesis but essential for autophagosome–lysosome fusion.
Abstract
Members of the Atg8 family of proteins are conjugated to autophagosomal membranes, where they have been proposed to drive autophagosome formation and selective sequestration of cargo. In mammals, the Atg8 family consists of six members divided into the LC3 and GABARAP subfamilies. To define Atg8 function, we used genome editing to generate knockouts of the LC3 and GABARAP subfamilies as well as all six Atg8 family members in HeLa cells. We show that Atg8s are dispensable for autophagosome formation and selective engulfment of mitochondria, but essential for autophagosome–lysosome fusion. We find that the GABARAP subfamily promotes PLEKHM1 recruitment and governs autophagosome–lysosome fusion, whereas the LC3 subfamily plays a less prominent role in these processes. Although neither GABARAPs nor LC3s are required for autophagosome biogenesis, loss of all Atg8s yields smaller autophagosomes and a slowed initial rate of autophagosome formation. Our results clarify the essential function of the Atg8 family and identify GABARAP subfamily members as primary contributors to PINK1/Parkin mitophagy and starvation autophagy.
Introduction
Autophagy is a catabolic pathway essential for maintaining cellular homeostasis through lysosome-mediated degradation of cytosolic material (Lamb et al., 2013). Starvation-induced autophagy is largely a nonselective process that recycles cellular material to provide energy and nutrients. Selective forms of autophagy target specific cargo for degradation, such as dysfunctional mitochondria during PINK1/Parkin-mediated mitophagy (Narendra et al., 2008; Matsuda et al., 2010) or invading bacteria during xenophagy (Gutierrez et al., 2004; Nakagawa et al., 2004; Ogawa et al., 2005). Central to autophagy is a double-membrane structure called an autophagosome, whose formation is driven by a core set of conserved proteins termed autophagy-related (Atg) proteins (Lamb et al., 2013). Atg proteins govern autophagosome initiation from isolation membranes and then expand these membranes to encapsulate cytoplasmic contents destined for lysosomal degradation.
Autophagy is initiated by the unc-51–like autophagy activating kinase 1/2 complex (ULK1/2–Atg13–FIP200–Atg101) and the transmembrane autophagy protein Atg9A, which are both recruited to the site of autophagosome formation. Subsequent recruitment of the class III phosphatidylinositol 3-kinase complex (Beclin-1–Atg14–Vps15–Vps34) results in generation of the lipid phosphatidylinositol-3-phosphate (PI3P). PI3P recruits the downstream effectors WIPI1/2 and DFCP1, and together with additional autophagy proteins, including Atg2A/B, Vmp1, and the Atg8s, remodel and expand isolation membranes to form the autophagosome.
The Atg8 family of ubiquitin-like proteins associate with autophagosomal membranes through conjugation to the lipid phosphatidylethanolamine. Atg8 lipidation occurs via two ubiquitin-like conjugation systems. The ubiquitin-like protein Atg12 is conjugated to Atg5 and together with Atg16L1 forms an E3-like ligase complex that conjugates Atg8s to phosphatidylethanolamine (Mizushima et al., 1998; Ichimura et al., 2000). In mammalian cells, there are six Atg8 orthologues that are divided into the LC3 and GABARAP subfamilies. Although the exact function of Atg8s during autophagosome biogenesis is unclear, they have been proposed to mediate the expansion and closure of autophagosomal membranes (Nakatogawa et al., 2007; Fujita et al., 2008; Weidberg et al., 2010, 2011; Itakura et al., 2012a; Manil-Ségalen et al., 2014; Sawa-Makarska et al., 2014; Stolz et al., 2014). Using RNAi to knock down each of the LC3 or GABARAP subfamilies, it was reported that they function nonredundantly during autophagosome biogenesis. LC3 subfamily members were shown to promote elongation of phagophore membranes before GABARAP-mediated maturation and possible sealing of the autophagosome (Weidberg et al., 2010). In contrast, a subsequent study in Caenorhabditis elegans reported that LC3 functions downstream of GABARAP during a developmental type of autophagy termed allophagy (Manil-Ségalen et al., 2014). In addition to autophagosome biogenesis, Atg8s can also function during selective autophagy (Stolz et al., 2014). In the canonical selective autophagy model, autophagy receptors that are resident on cargo or bound via ubiquitin chains can recruit isolation membranes through an LC3-interacting region (LIR) motif, which binds to LC3/GABARAP. The autophagy receptor and LC3/GABARAP interactions bridge the autophagy machinery with cargo, thereby fostering selective engulfment by the autophagosome (Birgisdottir et al., 2013; Stolz et al., 2014). Mutation of the LIR motif in autophagy receptors ablates LC3/GABARAP recruitment and blocks selective autophagy (Pankiv et al., 2007; Kirkin et al., 2009; Novak et al., 2010; von Muhlinen et al., 2012). Recent studies have revealed that PINK1/Parkin mitophagy functions via a different mechanism whereby autophagosome biogenesis is initiated on the surface of mitochondria rather than through the recruitment of preformed isolation membranes (Itakura et al., 2012a; Lazarou et al., 2015). The primary PINK1/Parkin mitophagy receptors, optineurin and NDP52 (Wong and Holzbaur, 2014; Lazarou et al., 2015), function by promoting ULK1 recruitment to damaged mitochondria to initiate autophagosome biogenesis (Lazarou et al., 2015).
To date, the contribution of individual Atg8 proteins and their subfamilies to different types of autophagy remains unclear. This can be attributed to the large number of Atg8 family members precluding studies into their function. To overcome this hurdle, we generated knockouts (KOs) of all six Atg8s and triple knockouts (TKOs) of each LC3 and GABARAP subfamily in HeLa cells. We discovered that Atg8s are not essential for autophagosome formation or selective engulfment of cargo during PINK1/Parkin mitophagy and starvation-induced autophagy. Instead, we found that Atg8s play a critical role during the late stages of autophagy by recruiting PLEKHM1 and driving the fusion of autophagosomes to lysosomes. Furthermore, we identified functional differences between LC3s and GABARAPs, revealing that GABARAPs are the primary drivers of the autophagy types tested. Overall, our findings update the current models of autophagosome biogenesis by defining nonredundant Atg8 function, yielding key insights into the mechanisms of autophagy.
Results
Assessing the contribution of Atg8s to PINK1/Parkin mitophagy and starvation autophagy
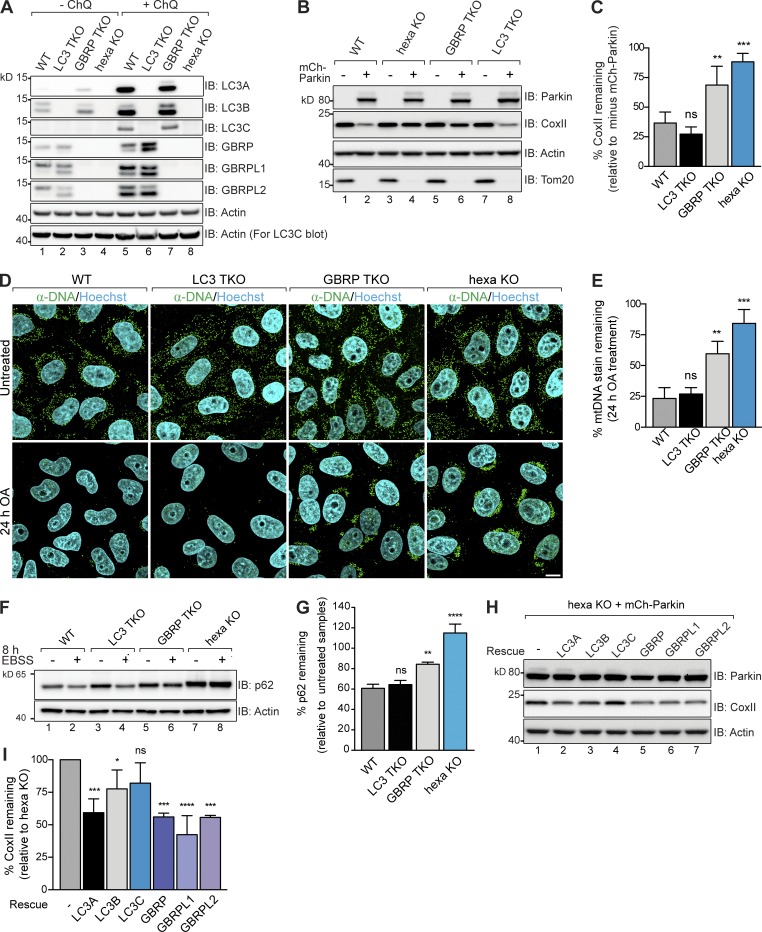
To clarify the role of mammalian Atg8 family members during both PINK1/Parkin mitophagy and starvation-induced autophagy, we used CRISPR/Cas9 genome editing to disrupt the genes encoding the six known Atg8s in HeLa cells (LC3A, LC3B, LC3C, GABARAP, GABARAPL1, and GABARAPL2; designated hexa KO). TKOs of the Atg8 subfamilies, LC3 (LC3 TKO) and GABARAP (GBRP TKO), were also generated to determine the role played by each subfamily. Knockout lines were confirmed by DNA sequencing of indels (Tables S1 and S3) and immunoblotting of LC3A, LC3B, LC3C, GABARAP (GBRP), GABARAPL1 (GBRPL1), and GABARAPL2 (GBRPL2) in cells treated either without or with chloroquine to accumulate lipidated LC3 and GBRP proteins (Fig. 1 A).
Figure 1.
Characterization of mammalian Atg8 family function during PINK1/Parkin mitophagy and starvation induced autophagy. (A) WT, LC3 TKO, GBRP TKO, and hexa KO (LC3/GBRP KO) lines were confirmed by immunoblotting (IB). Chloroquine (ChQ) treatment prevents lysosomal degradation of LC3s/GBRPs. For LC3C IB, 100 µg of lysates was analyzed. (B and C) The indicated cell lines with and without mCherry (mCh)–Parkin were analyzed by immunoblotting (B) and CoxII levels were quantified (C). (D) Representative images of WT, LC3 TKO, GBRP TKO and hexa KO expressing mCh-Parkin immunostained with mtDNA antibodies (green) and quantified for mitophagy (E). (F and G) WT, LC3 TKO, GBRP TKO, and hexa KO fully fed or starved for 8 h with Earle’s balanced salt solution (EBSS) were analyzed by immunoblotting (F), and p62 levels were quantified (G). (H and I) hexa KO cells expressing mCh-Parkin and individual untagged LC3s and GBRPs after 24-h OA treatment were analyzed by immunoblotting (H), and CoxII levels were quantified (I). Data in C, E, G, and I are mean ± SD from three independent experiments. *, P < 0.05; **, P < 0.005; ***, P < 0.001; ****, P < 0.0001 (one-way ANOVA). ns, not significant. Bar, 10 µm.
First, we analyzed PINK1/Parkin mitophagy in wild-type (WT), hexa KO, LC3 TKO, and GBRP TKO lines by measuring (a) degradation of cytochrome c oxidase subunit II (CoxII), a mitochondrial DNA (mtDNA)–encoded inner membrane protein (Fig. 1, B and C), and (b) clearance of mtDNA nucleoids (Fig. 1, D and E). To induce mitophagy, mitochondria were damaged using oligomycin and antimycin A. After 24-h oligomycin/antimycin A (OA) treatment, CoxII was robustly degraded and mtDNA nucleoids were cleared in WT and LC3 TKO cells expressing mCherry-Parkin, but not in hexa KO or GBRP TKO HeLa cell lines (Fig. 1, B–E), demonstrating a block in mitophagy. Immunoblotting confirmed that Atg8 family members were lipidated upon mitophagy induction in WT, LC3 TKO, and GBRP TKO cells and that the Parkin substrate Mfn1 was efficiently degraded in all KO cell lines, demonstrating that Parkin activity is not disrupted (Fig. S1 A). In addition, compensatory changes in the protein levels of LC3 and GBRP subfamily members were observed in TKO lines (Figs. 1 A and S1 A).
Next, we analyzed starvation-induced autophagy in WT and KO cell lines by measuring the degradation of p62, an autophagy substrate (Klionsky et al., 2016). WT, hexa KO, LC3 TKO, and GBRP TKO cells were incubated in Earle’s balanced salt solution (EBSS) for 8 h to induce starvation autophagy and then immunoblotted for p62 (Fig. 1, F and G). LC3 TKO cells degraded p62 similarly to WT cells, whereas GBRP TKO cells had a moderate but significant defect and hexa KOs had a complete block. Under nutrient-rich conditions, p62 levels were elevated in LC3 TKO, GBRP TKO, and hexa KO lines, indicating a decrease in autophagic flux and a requirement for both LC3 and GBRP subfamilies in basal autophagy (Fig. 1 F and Fig. S1, B and C). Chloroquine treatment confirmed that the elevated levels of p62 in LC3 TKO, GBRP TKO, and hexa KO were not caused by increased protein expression (Fig. S1, B and C). Together, the data show that GABARAPs play an important role for both PINK1/Parkin mitophagy and starvation autophagy, whereas the LC3 subfamily is dispensable for both. However, hexa KO cells had greater autophagy deficits than the GBRP TKOs (Fig. 1), indicating that LC3s can partially contribute to mitophagy and starvation in the absence of GABARAPs.
To determine the function of each Atg8 family member in isolation and assess whether a single LC3/GABARAP is sufficient to drive mitophagy or starvation autophagy, we generated hexa KO rescue cell lines expressing untagged LC3/GBRP proteins (Fig. S1 D). Equal expression of untagged LC3s and GABARAPs was achieved by FACS using GFP expressed from an internal ribosome entry site (IRES) as an expression level marker. PINK1/Parkin mitophagy and starvation autophagy were significantly rescued by all untagged GABARAP proteins (Fig. 1, H and I; and Fig. S1, E and F). LC3A was the most active of the LC3 subfamily members, followed by LC3B, which had low activity, whereas LC3C had little to no activity. Thus, the rescue data show that GABARAPs are more active than LC3s, corroborating the results observed with the LC3 and GBRP TKO lines (Fig. 1, B–G). Our results also reveal that individual Atg8 members are sufficient to drive autophagy. LC3 and GABARAP proteins are often expressed as N-terminal GFP fusions to analyze their function and track autophagosome formation (Klionsky et al., 2016). However, we found that GFP-tagged LC3s and GABARAPs were functionally impaired during mitophagy (Fig. S1, G and H), and GFP-GABARAPs do not robustly translocate to mitochondria (Lazarou et al., 2015). N-terminal fusion of GBRPL1 and GBRPL2 to the small HA tag instead does not inhibit their function (Fig. S1 I), and all three HA-tagged GABARAPs localized to structures encapsulating mitochondria during mitophagy (Fig. S2, A and B). It is therefore preferable to use small tags to study the function of Atg8 family members.
Atg8s are not essential for autophagosome formation but can regulate autophagosome size
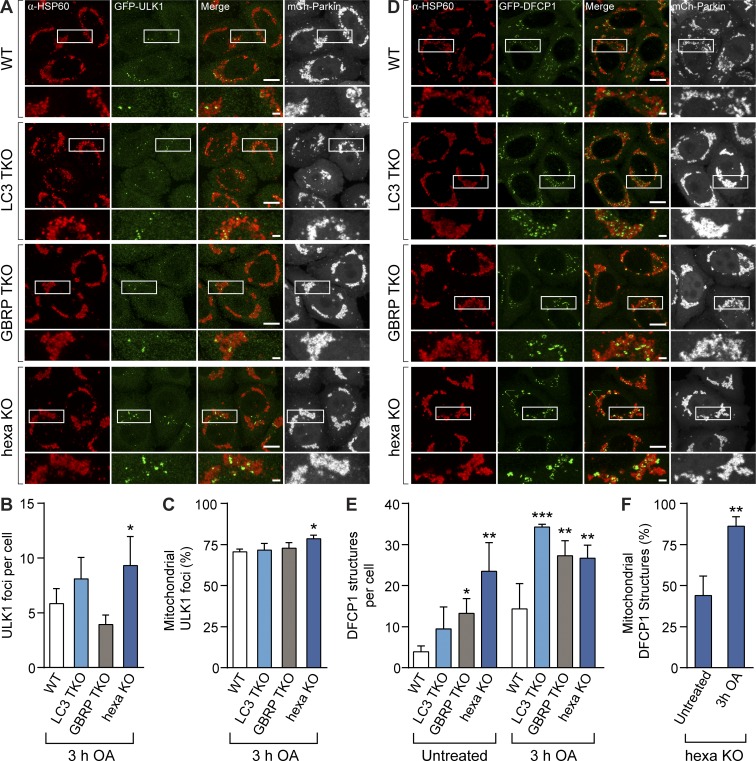
To identify the autophagy defect in GBRP TKOs and hexa KOs, we conducted a stepwise analysis of autophagosome biogenesis during mitophagy. Autophagosome formation on the mitochondrial surface is mediated by the mitophagy receptors optineurin and NDP52 (Wong and Holzbaur, 2014; Heo et al., 2015; Lazarou et al., 2015), which promote recruitment of the autophagy-initiating kinase ULK1 (Lazarou et al., 2015). Both optineurin and NDP52 were efficiently recruited in WT and hexa KO cells after 3-h OA treatment (Fig. S2, C–F), confirming that this early stage of mitophagy is unperturbed. Atg8 proteins, particularly GABARAPs, bind to ULK1 via an LIR motif and have been reported to both stabilize ULK1 complexes and promote ULK1 activation (Alemu et al., 2012; Kraft et al., 2012; Joachim and Tooze, 2016). We assessed ULK1-mediated autophagosome initiation by quantitative immunofluorescence microscopy of GFP-ULK1 foci. The formation of GFP-ULK1 foci and their recruitment to the surface of mitochondria was not significantly reduced in any of the KO cell lines when compared with WT cells (Fig. 2, A–C; and Fig. S2 G). The next stages of autophagosome biogenesis involve recruitment of the Vps34 lipid kinase complex. Vps34 enriches phagophore membranes with the lipid PI3P, which is required for autophagosome formation and recruitment of PI3P-binding proteins, including DFCP1 (Axe et al., 2008). Analysis of GFP-DFCP1–labeled autophagosome precursors formed during PINK1/Parkin mitophagy revealed that LC3 TKOs, GBRP TKOs, and hexa KOs were not defective in generating the precursors (Fig. 2, D and E; and Fig. S2 H). Indeed, all KO lines had significantly more GFP-DFCP1 structures than WT cells. Although the number of GFP-DFCP1 structures in hexa KOs was not markedly increased after PINK1/Parkin activation relative to the untreated controls, the proportion of GFP-DFCP1 structures around mitochondria increased significantly (Fig. 2 F). These results show that autophagosome initiation and nucleation steps can occur in GBRP TKO and hexa KO cells, indicating that the block in autophagy lies at a downstream step.
Figure 2.
Analysis of autophagosome initiation and nucleation events in Atg8 knockout lines during PINK1/Parkin mitophagy. (A and D) Representative images of WT, LC3 TKO, GBRP TKO, and hexa KO cells expressing mCh-Parkin and GFP-ULK1 (A) or GFP-DFCP1 (D), immunostained for HSP60 and GFP after 3-h OA treatment (untreated images shown in Fig. S2, G and H). Quantification by image analysis of ULK1 foci structures per cell (B), DFCP1 structures per cell (E), and the proportion of mitochondrially associated (C) ULK1 foci and (F) DFCP1 structures in WT, LC3 TKO, GBRP TKO, and hexa KO cells. Data in B, C, E, and F are mean ± SD from three independent experiments using measurements from >80 cells per sample. *, P < 0.05; **, P < 0.005; ***, P < 0.001 (one-way ANOVA). Bars: 10 µm; (insets) 2 µm.
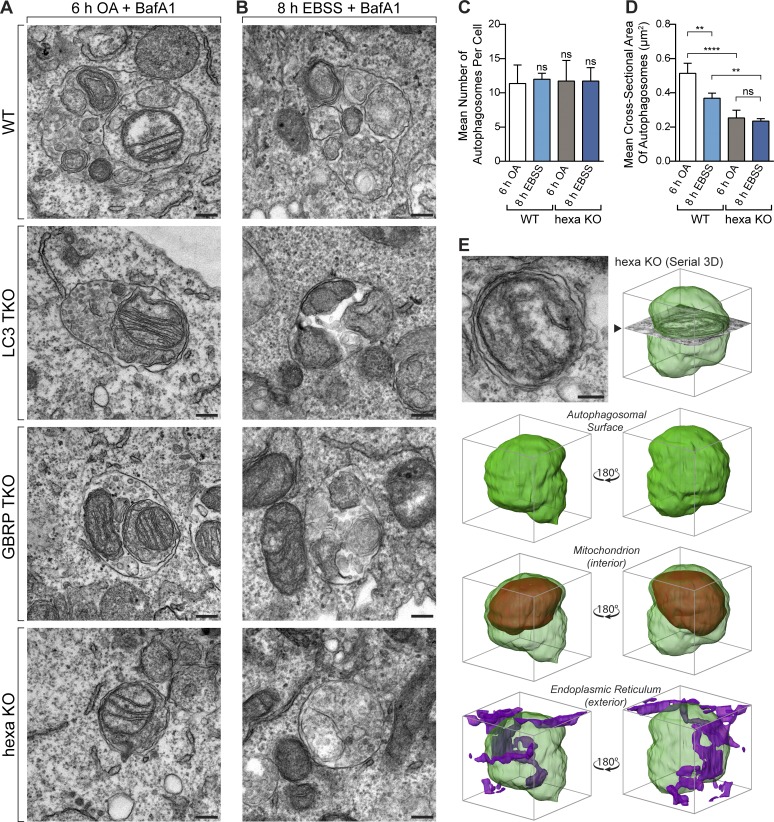
We used transmission electron microscopy (TEM) to identify where the downstream block in autophagy lies in GBRP TKOs and hexa KOs by analyzing autophagosomal structures in WT and KO cell lines during PINK1/Parkin mitophagy (Figs. 3 A and S3 D), and starvation autophagy (Figs. 3 B and S3 D). Cells were treated with bafilomycin A1 (BafA1) during both mitophagy and starvation conditions to prevent autophagosome turnover via lysosomal fusion and thereby enable analytical comparisons of autophagosome number and size. Similar to WT and LC3 TKOs, which do not have autophagy defects, both GBRP TKO and hexa KO lines contained fully formed autophagosomes, and mitochondria were correctly sequestered during mitophagy. Furthermore, hexa KO cells generated a similar number of autophagosomes compared with WT cells during mitophagy and starvation autophagy in the presence of BafA1 (Fig. 3 D), implying equivalent rates of biogenesis at this time point. Consistent with reported roles for Atg8s in expanding autophagosomal membranes (Nakatogawa et al., 2007; Weidberg et al., 2010), we observed smaller autophagosomes in hexa KO cells (Fig. 3, A and D). Notably, autophagosomes formed in WT cells during mitophagy were larger than starvation induced autophagosomes (Figs. 3 D and S3 D), but an equivalent difference was not detected in hexa KO cells, indicating a maximum achievable autophagosome size in the absence of Atg8s. To ensure that the observed structures in hexa KOs represent completely sealed autophagosomes, embedded samples were serial-sectioned and reconstructed in 3D (Fig. 3 E and Fig. S3, A–C). In each 3D reconstruction, mitochondria were localized within the delimiting membranes of a sealed autophagosomal compartment.
Figure 3.
Atg8s are dispensable for autophagosome formation but regulate autophagosome expansion. (A and B) Representative TEM images of autophagosomes containing mitochondria in WT, LC3 TKO, GBRP TKO, and hexa KO cells after incubation for 6 h with OA and BafA1 (A) or for 8 h with EBSS and BafA1 (B). Wider field-of-view images and untreated examples shown in Fig. S3 D. (C and D) TEM quantification of mean autophagosome number per cell (C) and the mean cross-sectional area of autophagosomes in WT and hexa KO cells treated with BafA1 and either OA for 6 h or starved with EBSS for 8 h (D). Scatterplot of measurements provided in Fig. S4 A. (E) 3D rendering of a reconstructed autophagosomal compartment (green) from a hexa KO cell after 6 h OA and BafA1 treatment, shown with sequestered mitochondrion (red) and endoplasmic reticulum (purple). (Source images and further examples shown in Fig. S3, A–C.) Data in C and D are mean ± SD from three independent double-blinded experiments, using measurements from exactly 12 randomly chosen cells (>100 autophagosomes) per sample. **, P < 0.005; ****, P < 0.0001 (one-way ANOVA). Bars, 200 nm.
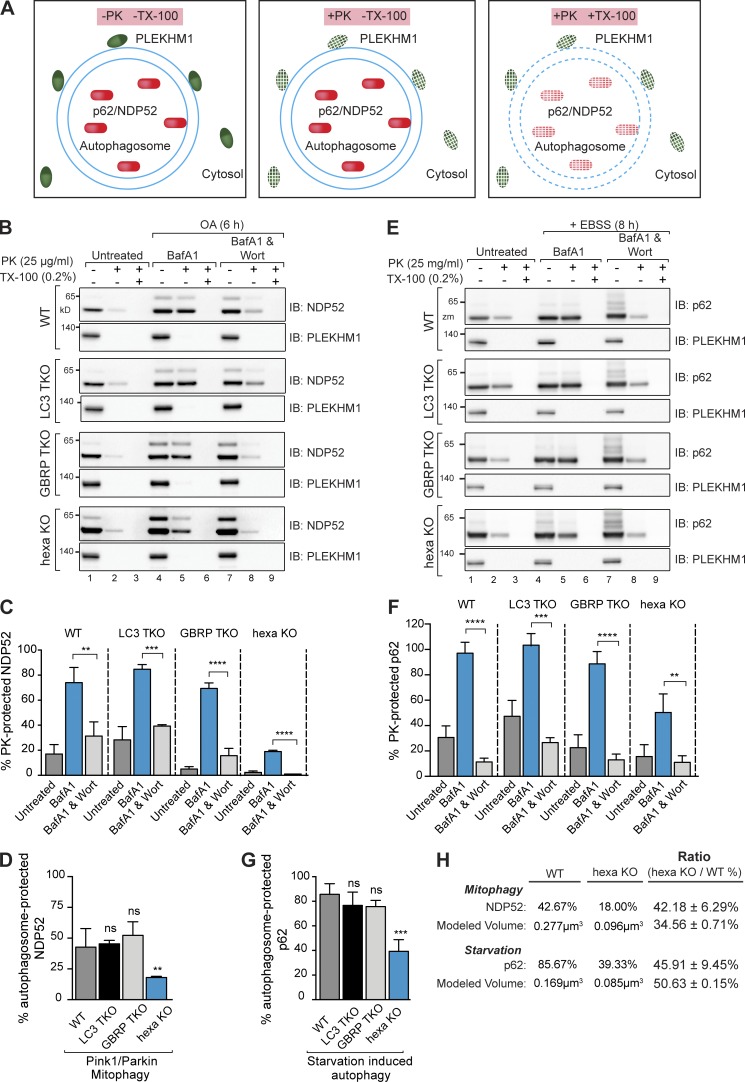
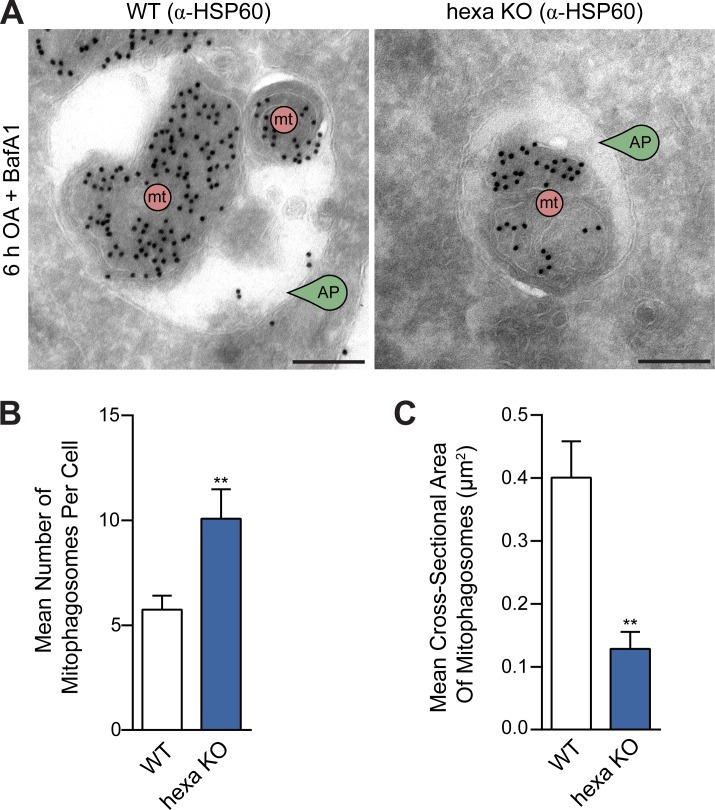
To further confirm that the autophagosomes are correctly sealed in all KO cell lines, a protease protection assay (McEwan et al., 2015a) was adapted to determine what proportion of the autophagic cargo is encapsulated by autophagosomes (see Fig. 4 A for a schematic of the assay). WT and KO cell lines were treated with BafA1 and either starved for 8 h or treated with OA for 6 h to accumulate autophagosomes. Control samples were incubated with wortmannin, which blocks autophagosome formation. Cell lysates were treated with external protease (Proteinase K [PK]) to determine the proportion of NDP52 (mitophagy) or p62 (starvation) protected within autophagosomes. The assay revealed that all KO cell lines formed sealed autophagosomes relative to wortmannin-treated controls during both mitophagy and starvation autophagy (Fig. 4, B, C, E, and F). Undegraded NDP52 and p62 in wortmannin and PK-treated samples (Fig. 4, B and E, lane 8) likely represents preexisting membrane-protected cargo (Fig. 4, B and E, lane 2). Hexa KO cells packaged less NDP52- and p62-associated cargo relative to WT cells (Fig. 4, D and G), possibly because of the presence of smaller autophagosomes in hexa KO cells (Fig. 3, A and D; and Fig. S4 A). Because the cargo capacity of an autophagosome is limited by its volume, we calculated and compared the effective volume of WT and hexa KO autophagosomes by Cavalieri’s principle (Fig. S4 C) using the cross-sectional areas determined in Fig. 3 D (Fig. 4 H). The calculated ratio of autophagosomal volume between WT and hexa KO during mitophagy (∼35%) and starvation autophagy (∼51%) was equivalent to the protease-protected ratio of NDP52 (∼42%) and p62 (∼46%), respectively (Fig. 4 H). Thus, hexa KO autophagosomes sequester less cargo because of their decreased capacity. To exclude the possibility that mitochondria are not correctly sequestered in hexa KO cells, we quantified mitophagosomes using HSP60-labeled immunogold TEM in WT and hexa KO cells undergoing PINK1/Parkin mitophagy. Cells were treated with OA for 6 h to activate PINK1/Parkin mitophagy and with BafA1 to prevent mitophagosome fusion with lysosomal compartments. Under these conditions, hexa KO cells formed more mitophagosomes than WT cells (Fig. 5, A and B), although they were smaller than those observed in WT cells (Figs. 5 C and S4 B), consistent with earlier results (Fig. 3). Collectively, our results support the conclusion that the reduced protection of NDP52 and p62 in hexa KOs (Fig. 4, D and G) is caused by decreased autophagosome capacity rather than defects in cargo sequestration or autophagosome closure.
Figure 4.
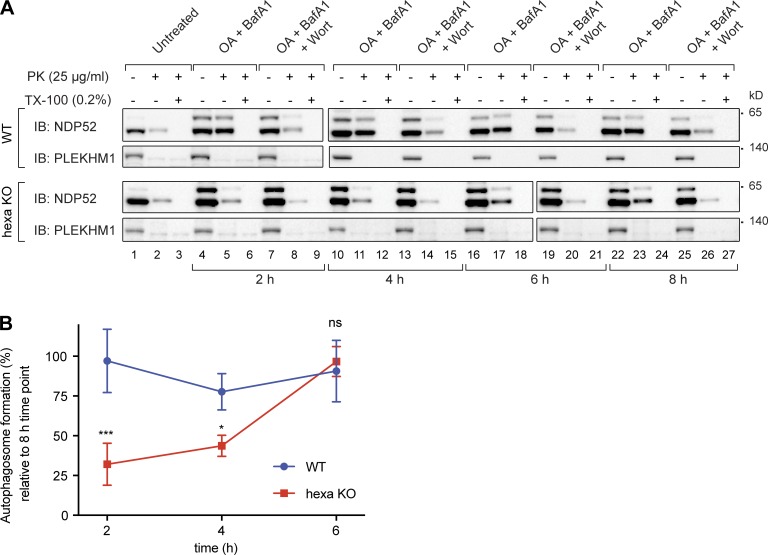
Formation of correctly sealed autophagosomes containing sequestered cargo in Atg8 knockout lines. (A) Schematic of the protease protection assay used to assess packaging of cargo in sealed autophagosomes. Protein cargoes in autophagosomes are protected from the addition of external proteinase K (PK; middle) unless Triton X-100 (TX-100) is present (right). (B–D) Lysates from WT, LC3 TKO, GBRP TKO, and hexa KO cells untreated or treated with OA and BafA1 in the presence or absence of wortmannin (Wort) were subjected to the PK protection assay. (B) Samples were subsequently analyzed by immunoblotting with NDP52. PLEKHM1 served as a cytosolic control. (C) The amount of PK-protected NDP52 for each condition (untreated, OA/BafA1, or OA/BafA1/Wort) was quantified (percentage of NDP52 amount in lanes 2, 5, and 8 relative to NDP52 amount in lanes 1, 4, and 7, respectively). (D) Quantification of the difference in the amount of PK-protected NDP52 in BafA1-treated samples (C, blue bars) versus BafA1/Wort-treated samples (light gray bars in C; represents the amount of NDP52 specifically protected within autophagosomes). (E–G) PK protection assay of WT, LC3 TKO, GBRP TKO, and hexa KO during starvation autophagy. (E) Lysates from the indicated cell lines untreated or treated with EBSS and BafA1 in the presence or absence of wortmannin were incubated with PK with or without 0.2% (vol/vol) Triton X-100 and subsequently immunoblotted with p62 antibodies. (F) Quantification of PK-protected p62 for each condition (untreated, EBSS/BafA1, or EBSS/BafA1/Wort) were performed. (G) The amount of autophagosome-protected p62 was also analyzed. (H) The mean volume of autophagosomes formed during mitophagy and starvation in WT and hexa KO cells was calculated (see Fig. S4 C for calculation) and compared with the proportion of autophagosome-protected NDP52 (mitophagy) and p62 (starvation) obtained from D and G, respectively. Data in C, D, F, and G are mean ± SD from three independent experiments. **, P < 0.005; ***, P < 0.001; ****, P < 0.0001 (one-way ANOVA). ns, not significant.
Figure 5.
Atg8s regulate autophagosome capacity, but not the selective sequestration of cargo, during PINK1/Parkin mitophagy. (A) Representative immunogold TEM images of HSP60 labeled WT and hexa KO cells after incubation with OA and BafA1 for 6 h. (B and C) Immunogold TEM quantification of the mean number of mitophagosomes per cell (B) and the mean cross-sectional area of mitophagosomes in WT and hexa KO cells (C) after treatment with OA and BafA1 for 6 h. (Scatterplot of measurements provided in Fig. S4 B.) Data in B and C are mean ± SD from three independent experiments, using measurements from exactly eight randomly chosen cells (>40 mitophagosomes measured per sample). **, P < 0.005 (Student’s t test). Bars, 200 nm.
Using the NDP52 protease-protection assay, we next assessed the kinetics of autophagosome formation during mitophagy in WT and hexa KO cells. A time course was performed, with the 8-h time point for each cell line serving as a reference point to determine the rate at which sealed autophagosomes form. Sealed autophagosomes formed rapidly within 2 h in WT cells (Fig. 6, A and B). In contrast, autophagosome formation at early time points was significantly lower in hexa KOs (2- and 4-h time points), followed by rapid generation of sealed autophagosomes by the 6-h time point. This result indicates that Atg8 proteins play an important role in the efficient production of autophagosomes, despite being dispensable for autophagosome formation.
Figure 6.
Autophagosome biogenesis is initially slow in the absence of LC3s and GABARAPs during Pink1/Parkin mitophagy. Lysates from WT and hexa KO cells untreated or treated with OA and BafA1 in the presence or absence of wortmannin (Wort) for indicated time points were subjected to the PK protection assay (see Fig. 4 A). (A) Samples were subsequently analyzed by immunoblotting with NDP52. All of the blots were transferred and exposed at the same time. PLEKHM1 served as a cytosolic control. (B) The amount of PK-protected NDP52 for each condition was first quantified (percentage of NDP52 amount PK only treated lanes relative to NDP52 amount in no PK/TX-100 ones). Then the difference in the amount of PK-protected NDP52 in BafA1 treated samples versus BafA1/Wort treated samples (represents the amount of autophagosome-protected NDP52 for each time point) was calculated. The autophagosome formation rates at 2, 4, and 6 h for each cell line were determined by the ratio of the autophagosome protected NDP52% at these time points to the autophagosome protected NDP52% at 8 h; i.e., the autophagosome-protected NDP52% at 8 h was considered to be 100%. Data in B are mean ± SD from three independent experiments. *, P < 0.05; ***, P < 0.001 (one-way ANOVA). ns, not significant.
GABARAPs are crucial for autophagosome–lysosome fusion
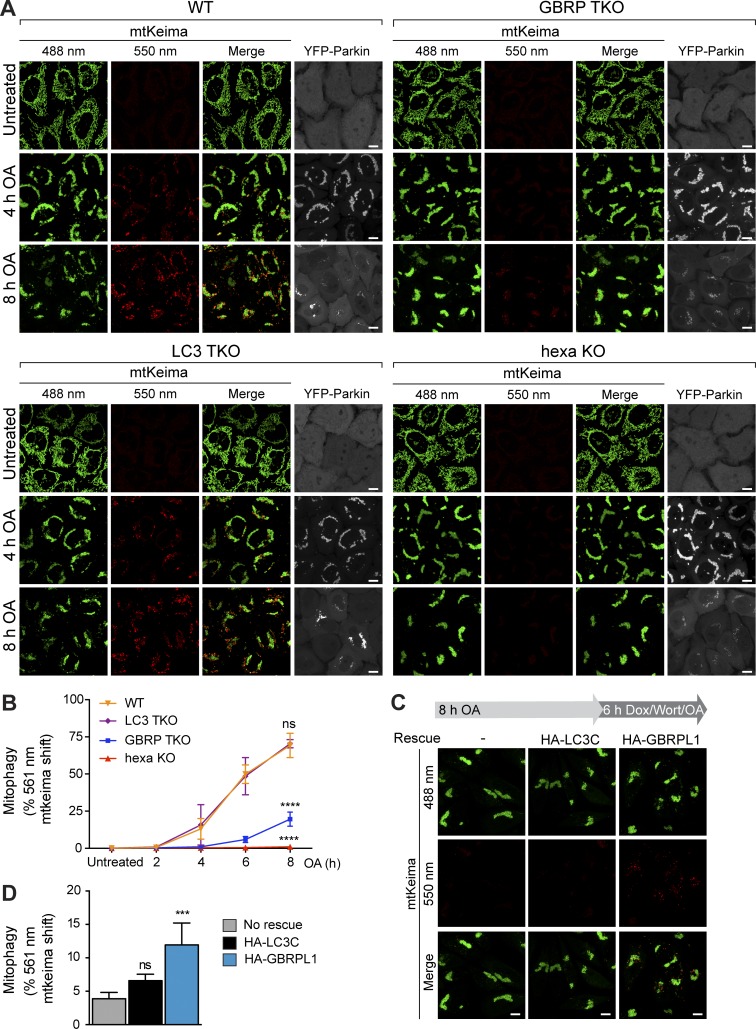
The primary autophagy defect in hexa KO cells is unlikely to be a result of reduced autophagosome capacity, because GBRP TKO cells sequester similar amounts of cargo as WTs (Fig. 4) but have significant autophagy defects (Fig. 1). We therefore asked whether the autophagy defect in GBRP TKO and hexa KO cells lies at the terminal step of autophagy, in which autophagosomes fuse with lysosomal compartments. Autophagosome–lysosome fusion was first assessed using WT and KO cell lines stably expressing YFP-Parkin and mitochondrially targeted mKeima (mtKeima), which undergoes a spectral shift in the low-pH environment of lysosomes (Katayama et al., 2011). WT and LC3 TKO cells displayed almost identical rates of mtKeima acidification during mitophagy (indicative of autophagosome–lysosome fusion), whereas GBRP TKO cells displayed significant defects and hexa KO cells had almost no detectable autophagosome–lysosome fusion at the time points tested (Fig. 7, A and B; and Fig. S5, A and B).
Figure 7.
Atg8s are crucial for correct acidification of sequestered cargo. (A and B) WT, LC3 TKO, GBRP TKO, and hexa KO expressing YFP-Parkin and mtKeima were either untreated or treated with OA for indicated times and analyzed for (A) lysosomal-positive mtKeima using fluorescence microscopy (550 nm mtKeima; see Fig. S5 A for 2- and 8-h images), and (B) FACS as the percentage of 561 nm mtKeima positive cells (see Fig. S5 B for FACS plots). (C and D) Hexa KO cells expressing mtKeima and doxycycline (Dox)–inducible HA-LC3C or HA-GBRPL1 were pretreated with OA for 8 h. After being incubated with Dox, Wort, and OA for 6 h, the cells were analyzed by confocal microscopy (C) and FACS (D) for lysosomal-positive mtKeima (see Fig. S5, C and D). Data in B and D are mean ± SD from three independent experiments. ***, P < 0.001; ****, P < 0.0001 (one-way ANOVA). ns, not significant. Bars, 10 µm.
To test whether expression of a GBRP can promote fusion between lysosomes and preformed autophagosomes, we generated hexa KO cell lines expressing mtKeima, YFP-Parkin, and vector control, HA-GBRPL1, or HA-LC3C under a doxycycline-inducible promoter (Fig. S5 C). The cells were treated with OA for 8 h to accumulate autophagosomes, followed by addition of wortmannin to prevent new autophagosome synthesis and doxycycline to induce HA-GBRPL1 or HA-LC3C expression. Consistent with the low autophagy activity observed for LC3C (Fig. 1), cells inducibly expressing HA-LC3C did not significantly promote autophagosome–lysosome fusion (Fig. 7, C and D; and Fig. S5 D). In contrast, HA-GBRPL1 expression induced a significant increase in autophagosome–lysosome fusion relative to the vector control as indicated by acidification of mtKeima (Fig. 7, C and D; and Fig. S5 D).
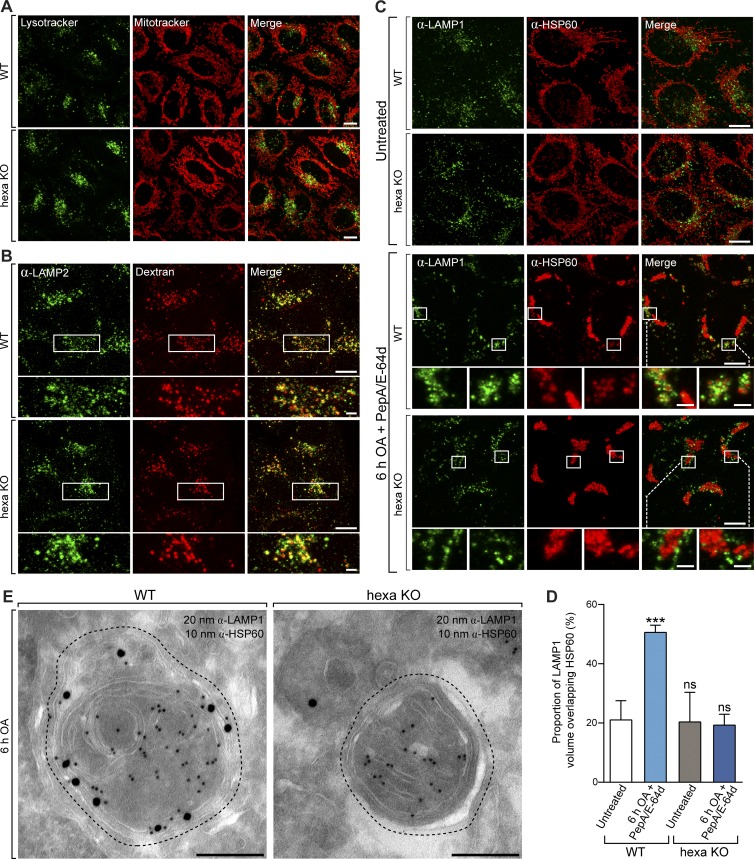
Given that the spectral shift of mtKeima is a measure of mitochondrial acidification within lysosomes, the defect in hexa KO cells may have been caused by a high lysosomal pH. We eliminated this possibility by incubating live cells with the acidophilic dye LysoTracker green, which equivalently stained the lysosomal compartments of WT and hexa KO cells (Fig. 8 A). The ability of lysosomes to fuse with endosomal compartments was also assessed in hexa KOs. WT and hexa KO cells were cultured for 24 h in the presence of fluorescently labeled dextran (Cascade blue–conjugated dextran), which passively enters the endolysosomal system via endocytosis (Fig. 8 B). Similar to WT cells, hexa KOs were still capable of fusing lysosomes with endosomal compartments (Fig. 8 B). Furthermore, lysosomes can relocate to perinuclear regions adjacent to mitochondria during mitophagy similarly in WT and hexa KO cells (Fig. S2 I). Collectively, these results demonstrate that lysosomal function and transport are not impaired in hexa KO cells.
Figure 8.
Atg8s are essential for autophagosome–lysosome fusion, but not lysosomal function. (A and B) Representative images of untreated WT and hexa KO cells stained with LysoTracker green and MitoTracker deep red (A) or loaded with Cascade blue–conjugated dextran (B) for 24 h before immunostaining for LAMP2. (C) Representative images of WT and hexa KO cells expressing untagged Parkin, immunostained for HSP60 and LAMP1 after incubation for 6 h in the presence or absence of OA and protease inhibitors (10 µg/ml Pepstatin A; 10 µg/ml E-64d). (D) Quantification by image analysis of the colocalization between LAMP1 and HSP60 after incubation for 6 h in the presence or absence of OA, Pepstatin A, and E-64d. (E) Representative immunogold TEM images of WT and hexa KO cells, double labeled for LAMP1 (20 nm gold) and HSP60 (10 nm gold) after incubation with OA alone for 6 h (compartment perimeters indicated by offset dashed line). Data in D are mean ± SD from three independent experiments. ***, P < 0.001 (one-way ANOVA). ns, not significant. Bars: (A–C) 10 µm; (A–C, insets) 2 µm; (E) 200 nm.
Despite the translocation of lysosomes to regions adjacent to mitochondria during mitophagy, colocalization between mitochondria and lysosomes was not observed in either WT or hexa KOs (Fig. S2 I). We attribute this lack of colocalization to the rapid degradation of mitochondria after their delivery to lysosomal compartments. Therefore, to further assess autophagosome–lysosome fusion, WT and hexa KO cells stably expressing Parkin were either untreated or treated for 6 h with OA in the presence of the protease inhibitors (Pepstatin A and E-64d) to block lysosomal degradation. The overlap between LAMP1 and HSP60 staining was measured in untreated WTs and hexa KOs to provide a baseline measure of random colocalization (Fig. 8 C). Upon mitophagy activation in the presence of protease inhibitors, a significant increase in LAMP1/HSP60 colocalization was observed in WT, but not hexa KO cells (Fig. 8, C and D). Double-labeled immunogold TEM experiments were also performed to analyze autophagosome–lysosome fusion (Figs. 8 E and S3 E). WT and hexa KO cells were treated with OA alone for 6 h and stained with antibodies against HSP60 and LAMP1. Mitophagosomal structures in WT cells were stained positively for both LAMP1 and HSP60, whereas HSP60-positive mitophagosomes in hexa KOs were devoid of LAMP1 (Fig. 8 E). In WT cells, HSP60 staining was also detected in late-stage endolysosomal compartments that lacked evidence of mitochondrial morphology (Fig. S3 E). HSP60 staining was absent from the endolysosomal compartments of hexa KO cells (Fig. S3 E), providing additional evidence that hexa KOs cannot proceed beyond autophagosome formation. We conclude that the Atg8 family is not essential for autophagosome formation, closure, or cargo sequestration but is essential for autophagosome–lysosome fusion.
PLEKHM1 is preferentially recruited by GABARAPs during PINK1/Parkin mitophagy
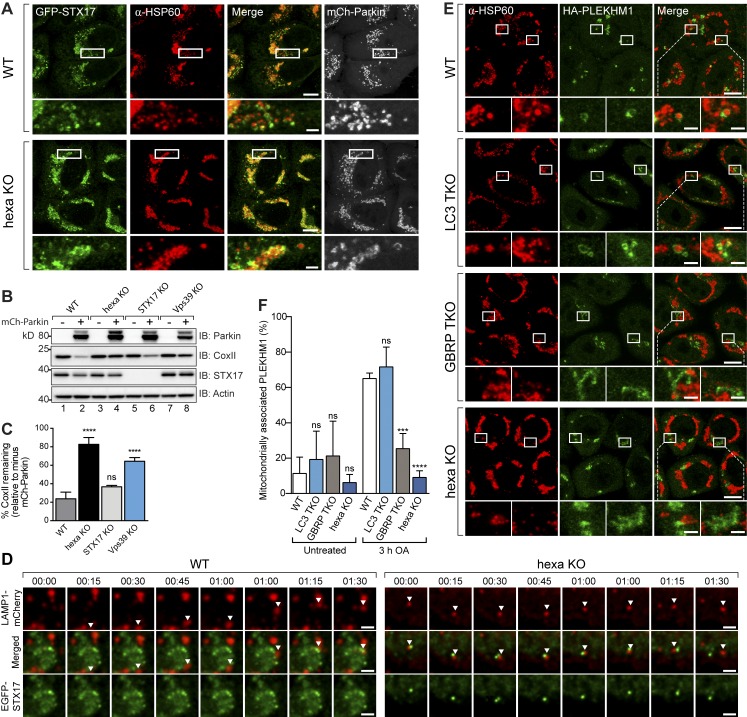
The autophagosomal soluble N-ethylmaleimide-sensitive factor syntaxin 17 (STX17) has been shown to mediate autophagosome–lysosome fusion (Itakura et al., 2012b; Hamasaki et al., 2013; Diao et al., 2015). Therefore, we asked whether STX17 fails to localize to structures encapsulating mitochondria during mitophagy in GBRP TKOs and hexa KOs. GFP-STX17 localized around mitochondria comparably in WT and all KO cell lines during mitophagy (Fig. 9 A; and Fig. S2, J and K), and KO of STX17 did not block mitophagy (Fig. 9, B and C). Furthermore, GFP-STX17–labeled structures analyzed dynamically in live cells failed to fuse with mCherry-LAMP1 in hexa KOs (Fig. 9 D, right) but did fuse with mCherry-LAMP1 in WT cells (Fig. 9 D, left). Although STX17’s importance during autophagy is not discounted (Itakura et al., 2012b; Diao et al., 2015), these results show that STX17 is unlikely to account for the autophagosome–lysosome fusion defect observed in GRBP TKO and hexa KO cells specifically during PINK1/Parkin mitophagy. Our finding is also in agreement with a recent study showing that STX17 is not required for PINK1/Parkin mitophagy but is important for fusion of mitochondrial-derived vesicles with lysosomes (McLelland et al., 2016).
Figure 9.
Recruitment of PLEKHM1 by GABARAPs promotes autophagosome–lysosome fusion. (A) Representative images of WT and hexa KO cells expressing mCh-Parkin and GFP-STX17, immunostained for HSP60 and GFP after 3 h OA treatment. (B and C) Lysates from WT, hexa KO, STX17 KO, and Vps39 KO treated for 24 h with OA were analyzed by immunoblotting and (C) CoxII levels were quantified. (D) Deconvolved live-cell time-series images of WT and hexa KO cells stably expressing untagged Parkin and transiently expressing GFP-STX17 and mCherry-LAMP1, showing interactions between STX17 structures and lysosomes (arrowhead) after 6-h OA treatment. (E) Representative images of WT, LC3 TKO, GBRP TKO, and hexa KO cells expressing mCh-Parkin and HA-PLEKHM1, immunostained for HSP60 and HA after 3-h OA treatment. (mCh-Parkin channels and untreated examples are shown in Fig. S2, L and M). (F) Quantification of the mitochondrially associated proportion of PLEKHM1 in WT, LC3 TKO, GBRP TKO, and hexa KO cells. Data in C and F are mean ± SD from three independent experiments. ***, P < 0.001; ****, P < 0.0001 (one-way ANOVA). ns, not significant. Bars: (A and E) 10 µm; (A and E, insets) 2 µm; (D) 2 µm.
PLEKHM1 has been shown to regulate autophagosome–lysosome fusion during starvation autophagy, xenophagy, and aggrephagy (McEwan et al., 2015a,b). LC3s and GABARAPs recruit PLEKHM1 to autophagosomes via a LIR motif, where PLEKHM1’s association with the homotypic fusion and protein sorting (HOPS) complex promotes autophagosome–lysosome fusion (McEwan et al., 2015a,b). Analysis of HA-PLEKHM1 during mitophagy revealed that it forms structures encapsulating mitochondria in WT and LC3 TKO cells, which were significantly reduced in GBRP TKO cells and even more so in hexa KO cells (Fig. 9, E and F; and Fig. S2, L and M). Although in vitro binding data indicate that PLEKHM1 interacts with all mammalian Atg8 family members (McEwan et al., 2015a), our results show that in cells, HA-PLEKHM1 is preferentially recruited by GABARAPs (Fig. 9, E and F). Knockout of Vps39, a core subunit of the HOPS complex, blocks mitophagy (Fig. 9, B and C), supporting the importance of HOPS recruitment mediated by GABRAP-PLEKHM1 interactions in driving autophagosome–lysosome fusion. Attempts at knocking out PLEKHM1 using multiple different CRISPR guides proved to be unsuccessful, perhaps owing to the presence of a PLEKHM1 pseudogene within the human genome.
Discussion
Current models of autophagy depict members of the Atg8 family as essential players of autophagosome biogenesis that expand and seal autophagosomal membranes and drive sequestration of selective cargo. In this study, we made the unexpected discovery that Atg8s are not essential for autophagosome formation but are crucial for autophagosome–lysosome fusion. Our results also support a role for Atg8s in regulating autophagosome size, corroborating previous studies (Nakatogawa et al., 2007; Weidberg et al., 2010; Yamano et al., 2014; Landajuela et al., 2016). Despite a reported lack of autophagosomes in Atg8 KO yeast, small autophagosome-like structures were observed (Kirisako et al., 1999). Some of these structures were described as being morphologically indistinguishable from autophagosomes. However, in contrast to our findings showing that hexa KOs form equivalent or greater numbers of autophagosomes than WT cells, the autophagosome-like structures in Atg8 KO yeast were reportedly rare (Kirisako et al., 1999).
Atg8s have been shown to mediate membrane fusion and hemifusion in vitro (Nakatogawa et al., 2007; Weidberg et al., 2011; Wu et al., 2015), whereas genetic ablation of the Atg8 lipidation machinery results in the accumulation of unsealed autophagosomes (Komatsu et al., 2005; Fujita et al., 2008; Sou et al., 2008; Itakura et al., 2012a). Collectively, these observations led to the conclusion that Atg8s are also required for autophagosome closure. Our contrasting results show that Atg8s are not essential for sealing autophagosomes, indicating that other factors may mediate this process. Atg2A and Atg2B are leading candidates because their simultaneous knockdown in cells leads to the accumulation of unsealed autophagosomes (Velikkakath et al., 2012). Although autophagosome closure is typically described as a fusion process, a recent study has put forward an alternative mechanism in which autophagosomes are sealed via membrane scission (Knorr et al., 2015). Because Atg8s have been shown mediate membrane fusion, but not membrane scission, our results are in agreement with this model. Whether Atg2A and Atg2B can contribute to membrane scission events remains to be determined. Given that hexa KO cells can form sealed autophagosomes, the presence of unsealed autophagosomes in cells lacking Atg8 lipidation suggests that conjugation factors, including Atg5 and Atg3, may have additional functions. For example, Atg14 can promote autophagosome–lysosome fusion in addition to its well-characterized role in the PI3K complex (Diao et al., 2015). Alternatively, it is possible that other ubiquitin-like proteins outside of the Atg8 family may also be conjugated to autophagosomal membranes and can contribute to autophagosome formation.
LC3B is the most widely studied Atg8 protein and is used as a primary marker of autophagosomes and autophagy (Klionsky et al., 2016). We have found that LC3B, as well as the LC3 subfamily overall, was not essential for starvation-induced autophagy and PINK1/Parkin mitophagy. However, LC3 TKO cells showed defects in autophagic flux. This indicates that LC3s are important for constitutive autophagy that will likely play a role in maintaining cellular homeostasis. The GABARAP subfamily was identified as the primary driver of PINK1/Parkin mitophagy and starvation autophagy in this study. Rescue of hexa KO cells with each individual GABARAP was sufficient to restore autophagy, but not to WT levels. Therefore, GABARAPs may function cooperatively either with each other or with LC3 subfamily members to improve autophagy efficiency. The clear importance for GABARAPs during PINK1/Parkin mitophagy correlates with their high expression levels in the central nervous system (Xin et al., 2001) and the role of PINK1 and Parkin in Parkinson’s disease (Kitada et al., 1998; Valente et al., 2004). Given the major role played by GABARAPs, it is important to analyze their function when assessing autophagy pathways. The avoidance of bulky tags that decrease Atg8 function, such as GFP, is also a significant factor to consider.
Despite the low activity of LC3C in mitophagy and starvation autophagy, it is critical for Salmonella clearance during xenophagy (von Muhlinen et al., 2012). This suggests more specialized roles for LC3s in alternate autophagy pathways and indicates a broader role for the LC3 subfamily in host defense pathways. A key question arises: why are LC3s mechanistically different from GABARAPs? Structurally, LC3s and GABARAPs share a ubiquitin-like core but differ substantially at their N-terminal α helices (Rogov et al., 2014). The N-terminal helices of Atg8s have been shown to mediate membrane fusion, with GABARAPs having higher in vitro activity than LC3s (Weidberg et al., 2011; Wu et al., 2015; Landajuela et al., 2016). The N termini may also influence how Atg8s interact with other proteins, potentially explaining why the LC3s and GABARAPs do not have the same autophagic activity. Analysis of the Atg8 interactome led to the identification of several factors that interact specifically with one subfamily, but not the other (Behrends et al., 2010). PLEKHM1 was recently identified to drive autophagosome–lysosome fusion through recruitment of the HOPS complex during both selective and nonselective autophagy (McEwan et al., 2015a,b). Both LC3s and GABARAPs were shown to interact with PLEKHM1 using coimmunoprecipitations and in vitro binding assays. However, we have found that in the cellular environment, PLEKHM1 is preferentially recruited by GABARAPs during PINK1/Parkin mitophagy to drive autophagosome–lysosome fusion. PI4K2α is another factor that has been shown to promote autophagosome–lysosome fusion via its interaction with GABARAPs (Wang et al., 2015). However, the mechanism of PI4K2α function during autophagy is unclear, because overexpression of PI4K2α can compensate for the absence of GABARAPs.
LIR-mediated interactions with Atg8 proteins have been shown in many studies to be crucial for selective autophagy by incorporating specific cargo into autophagosomes (Birgisdottir et al., 2013; Stolz et al., 2014). Here, we show that Atg8s are dispensable for mitochondrial sequestration during PINK1/Parkin mitophagy. However, PINK1/Parkin mitophagy may be functionally distinct from other selective autophagy pathways because autophagosome formation is initiated on the surface of mitochondria rather than via the recruitment of preformed isolation membranes (Itakura et al., 2012a; Lazarou et al., 2015). The selectivity is achieved by S65 phosphorylated ubiquitin chains placed on the surface of mitochondria by PINK1 and Parkin (Kane et al., 2014; Kazlauskaite et al., 2014; Koyano et al., 2014). The S65 phosphorylated ubiquitin chains are recognized by the autophagy receptors optineurin and NDP52 that initiate mitophagy through the activity of ULK1 (Lazarou et al., 2015; Richter et al., 2016). Other types of selective autophagy may still require the recruitment of preformed autophagosome precursors via Atg8-LIR–mediated interactions. For example, Nix-mediated mitophagy does not involve ubiquitin-binding autophagy receptors such as optineurin and NDP52 and critically depends on LIR-mediated interactions with Atg8s (Sandoval et al., 2008; Novak et al., 2010). Thus, two types of selective autophagy may exist, where the mechanism of signaling and initiation may dictate whether preformed autophagosomes are recruited or autophagosomes are initiated on the surface of the cargo. During PINK1/Parkin mitophagy, LIR-mediated interactions between autophagy receptors such as optineurin and NDP52 can enable recruitment of Atg8s to autophagosomes to promote autophagosome growth and formation efficiency. Subsequent LIR-mediated recruitment of PLEKHM1 by Atg8s can complete the autophagy pathway by promoting autophagosome–lysosome fusion (McEwan et al., 2015a,b).
By clarifying the function of Atg8s during PINK1/Parkin mitophagy and starvation autophagy, we have revealed functional distinctions within the Atg8 family and identified their crucial role in autophagosome–lysosome fusion. Regulation of autophagosome capacity by Atg8s is predicted to play a prominent role during selective autophagy in which large cargo must be encapsulated. Given that Atg8s are not essential for autophagosome closure in mammalian cells, our work highlights the need for further studies interrogating the mechanisms of autophagosome biogenesis. Indeed, during revision of this manuscript, an interesting study was published revealing that autophagosome-like structures can be generated in the absence of Atg conjugation systems (Tsuboyama et al., 2016), corroborating our observations. However, the Stx17-positive autophagosomal structures could fuse with lysosomes, albeit at a slightly slower rate, and a significant delay in degradation of the inner autophagosomal membrane was also observed. Thus, it is possible that conjugation of Atg8s to lipid may not be essential for their role in autophagosome–lysosome fusion but is likely to be important for regulating autophagosome size. We anticipate that additional factors required for autophagosome formation remain to be identified and/or characterized.
Materials and methods
Cell culture, antibodies, and reagents
HeLa cells were cultured in DMEM supplemented with 10% (vol/vol) FBS (Gemini Bio Products), 25 mM Hepes, and GlutaMAX (Thermo Fisher Scientific). The transfection reagents used include Lipofectamine LTX (Thermo Fisher Scientific) and X-tremeGENE 9 (Roche). Cells were stained using Hoechst33342 (Thermo Fisher Scientific), MitoTracker deep red FM (Thermo Fisher Scientific), LysoTracker green DND-26 (Thermo Fisher Scientific), and Cascade blue–conjugated dextran (10,000 mol wt, anionic, lysine fixable; Thermo Fisher Scientific). The following rabbit monoclonal and polyclonal antibodies were used: HA (Cell Signaling Technology), GABARAPL1 (Abcam), GABARAPL2 (Abcam), PLEKHM1 (Abcam), LC3A (Cell Signaling Technology), LC3B (Cell Signaling Technology), LC3C (Cell Signaling Technology), LAMP1 (Cell Signaling Technology), GABARAP (Cell Signaling Technology), NDP52 (Cell Signaling Technology), OPTN (Proteintech), and Tom20 (Santa Cruz Biotechnology, Inc.). MFN1 and B17.2L antibodies were generated previously (Lazarou et al., 2007, 2015). The polyclonal MFN1 and B17.2L antibodies were generated in rabbits using recombinant GST-Mfn1 (aa 667–741) and full-length B17.2L immunogens, respectively. The following mouse monoclonal antibodies were used: CoxII (Abcam), Hsp60 (Abcam), FLAG (Sigma-Aldrich), LAMP2 (Abcam), DNA (Progen), Actin (Abcam), p62 (Abnova), Parkin (Santa Cruz Biotechnology, Inc.), and Tom20 (Santa Cruz Biotechnology, Inc.). Chicken anti-GFP (Thermo Fisher Scientific) was also used. See Table S2 for catalog numbers.
Generation of KO lines using CRISPR/Cas9 gene editing
All of the KO cell lines listed in Table S3 were generated using CRISPR guide RNAs (gRNAs) that target a common exon of all splicing variants of a gene of interest. CRISPR constructs were created by ligating annealed oligonucleotides (Sigma-Aldrich) that contain CRISPR sequences into BbsI-linearized pSpCas9(BB)-2A-GFP vector (Ran et al., 2013; a gift from F. Zhang, Broad Institute, Cambridge, MA; plasmid 48138; Addgene). gRNA constructs were transfected into HeLa cells for 24 h, and GFP-positive cells were individually sorted by FACS into 96-well plates. Single-cell colonies were expanded before being screened for the loss of the targeted gene product by immunoblotting. To confirm the presence of frameshift indels in the genes of interest in KO clones identified by immunoblotting, genomic DNA was isolated and PCR was performed to amplify the targeted regions (See Table S1 for amplifying primers) that were subsequently cloned into a pGEM4Z vector for sequencing analysis. Where antibodies were not available, putative KO clones were identified using three-primer PCR (Yu et al., 2014) and then sequenced to determine the presence of frameshift indels. Multiple KO lines were generated by sequential transfections of one or multiple gRNA constructs. For generation of LC3 TKO, LC3A and LC3B CRISPRs were first introduced into WT HeLa cells to create LC3A/B DKO that was transfected with the LC3C CRISPR. Similarly, GBRP TKO was generated by transfecting GBRP and GBRPL1 CRISPRs followed by the addition of a GBRPL2 CRISPR into the GBRP/GBRPL1 DKO. The GBRP/GBRPL1 DKO line was also chosen to make the hexa KO line. LC3A and LC3B CRISPRs were used to produce LC3A/LC3B/GBRP/GBRPL1 tetra KO that was subsequently transfected with LC3C and GBRPL2 CRISPRs to ultimately generate the cell line with all Atg8 genes disrupted.
Cloning and generation of stable cell lines
pMRXIP GFP-STX17 (Itakura et al., 2012b; plasmid 45909; Addgene) and pCold-TF-hPLEKHM1 (Tabata et al., 2010; plasmid 64146; Addgene) were gifts from N. Mizushima (The University of Tokyo, Tokyo, Japan) and T. Yoshimori (Osaka University, Osaka, Japan), respectively. The following plasmids were generated by ligating open reading frames amplified by PCR into linearized pMX-IRES-YFP (a gift from G. Dewson, The Walter and Eliza Hall Institute of Medical Research, Melbourne, Victoria, Australia), pBMNZ, and pLVX-Tetone-puro (Takara Bio Inc.) using the Gibson Cloning kit (New England Biolabs, Inc.) according to the manufacturer’s instructions: pMX; -LC3A, -LC3B, -LC3C, -GABARAPL1, -HA-GABARAPL1, -GABARAPL2, -HA-GABARAPL2, pBMN; -GABARAP, -PLEKHM1-HA, and pLVX-Tetone-puro; -HA-LC3C, -HA-GABARAPL1. All constructs were sequence verified. The GFP-tagged plasmids of pBMN-mEGFP, -OPTN, -NDP52, -LC3s and -GBRPs, -DFCP1, -WIPI1, -ULK1, and pBMN-mCherry-Parkin and pBMN-YFP-Parkin were described previously (Lazarou et al., 2015). Stably transfected cell lines were generated using retroviral (for pBMN and pMX-IRES-YFP constructs) and lentiviral (for pLVX-Tetone-puro) systems as described previously (Lazarou et al., 2015), and protein expression levels were equalized among cell lines by FACS.
Translocation and mitophagy and starvation treatments
For translocation and mitophagy experiments, cells were either left untreated or treated with 10 µM oligomycin (EMD Millipore), 4 µM antimycin A in fresh growth medium for different periods of time as indicated in the figure legends. Long-treatment time-point samples were additionally treated with 10 µM QVD (ApexBio). For starvation experiments, cells were incubated in fresh growth medium for 1 h before 8-h starvation using EBSS (Thermo Fisher Scientific).
Immunoblotting
HeLa cells were cultured in six-well plates for 24 h before incubation with fresh culture medium, EBSS starvation medium, or fresh medium containing 10 µM oligomycin (EMD Millipore), 4 µM antimycin A (Sigma-Aldrich), and 10 μΜ QVD (Assay Matrix) for the times indicated. Cells were lysed in 1× LDS sample buffer (Thermo Fisher Scientific) supplemented with 100 mM DTT (Sigma-Aldrich) and heated to 99°C with shaking for 7–10 min. Approximately 25–50 µg of protein per sample was separated on 4–12% Bis-Tris gels (Thermo Fisher Scientific) according to the manufacturer’s instructions, electrotransferred to polyvinyldifluoride membranes, and then immunoblotted using antibodies as indicated.
PK protection assay
HeLa cells were seeded into six-well plates and left untreated or treated with 10 µM oligomycin, 4 µM antimycin A, 10 µM QVD, and 100 nM bafilomycin A1 (Sigma-Aldrich and Bioaustralis) in the absence or presence of 1 µM wortmannin (Bioaustralis) in fresh growing medium for 6 h (Fig. 4, B and E) or various time points (Fig. 6). For starvation, cells were treated with 100 nM bafilomycin A1 in the absence or presence of 1 µM wortmannin in EBSS for 8 h. After treatment, cells were homogenized in 20 mM Hepes, pH 7.6, 220 mM mannitol, 70 mM sucrose, and 1 mM EDTA. After a centrifugation at 500 g at 4°C for 5 min, the postnuclear supernatant was collected and equally divided into three Eppendorf tubes. One of the samples was left untreated, whereas the other two were incubated with 25 µg/ml PK only or both PK and Triton X-100 (TX-100; 0.2% [vol/vol]) for 10 min on ice (see Fig. 4 A). PK wasn then inhibited by the addition of 1 mM PMSF and incubation for 10 min on ice. All samples were then subjected to TCA precipitation, and protein pellets were resuspended in the same volume of 1 x LDS sample buffer and prepared as described in the Immunoblotting section. Approximately 40–60 µg of each sample was analyzed by immunoblotting.
Specimen preparation for conventional TEM
Cell monolayers were fixed overnight at 4°C with 0.1 M sodium cacodylate–buffered 2% (wt/vol) glutaraldehyde. Fixed samples were rinsed with sodium cacodylate and postfixed with ferricyanide-reduced osmium tetroxide (1% [wt/vol] OsO4, 1.5% [wt/vol] K3[Fe(CN)6], and 0.065 M cacodylate buffer) for 2 h at 4°C. The postfixed samples were scraped and pelleted in agarose, rinsed with distilled water, and then stored overnight in 70% (wt/vol) ethanol. Dehydration was performed by graduated ethanol series (80%, 90%, 95%, 100%, and 100% [wt/vol]; 10 min each) and propylene oxide (100% and 100% [wt/vol]; 5 min each). Samples were infiltrated with Araldite 502/Embed 812 by graduated concentration series in propylene oxide (25% [vol/vol] for 1 h, 33% [vol/vol] for 1 h, 50% overnight, 66% [vol/vol] for 4 h, 75% [vol/vol] for 4 h, 100% [vol/vol] overnight, and 100% [vol/vol] for 5 h) and then polymerized at 60°C for 48 h. Embedded samples were sectioned using an Ultracut UCT ultramicrotome (Leica Biosystems) equipped with a 45° diamond knife (Diatome) to cut 75-nm ultrathin sections. The grids were stained at room temperature using 2% (wt/vol) aqueous uranyl acetate (5 min) and Reynolds lead citrate (3 min) before routine imaging. All TEM imaging was performed at 80 kV on a Hitachi H-7500 TEM using a Gatan 791 MultiScan side-mount CCD camera and DigitalMicrograph (Version 1.71.38) acquisition software.
Specimen preparation for immunogold-labeled TEM
Cell monolayers were fixed overnight at 4°C with 0.1 M phosphate-buffered 4% (wt/vol) paraformaldehyde. The fixed samples were scraped and pelleted in 12% (wt/vol) gelatin in 0.1 M phosphate buffer at 37°C, which was allowed to set at 4°C before being cut into small cubes measuring ∼0.5 mm on each edge. The gelatin embedded cells were infiltrated with 2.3 M sucrose in 0.1 M phosphate buffer at 4°C overnight on a rocker. The sucrose infiltrated gelatin blocks were mounted on cryopins and then frozen in liquid nitrogen for cryoultramicrotomy. Frozen samples were trimmed at −100°C and sectioned at −120°C using a Cryoelectron microscopy UC7 ultramicrotome (Leica Biosystems) equipped with a 45° diamond cryotrimming knife (Diatome) and a 35° diamond cryoimmuno knife (Diatome). Cryosections were retrieved by pick-up loop with a droplet of phosphate-buffered 1% (wt/vol) methyl cellulose and 1.15 M sucrose and then deposited on carbon-coated formvar grids for immunolabeling. The grids were prepared for immunolabeling by melting upside down on a layer of 2% (wt/vol) gelatin in PBS at 37°C and then rinsing the grids on droplets of 0.02 M glycine in PBS (5 min). The grids were blocked with 1% (wt/vol) BSA in PBS (5 min) before immunolabeling. For single labeling of HSP60, samples were incubated with the primary antibody diluted 1:300 in 1% (wt/vol) BSA in PBS (room temperature; 1 h), rinsed with 0.1% (wt/vol) BSA in PBS (5 × 2 min), incubated with a bridging antibody (1:1,000 rabbit anti–mouse; Rockland Immunochemicals) diluted in 1% (wt/vol) BSA in PBS (room temperature; 30 min), and then rinsed (5 × 2 min) with 0.1% (wt/vol) BSA in PBS. Single-labeled samples were probed with protein A–conjugated 15-nm gold particles (Department of Cell Biology, University Medical Center (UMC) Utrecht) diluted 1:50 in 1% (wt/vol) BSA in PBS (room temperature; 30 min). For double labeling of LAMP1 and HSP60, samples were incubated with 1:200 LAMP1 antibody in 1% (wt/vol) BSA in PBS (room temperature; 30 min), rinsed (5 × 2 min) with 0.1% (wt/vol) BSA in PBS, and then probed with protein A–conjugated 20-nm gold particles (Department of Cell Biology, UMC Utrecht) diluted 1:60 in 1% (wt/vol) BSA in PBS (room temperature; 30 min). The samples were then rinsed with 0.1% (wt/vol) BSA in PBS (5 × 2 min) and PBS (5 × 2 min) before inactivating the LAMP1 antibody with 1% (wt/vol) glutaraldehyde in PBS (room temperature; 5 min). The inactivated samples were then immunolabeled for HSP60 as described for single labeling, before probing with protein A–conjugated 10-nm gold particles (Department of Cell Biology, UMC Utrecht) diluted 1:60 in 1% (wt/vol) BSA in PBS (room temperature; 30 min). After immunolabeling, the samples were rinsed with PBS (5 × 2 min) and postfixed with 1% (wt/vol) glutaraldehyde in PBS (room temperature; 5 min). Finally, the samples were rinsed with distilled water (5 × 2 min), stained with 2% (wt/vol) uranyloxaalacetate (pH 7; room temperature; 5 min) and then 0.4% (wt/vol) uranyl acetate in 1.8% (wt/vol) aqueous methyl cellulose (pH 4; 4°C; 10 min), and dried in a thin film of the final stain in a wire pick-out loop.
Quantification of autophagosome size by electron microscopy
Stained TEM grids were renamed and reorganized to mask their identities from the microscopist and image analyst. One ultrathin section per sample was surveyed, and 12 cells were randomly selected at a low magnification (2,000×) for further analysis. Cells were excluded from selection if they were directly adjacent to a previously selected cell, partially obstructed by a grid bar, or lacking a visible nucleus. Valid cells were imaged at high magnification (25,000×) to catalog all visible structures within the cell. Autophagosomal compartments were identified and manually outlined in the high-magnification images using the freehand selection tool in ImageJ (version 1.50g). The cross-sectional area of each autophagosome was measured using a batch processing macro written for ImageJ. Data were acquired from three independent experiments. For statistical analysis of immunogold-labeled samples, eight cells were randomly selected using the cell selection procedure described for resin-embedded samples, without masking of sample identities.
Serial sectioning and 3D autophagosome reconstruction
Embedded samples were sectioned using an Ultracut UCT ultramicrotome equipped with a 35° diamond knife (Diatome) to acquire 32 serial sections with a mean thickness of 90 nm, which were stained as described in the Specimen preparation for conventional TEM section. TEM images were acquired of sections from the middle of the series to identify autophagosomal structures before relocation and imaging of the same structures in adjacent sections (between the fifth and 22nd sections). The TEM images were corrected for lens distortion using Distortion Correction plugin for FIJI (version 1.50g) and manually aligned into an image stack using GIMP (GNU Image Manipulation Program, version 2.8.16). The outermost membranes in images of each autophagosomal structure were manually segmented to using the TrakEM2 plugin for FIJI. The segmented data were interpolated (cubic interpolation; 8× interslice) and rendered as surfaces from two orthometric viewpoints using the Amira software package (FEI Technologies).
Live-cell confocal microscopy and immunofluorescence
Live-cell imaging samples were prepared by culturing cells in poly-d-lysine–coated 35-mm FluoroDishes (FD35; World Precision Instruments) for 48 h before replacing the culture medium with a phenol-free equivalent (DMEM, 4.5 g/L d-glucose, 10% FCS, 5 mM Hepes, pH 7.4, and 2 mM GlutaMAX; Thermo Fisher Scientific) containing 50 nM MitoTracker deep red FM (Thermo Fisher Scientific) and 100 nM LysoTracker green DND-26 (Thermo Fisher Scientific). Live-cell imaging of cells expressing GFP-Stx17 was performed 24 h after transient transfection with pcDNA3.1-LAMP1-mCherry (Van Engelenburg and Palmer, 2010; a gift from A. Palmer, University of Colorado, Boulder, CO; plasmid 45147; Addgene), at 37°C by time-series optical sectioning (15 s interval for 60 min; 8 sections with 0.738 µm axial sampling; 3 A.U. pinhole) using an inverted SP8 confocal laser scanning microscope equipped with a 40×/1.10 NA objective (Water immersion, HC PLAPO, CS2; Leica Biosystems). The live-cell images were acquired using an HyD Hybrid Detector (Leica Biosystems) through the Leica Application Suite X (LASX v2.0.1), and deconvolved (fast classic maximum likelihood estimation; 5 signal to noise ratio; 20 iterations; 0.05 quality threshold) using Huygens Professional (v15.10; Scientific Volume Imaging). For immunofluorescence, cells were cultured on poly-d-lysine–coated glass coverslips for 48 h before experimental treatment. For endosome analysis experiments, cells were cultured during the final 24 h in the presence of medium containing 0.5 mg/ml lysine-fixable 10,000-mol-wt dextran conjugated to Cascade blue (Thermo Fisher Scientific), as indicated in the figure legends. Samples were fixed with 4% (wt/vol) PFA in 0.1 M phosphate buffer (15 min), rinsed three times with PBS, permeabilized with 0.1% (vol/vol) Triton X-100 in PBS (10 min), and then blocked with 3% (vol/vol) goat serum in 0.1% (vol/vol) Triton X-100/PBS (10 min). The samples were incubated with primary antibodies (as indicated in the figure legends) diluted in 3% (vol/vol) goat serum in 0.1% (vol/vol) Triton X-100/PBS for 2 h at room temperature, rinsed three times with PBS, and then incubated at room temperature for 1 h with secondary antibodies conjugated to Alexa Fluor 488, 555, 633, or 647 (Thermo Fisher Scientific). The coverslips were rinsed once with 0.1% (vol/vol) Triton X-100/PBS and three times with PBS, stained with 1 µM Hoechst 33342 (Thermo Fisher Scientific) in PBS (5 min), and then mounted using a Tris-buffered DABCO-glycerol mounting medium. All samples were imaged in 3D by optical sectioning using an inverted SP8 confocal laser scanning microscope equipped with an 63×/1.40 NA objective (oil immersion, HC PLAPO, CS2; Leica Biosystems), with a minimum z-stack range of 1.8 µm and a maximum voxel size of 90 nm laterally (x, y) and 300 nm axially (z). All figure images were acquired at ambient room temperature using an HyD Hybrid Detector (Leica Biosystems) and the Application Suite X (LASX v2.0.1). All images are displayed as z-stack maximum projections.
Confocal image analysis
All 3D image data were processed and analyzed using automated image segmentation in Imaris (V8.0; Bitplane). For the analysis of mitophagy by mtDNA/Hoechst33342 immunostaining, images were segmented using 300-nm-diameter background subtraction, 90-nm surface smoothing, and a manual intensity threshold of 12 (arbitrary). Segmented DNA structures were defined as mtDNA unless their intensity in the Hoechst33342 channel exceeded a sum total of 685 (arbitrary). The mean mtDNA volume per cell after treatment was normalized to the untreated values to quantify the nondegraded percentage of mtDNA. DFCP1 structures were detected by 90-nm-diameter background subtraction, 90-nm surface smoothing, and a manual intensity threshold of 3 (arbitrary). ULK1 foci were detected by 320-nm-diameter background subtraction, 183-nm surface smoothing, and a manual intensity threshold of 12 (arbitrary). PLEKHM1 structures were detected by 320-nm-diameter background subtraction, 183-nm surface smoothing, a manual intensity threshold of 7 (arbitrary), and seeded region growing segmentation (1.25-µm-diameter seed). LAMP1-positive lysosomes were detected by 240-nm-diameter background subtraction, 183-nm surface smoothing, a manual intensity threshold of 2.5 (arbitrary), and seeded region growing segmentation (0.5-µm-diameter seed). Segmented structures <10 voxels were excluded to remove shot noise. The number of cells per image was quantified manually. Segmented objects were defined as mitochondrially associated if the standard deviation of their intensity in the mitochondrial channel exceeded 25 (arbitrary) for ULK1 foci and DFCP1 structures or 2.5 (arbitrary) for PLEKHM1 structures. LAMP1 structures were deemed to be HSP60 positive if their intensity in the mitochondrial channel fulfilled two criteria: a mean greater than 10 (arbitrary) and a minimum value less than 1 (arbitrary; to remove LAMP1 structures surrounded by mitochondria). The mitochondrial proportion was calculated for the total number of ULK1 foci and DFCP1 structures. LAMP1 and PLEKHM1 structures were too densely packed to resolve individual structures for counting. Instead, the mitochondrial proportion was calculated for the total volume of subdivided components, as detected by seeded region growing segmentation.
mtKeima autophagosome–lysosome fusion mitophagy assay
mtKeima (a gift from R. Youle, National Institutes of Health, Bethesda, MD) was cloned into a pCHAC-MCS1-IRES-MCS2 and transduced using retrovirus into WT, LC3 TKO, GBRP TKO, hexa KO, and hexa KOs. YFP-Parkin was also stably expressed in these cells using retrovirus transduction (pBMN-YFP-Parkin) as described in the Cloning and generation of stable cell lines section. WT, LC3 TKO, GBRP TKO, and hexa KO cells stably expressing mCherry-Parkin and mtKeima were treated either without or with 10 µM oligomycin and 4 µM antimycin A in fresh growth medium for different periods of time as indicated in figure legends. Doxycycline-inducible HA-LC3C or HA-GABARAPL1 pLVX-Tetone-puro lentiviral constructs were transduced into hexa KO/mCherry-Parkin/mtKeima using lentivirus. The cells were treated with 10 µM oligomycin, 4 µM antimycin A for 8 h and then supplemented with 1 µM wortmannin and either 5 ng/ml (HA-LC3C) or 1 µg/ml (HA-GABARAPL1) doxycycline to induce protein expression. After treatment, live cells were analyzed using confocal microscopy (as described in the Live-cell confocal microscopy and immunofluorescence section) using a 63×/1.40 NA objective (oil immersion, HC PLAPO, CS2; Leica Biosystems). For FACS analysis, trypsinized cells were resuspended in sorting buffer (10% vol/vol FBS and 0.5 mM EDTA in PBS) and analyzed using the FAVSDiva software on a LSR Fortessa X-20 cell sorter (BD). Measurements of lysosomal mtKeima were made using dual-excitation ratiometric pH measurements with 488-nm (pH 7) and 561-nm (pH 4) lasers with 695/40 nm and 670/30 nm detector filters, respectively. For each sample, 50,000 events were collected. A control mtKeima ratio gate was drawn around untreated samples from each cell line and was used to determine lysosomal mtKeima in treated samples; cell populations vertically shifted above the gate were classified as undergoing autophagosome–lysosome fusion and mitophagy. Data were analyzed using FlowJo (v10.0.8r1).
Statistical calculations
All statistical comparisons were conducted on data originating from three or more biologically independent experimental replicates (as indicated in the figure legends), with similar data variances observed between groups. Comparisons between groups were planned before statistical testing, and target effect sizes were not predetermined. No statistical methods were used to determine sample size. Experimental blinding and randomization were only performed where indicated. All statistical data were calculated and graphed using GraphPad Prism 6. Statistical comparisons between two groups were performed by t test. Comparisons between three or more groups were performed by one-way analysis of variance (ANOVA) with post hoc testing by unprotected Fisher's least significant difference test. P-values exceeding 0.05 were considered nonsignificant.
Online supplemental material
Fig. S1 shows biochemical analyses and verification of Atg8 KO and rescue cell lines. Fig. S2 shows supplementary confocal microscopy as referenced in the main figure legends. Fig. S3 shows all supplementary electron microscopy as referenced in the main figure legends. Fig. S4 shows raw measurements and statistical analyses of autophagosome and mitophagosome size. Fig. S5 shows supplementary confocal microscopy and FACS plots of mtKeima. Table S1 shows the genotyping primers used for sequencing analysis of the KO lines. Table S2 details the antibodies used in this study. Table S3 details the CRISPR sequences, indels, and genotyping results of all KO cell lines in this study.
Supplementary Material
Acknowledgments
We thank Richard Youle for sharing plasmids and reagents, the Monash Flow cytometry Platform (FlowCore) and the Monash Micro Imaging Platform, Adam Fogel for critical reading of the manuscript, and Mike Ryan for reagents and helpful discussions.
This work was supported by the National Health and Medical Research Council (grants GNT1063781 and GNT1106471).
The authors declare no competing financial interests.
Author contributions: T.N. Nguyen, B.S. Padman, and M. Lazarou conceived the projects; T.N. Nguyen, B.S. Padman, J. Usher, V. Oorschot, G. Ramm, and M. Lazarou designed and performed experiments; M. Lazarou wrote the manuscript; and all authors contributed to editing the manuscript.
Footnotes
Abbreviations used:
- ANOVA
- analysis of variance
- Atg
- autophagy related
- BafA1
- bafilomycin
- CoxII
- cytochrome c oxidase subunit II
- EBSS
- Earle’s balanced salt solution
- gRNA
- guide RNA
- HOPS
- homotypic fusion and protein sorting
- IRES
- internal ribosome entry site
- LIR
- LC3-interacting region
- KO
- knockout
- mtDNA
- mitochondrial DNA
- mtKeima
- mitochondrially targeted mKeima
- OA
- oligomycin/antimycin A
- PI3P
- phosphatidylinositol-3-phosphate
- PK
- Proteinase K
- STX
- syntaxin
- TEM
- transmission electron microscopy
- TKO
- triple knockout
- WT
- wild type
References
- Alemu E.A., Lamark T., Torgersen K.M., Birgisdottir A.B., Larsen K.B., Jain A., Olsvik H., Øvervatn A., Kirkin V., and Johansen T.. 2012. ATG8 family proteins act as scaffolds for assembly of the ULK complex: sequence requirements for LC3-interacting region (LIR) motifs. J. Biol. Chem. 287:39275–39290. 10.1074/jbc.M112.378109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Axe E.L., Walker S.A., Manifava M., Chandra P., Roderick H.L., Habermann A., Griffiths G., and Ktistakis N.T.. 2008. Autophagosome formation from membrane compartments enriched in phosphatidylinositol 3-phosphate and dynamically connected to the endoplasmic reticulum. J. Cell Biol. 182:685–701. 10.1083/jcb.200803137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behrends C., Sowa M.E., Gygi S.P., and Harper J.W.. 2010. Network organization of the human autophagy system. Nature. 466:68–76. 10.1038/nature09204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birgisdottir A.B., Lamark T., and Johansen T.. 2013. The LIR motif: crucial for selective autophagy. J. Cell Sci. 126:3237–3247. 10.1242/jcs.126128 [DOI] [PubMed] [Google Scholar]
- Diao J., Liu R., Rong Y., Zhao M., Zhang J., Lai Y., Zhou Q., Wilz L.M., Li J., Vivona S., et al. 2015. ATG14 promotes membrane tethering and fusion of autophagosomes to endolysosomes. Nature. 520:563–566. 10.1038/nature14147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita N., Hayashi-Nishino M., Fukumoto H., Omori H., Yamamoto A., Noda T., and Yoshimori T.. 2008. An Atg4B mutant hampers the lipidation of LC3 paralogues and causes defects in autophagosome closure. Mol. Biol. Cell. 19:4651–4659. 10.1091/mbc.E08-03-0312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutierrez M.G., Master S.S., Singh S.B., Taylor G.A., Colombo M.I., and Deretic V.. 2004. Autophagy is a defense mechanism inhibiting BCG and Mycobacterium tuberculosis survival in infected macrophages. Cell. 119:753–766. 10.1016/j.cell.2004.11.038 [DOI] [PubMed] [Google Scholar]
- Hamasaki M., Furuta N., Matsuda A., Nezu A., Yamamoto A., Fujita N., Oomori H., Noda T., Haraguchi T., Hiraoka Y., et al. 2013. Autophagosomes form at ER-mitochondria contact sites. Nature. 495:389–393. 10.1038/nature11910 [DOI] [PubMed] [Google Scholar]
- Heo J.M., Ordureau A., Paulo J.A., Rinehart J., and Harper J.W.. 2015. The PINK1-PARKIN mitochondrial ubiquitylation pathway drives a program of OPTN/NDP52 recruitment and TBK1 activation to promote mitophagy. Mol. Cell. 60:7–20. 10.1016/j.molcel.2015.08.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ichimura Y., Kirisako T., Takao T., Satomi Y., Shimonishi Y., Ishihara N., Mizushima N., Tanida I., Kominami E., Ohsumi M., et al. 2000. A ubiquitin-like system mediates protein lipidation. Nature. 408:488–492. 10.1038/35044114 [DOI] [PubMed] [Google Scholar]
- Itakura E., Kishi-Itakura C., Koyama-Honda I., and Mizushima N.. 2012a Structures containing Atg9A and the ULK1 complex independently target depolarized mitochondria at initial stages of Parkin-mediated mitophagy. J. Cell Sci. 125:1488–1499. 10.1242/jcs.094110 [DOI] [PubMed] [Google Scholar]
- Itakura E., Kishi-Itakura C., and Mizushima N.. 2012b The hairpin-type tail-anchored SNARE syntaxin 17 targets to autophagosomes for fusion with endosomes/lysosomes. Cell. 151:1256–1269. 10.1016/j.cell.2012.11.001 [DOI] [PubMed] [Google Scholar]
- Joachim J., and Tooze S.A.. 2016. GABARAP activates ULK1 and traffics from the centrosome dependent on Golgi partners WAC and GOLGA2/GM130. Autophagy. 12:892–893. 10.1080/15548627.2016.1159368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kane L.A., Lazarou M., Fogel A.I., Li Y., Yamano K., Sarraf S.A., Banerjee S., and Youle R.J.. 2014. PINK1 phosphorylates ubiquitin to activate Parkin E3 ubiquitin ligase activity. J. Cell Biol. 205:143–153. 10.1083/jcb.201402104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katayama H., Kogure T., Mizushima N., Yoshimori T., and Miyawaki A.. 2011. A sensitive and quantitative technique for detecting autophagic events based on lysosomal delivery. Chem. Biol. 18:1042–1052. 10.1016/j.chembiol.2011.05.013 [DOI] [PubMed] [Google Scholar]
- Kazlauskaite A., Kondapalli C., Gourlay R., Campbell D.G., Ritorto M.S., Hofmann K., Alessi D.R., Knebel A., Trost M., and Muqit M.M.. 2014. Parkin is activated by PINK1-dependent phosphorylation of ubiquitin at Ser65. Biochem. J. 460:127–139. 10.1042/BJ20140334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirisako T., Baba M., Ishihara N., Miyazawa K., Ohsumi M., Yoshimori T., Noda T., and Ohsumi Y.. 1999. Formation process of autophagosome is traced with Apg8/Aut7p in yeast. J. Cell Biol. 147:435–446. 10.1083/jcb.147.2.435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirkin V., Lamark T., Sou Y.S., Bjørkøy G., Nunn J.L., Bruun J.A., Shvets E., McEwan D.G., Clausen T.H., Wild P., et al. 2009. A role for NBR1 in autophagosomal degradation of ubiquitinated substrates. Mol. Cell. 33:505–516. 10.1016/j.molcel.2009.01.020 [DOI] [PubMed] [Google Scholar]
- Kitada T., Asakawa S., Hattori N., Matsumine H., Yamamura Y., Minoshima S., Yokochi M., Mizuno Y., and Shimizu N.. 1998. Mutations in the parkin gene cause autosomal recessive juvenile parkinsonism. Nature. 392:605–608. 10.1038/33416 [DOI] [PubMed] [Google Scholar]
- Klionsky D.J., Abdelmohsen K., Abe A., Abedin M.J., Abeliovich H., Acevedo Arozena A., Adachi H., Adams C.M., Adams P.D., Adeli K., et al. 2016. Guidelines for the use and interpretation of assays for monitoring autophagy (3rd edition). Autophagy 12:1–222. 10.1080/15548627.2015.1100356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knorr R.L., Lipowsky R., and Dimova R.. 2015. Autophagosome closure requires membrane scission. Autophagy. 11:2134–2137. 10.1080/15548627.2015.1091552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komatsu M., Waguri S., Ueno T., Iwata J., Murata S., Tanida I., Ezaki J., Mizushima N., Ohsumi Y., Uchiyama Y., et al. 2005. Impairment of starvation-induced and constitutive autophagy in Atg7-deficient mice. J. Cell Biol. 169:425–434. 10.1083/jcb.200412022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koyano F., Okatsu K., Kosako H., Tamura Y., Go E., Kimura M., Kimura Y., Tsuchiya H., Yoshihara H., Hirokawa T., et al. 2014. Ubiquitin is phosphorylated by PINK1 to activate parkin. Nature. 510:162–166. 10.1038/nature13392 [DOI] [PubMed] [Google Scholar]
- Kraft C., Kijanska M., Kalie E., Siergiejuk E., Lee S.S., Semplicio G., Stoffel I., Brezovich A., Verma M., Hansmann I., et al. 2012. Binding of the Atg1/ULK1 kinase to the ubiquitin-like protein Atg8 regulates autophagy. EMBO J. 31:3691–3703. 10.1038/emboj.2012.225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamb C.A., Yoshimori T., and Tooze S.A.. 2013. The autophagosome: origins unknown, biogenesis complex. Nat. Rev. Mol. Cell Biol. 14:759–774. 10.1038/nrm3696 [DOI] [PubMed] [Google Scholar]
- Landajuela A., Hervás J.H., Antón Z., Montes L.R., Gil D., Valle M., Rodriguez J.F., Goñi F.M., and Alonso A.. 2016. Lipid geometry and bilayer curvature modulate LC3/GABARAP-mediated model autophagosomal elongation. Biophys. J. 110:411–422. 10.1016/j.bpj.2015.11.3524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazarou M., McKenzie M., Ohtake A., Thorburn D.R., and Ryan M.T.. 2007. Analysis of the assembly profiles for mitochondrial- and nuclear-DNA-encoded subunits into complex I. Mol. Cell. Biol. 27:4228–4237. 10.1128/MCB.00074-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazarou M., Sliter D.A., Kane L.A., Sarraf S.A., Wang C., Burman J.L., Sideris D.P., Fogel A.I., and Youle R.J.. 2015. The ubiquitin kinase PINK1 recruits autophagy receptors to induce mitophagy. Nature. 524:309–314. 10.1038/nature14893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manil-Ségalen M., Lefebvre C., Jenzer C., Trichet M., Boulogne C., Satiat-Jeunemaitre B., and Legouis R.. 2014. The C. elegans LC3 acts downstream of GABARAP to degrade autophagosomes by interacting with the HOPS subunit VPS39. Dev. Cell. 28:43–55. (published erratum appears in Dev. Cell 2014. 30:110) 10.1016/j.devcel.2013.11.022 [DOI] [PubMed] [Google Scholar]
- Matsuda N., Sato S., Shiba K., Okatsu K., Saisho K., Gautier C.A., Sou Y.S., Saiki S., Kawajiri S., Sato F., et al. 2010. PINK1 stabilized by mitochondrial depolarization recruits Parkin to damaged mitochondria and activates latent Parkin for mitophagy. J. Cell Biol. 189:211–221. 10.1083/jcb.200910140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McEwan D.G., Popovic D., Gubas A., Terawaki S., Suzuki H., Stadel D., Coxon F.P., Miranda de Stegmann D., Bhogaraju S., Maddi K., et al. 2015a PLEKHM1 regulates autophagosome-lysosome fusion through HOPS complex and LC3/GABARAP proteins. Mol. Cell. 57:39–54. 10.1016/j.molcel.2014.11.006 [DOI] [PubMed] [Google Scholar]
- McEwan D.G., Richter B., Claudi B., Wigge C., Wild P., Farhan H., McGourty K., Coxon F.P., Franz-Wachtel M., Perdu B., et al. 2015b PLEKHM1 regulates Salmonella-containing vacuole biogenesis and infection. Cell Host Microbe. 17:58–71. 10.1016/j.chom.2014.11.011 [DOI] [PubMed] [Google Scholar]
- McLelland G.L., Lee S.A., McBride H.M., and Fon E.A.. 2016. Syntaxin-17 delivers PINK1/parkin-dependent mitochondrial vesicles to the endolysosomal system. J. Cell Biol. 214:275–291. 10.1083/jcb.201603105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizushima N., Noda T., Yoshimori T., Tanaka Y., Ishii T., George M.D., Klionsky D.J., Ohsumi M., and Ohsumi Y.. 1998. A protein conjugation system essential for autophagy. Nature. 395:395–398. 10.1038/26506 [DOI] [PubMed] [Google Scholar]
- Nakagawa I., Amano A., Mizushima N., Yamamoto A., Yamaguchi H., Kamimoto T., Nara A., Funao J., Nakata M., Tsuda K., et al. 2004. Autophagy defends cells against invading group A Streptococcus. Science. 306:1037–1040. 10.1126/science.1103966 [DOI] [PubMed] [Google Scholar]
- Nakatogawa H., Ichimura Y., and Ohsumi Y.. 2007. Atg8, a ubiquitin-like protein required for autophagosome formation, mediates membrane tethering and hemifusion. Cell. 130:165–178. 10.1016/j.cell.2007.05.021 [DOI] [PubMed] [Google Scholar]
- Narendra D., Tanaka A., Suen D.F., and Youle R.J.. 2008. Parkin is recruited selectively to impaired mitochondria and promotes their autophagy. J. Cell Biol. 183:795–803. 10.1083/jcb.200809125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novak I., Kirkin V., McEwan D.G., Zhang J., Wild P., Rozenknop A., Rogov V., Löhr F., Popovic D., Occhipinti A., et al. 2010. Nix is a selective autophagy receptor for mitochondrial clearance. EMBO Rep. 11:45–51. 10.1038/embor.2009.256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogawa M., Yoshimori T., Suzuki T., Sagara H., Mizushima N., and Sasakawa C.. 2005. Escape of intracellular Shigella from autophagy. Science. 307:727–731. 10.1126/science.1106036 [DOI] [PubMed] [Google Scholar]
- Pankiv S., Clausen T.H., Lamark T., Brech A., Bruun J.A., Outzen H., Øvervatn A., Bjørkøy G., and Johansen T.. 2007. p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. J. Biol. Chem. 282:24131–24145. 10.1074/jbc.M702824200 [DOI] [PubMed] [Google Scholar]
- Ran F.A., Hsu P.D., Wright J., Agarwala V., Scott D.A., and Zhang F.. 2013. Genome engineering using the CRISPR-Cas9 system. Nat. Protoc. 8:2281–2308. 10.1038/nprot.2013.143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter B., Sliter D.A., Herhaus L., Stolz A., Wang C., Beli P., Zaffagnini G., Wild P., Martens S., Wagner S.A., et al. 2016. Phosphorylation of OPTN by TBK1 enhances its binding to Ub chains and promotes selective autophagy of damaged mitochondria. Proc. Natl. Acad. Sci. USA. 113:4039–4044. 10.1073/pnas.1523926113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogov V., Dötsch V., Johansen T., and Kirkin V.. 2014. Interactions between autophagy receptors and ubiquitin-like proteins form the molecular basis for selective autophagy. Mol. Cell. 53:167–178. 10.1016/j.molcel.2013.12.014 [DOI] [PubMed] [Google Scholar]
- Sandoval H., Thiagarajan P., Dasgupta S.K., Schumacher A., Prchal J.T., Chen M., and Wang J.. 2008. Essential role for Nix in autophagic maturation of erythroid cells. Nature. 454:232–235. 10.1038/nature07006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawa-Makarska J., Abert C., Romanov J., Zens B., Ibiricu I., and Martens S.. 2014. Cargo binding to Atg19 unmasks additional Atg8 binding sites to mediate membrane-cargo apposition during selective autophagy. Nat. Cell Biol. 16:425–433. 10.1038/ncb2935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sou Y.S., Waguri S., Iwata J., Ueno T., Fujimura T., Hara T., Sawada N., Yamada A., Mizushima N., Uchiyama Y., et al. 2008. The Atg8 conjugation system is indispensable for proper development of autophagic isolation membranes in mice. Mol. Biol. Cell. 19:4762–4775. 10.1091/mbc.E08-03-0309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stolz A., Ernst A., and Dikic I.. 2014. Cargo recognition and trafficking in selective autophagy. Nat. Cell Biol. 16:495–501. 10.1038/ncb2979 [DOI] [PubMed] [Google Scholar]
- Tabata K., Matsunaga K., Sakane A., Sasaki T., Noda T., and Yoshimori T.. 2010. Rubicon and PLEKHM1 negatively regulate the endocytic/autophagic pathway via a novel Rab7-binding domain. Mol. Biol. Cell. 21:4162–4172. 10.1091/mbc.E10-06-0495 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuboyama K., Koyama-Honda I., Sakamaki Y., Koike M., Morishita H., and Mizushima N.. 2016. The ATG conjugation systems are important for degradation of the inner autophagosomal membrane. Science. 10.1126/science.aaf6136 [DOI] [PubMed] [Google Scholar]
- Valente E.M., Abou-Sleiman P.M., Caputo V., Muqit M.M., Harvey K., Gispert S., Ali Z., Del Turco D., Bentivoglio A.R., Healy D.G., et al. 2004. Hereditary early-onset Parkinson’s disease caused by mutations in PINK1. Science. 304:1158–1160. 10.1126/science.1096284 [DOI] [PubMed] [Google Scholar]
- Van Engelenburg S.B., and Palmer A.E.. 2010. Imaging type-III secretion reveals dynamics and spatial segregation of Salmonella effectors. Nat. Methods. 7:325–330. 10.1038/nmeth.1437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velikkakath A.K., Nishimura T., Oita E., Ishihara N., and Mizushima N.. 2012. Mammalian Atg2 proteins are essential for autophagosome formation and important for regulation of size and distribution of lipid droplets. Mol. Biol. Cell. 23:896–909. 10.1091/mbc.E11-09-0785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Muhlinen N., Akutsu M., Ravenhill B.J., Foeglein Á., Bloor S., Rutherford T.J., Freund S.M., Komander D., and Randow F.. 2012. LC3C, bound selectively by a noncanonical LIR motif in NDP52, is required for antibacterial autophagy. Mol. Cell. 48:329–342. 10.1016/j.molcel.2012.08.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H., Sun H.Q., Zhu X., Zhang L., Albanesi J., Levine B., and Yin H.. 2015. GABARAPs regulate PI4P-dependent autophagosome:lysosome fusion. Proc. Natl. Acad. Sci. USA. 112:7015–7020. 10.1073/pnas.1507263112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weidberg H., Shvets E., Shpilka T., Shimron F., Shinder V., and Elazar Z.. 2010. LC3 and GATE-16/GABARAP subfamilies are both essential yet act differently in autophagosome biogenesis. EMBO J. 29:1792–1802. 10.1038/emboj.2010.74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weidberg H., Shpilka T., Shvets E., Abada A., Shimron F., and Elazar Z.. 2011. LC3 and GATE-16 N termini mediate membrane fusion processes required for autophagosome biogenesis. Dev. Cell. 20:444–454. 10.1016/j.devcel.2011.02.006 [DOI] [PubMed] [Google Scholar]
- Wong Y.C., and Holzbaur E.L.. 2014. Optineurin is an autophagy receptor for damaged mitochondria in parkin-mediated mitophagy that is disrupted by an ALS-linked mutation. Proc. Natl. Acad. Sci. USA. 111:E4439–E4448. 10.1073/pnas.1405752111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu F., Watanabe Y., Guo X.Y., Qi X., Wang P., Zhao H.Y., Wang Z., Fujioka Y., Zhang H., Ren J.Q., et al. 2015. Structural basis of the differential function of the two C. elegans Atg8 homologs, LGG-1 and LGG-2, in autophagy. Mol. Cell. 60:914–929. 10.1016/j.molcel.2015.11.019 [DOI] [PubMed] [Google Scholar]
- Xin Y., Yu L., Chen Z., Zheng L., Fu Q., Jiang J., Zhang P., Gong R., and Zhao S.. 2001. Cloning, expression patterns, and chromosome localization of three human and two mouse homologues of GABA(A) receptor-associated protein. Genomics. 74:408–413. 10.1006/geno.2001.6555 [DOI] [PubMed] [Google Scholar]
- Yamano K., Fogel A.I., Wang C., van der Bliek A.M., and Youle R.J.. 2014. Mitochondrial Rab GAPs govern autophagosome biogenesis during mitophagy. eLife. 3:e01612 10.7554/eLife.01612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu C., Zhang Y., Yao S., and Wei Y.. 2014. A PCR based protocol for detecting indel mutations induced by TALENs and CRISPR/Cas9 in zebrafish. PLoS One. 9:e98282 10.1371/journal.pone.0098282 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.