Martens previews work by Nguyen et al. analyzing the essential functions of ATG8 family proteins LC3/GABARAPs in autophagy.
Abstract
The ATG8 family LC3/GABARAP proteins are attached to the membrane of nascent autophagosomes, but their functions during autophagy are unclear. In this issue, Nguyen et al. (2016. J. Cell Biol. https://doi.org/10.1083/jcb.201607039) show that LC3/GABARAP proteins are not essential for autophagosome formation but are critical for autophagosome–lysosome fusion.
Macroautophagy (hereafter autophagy) is a major pathway for endo-lysosomal degradation of cellular cargo sequestered within double-membrane organelles called autophagosomes. Upon induction of autophagy, autophagosomes form de novo and initially appear as small membrane structures referred to as isolation membranes or phagophores. The isolation membranes expand, gradually enclosing a part of the cytoplasm, and eventually close to give rise to autophagosomes. Subsequently, the outer membrane of the autophagosome fuses with the lysosome membrane, and the autophagosome inner membrane and autophagosome cargo are degraded. When induced by starvation, autophagy is largely nonselective with regard to the cargo enclosed in autophagosomes. In contrast, specific intracellular cargo such as damaged mitochondria can selectively trigger the formation of autophagosomes for its degradation (Zaffagnini and Martens, 2016) in a process called selective autophagy (or mitophagy, in the case of damaged mitochondria). The formation of autophagosomes is generally thought to require the action of a conserved machinery that includes the ULK1/Atg1 complex, the class III phosphatidylinositol 3-kinase complex 1, ATG9, the WIPIs, and the ATG12 and LC3/GABARAP conjugation systems. All of these components localize to the isolation membrane at some stage of autophagosome formation. In addition to these conserved core components, other factors such as cargo receptors are required for selective autophagy (Zaffagnini and Martens, 2016).
LC3/GABARAP family proteins are conserved ubiquitin-like proteins that are conjugated to the headgroup of the membrane lipid phosphatidylethanolamine (PE) during autophagosome formation (Ichimura et al., 2000) in a reaction that requires, among other proteins, ATG3, ATG5, and ATG7 (Fig. 1). Whereas there is a single LC3/GABARAP-like protein in Saccharomyces cerevisiae (Atg8), there are seven of them in humans, which are subdivided into the LC3 and GABARAP families. One of these proteins, LC3B, is widely used as marker for isolation membranes and autophagosomes. Autophagosome formation is almost completely abolished in Atg8-deficient S. cerevisiae cells (Kirisako et al., 1999) and deletion of genes required for LC3/GABARAP conjugation to PE blocks autophagosome biogenesis in human cells (Sou et al., 2008). These findings led to the assumption that the conjugation of LC3/GABARAP proteins to PE is essential for autophagosome biogenesis. However, the precise mechanism of action of ATG8 family proteins is unclear. In addition, mammalian LC3/GABARAPs recruit adaptor proteins including PLEKHM1 to fully formed autophagosomes to facilitate autophagosome–lysosome fusion (Stolz et al., 2014; McEwan et al., 2015), suggesting that they have an important role downstream of autophagosome formation. Furthermore, LC3s and GABARAPs function during selective autophagy via their interaction with cargo receptors, which in turn link the LC3/GABARAP-coated isolation membrane to specific cargo material (Zaffagnini and Martens, 2016).
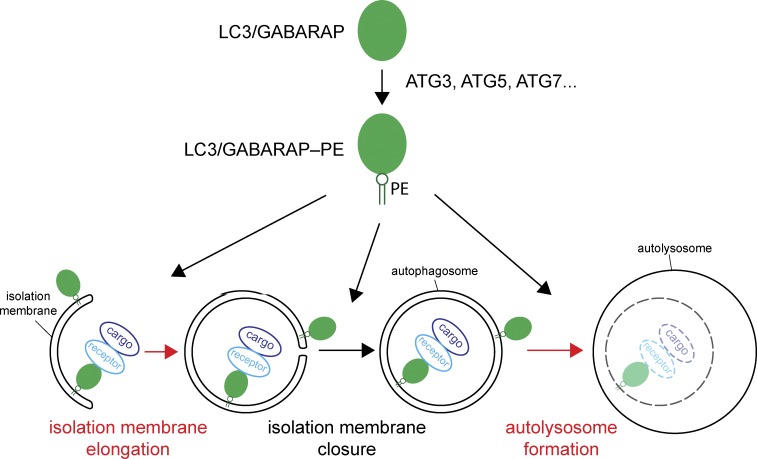
Figure 1.
How LC3/GABARAPs regulate autophagy. LC3/GABARAP proteins are initially synthesized as soluble precursors and become conjugated to PE in the isolation membrane during a series of reactions requiring the activity of several enzymes, including ATG3, ATG5, and ATG7. Several steps of autophagosome formation and their fusion with lysosomes have been proposed to require the action of LC3/GABARAPs. These steps are the promotion of isolation membrane elongation, isolation membrane closure, and the fusion of autophagosomes with lysosomes to form autolysosomes. LC3/GABARAPs have also been shown to link cargo material to the isolation membrane to confer selectivity to autophagy. This interaction is mediated by cargo receptors. The steps that were found to require LC3/GABARAPS in the work of Nguyen et al. (2016) are highlighted in red.
Because up to seven LC3/GABARAPs are expressed in human cells, determining the function of each protein is challenging, and many of the previous experiments addressing the essential functions of ATG8 family members were indirect. For example, researchers addressed the contribution of ATG8 proteins by mutating the specific sequences in ATG8 partners that are known to mediate the interaction with ATG8 proteins (these sequences are known as LIR motifs). Another approach used was to delete the enzymes that are required for the conjugation of LC3/GABARAPs to PE and to analyze the effects of these manipulations on autophagy. If LIR motifs have roles independent of LC3/GABARAP binding and if the deleted enzymes have functions distinct from LC3/GABARAP conjugation, the data interpretation becomes less straightforward. These lines of investigation also fail to address the potential functional redundancy of individual LC3/GABARAPs or of the two subfamilies. In this issue, Nguyen et al. generated CRISPR/Cas9-mediated knockouts of all LC3 and GABARAP proteins, allowing them to analyze the essential functions of ATG8 subfamilies and specific members.
Nguyen et al. (2016) set out to determine which of the many functions of the LC3/GABARAPs are essential and which of these proteins and subfamilies are most important for autophagy. The authors knocked out the LC3 and GABARAP subfamilies, respectively, as well as all of these proteins in HeLa cells using the CRISPR/Cas9 technology. The researchers studied PINK/Parkin-dependent mitophagy and starvation-induced autophagy and made several fascinating observations. They first confirmed an important function of the LC3/GABARAPs for both types of autophagy, as assessed by detection of the degradation of mitochondrial DNA and CoxII for mitophagy, and the degradation of the autophagy substrate p62 for starvation-induced autophagy. The authors went on to determine at which step autophagy was blocked by analyzing autophagosomal structures via fluorescence and electron microscopy. They combined these imaging data with a protease protection assay evaluating whether autophagosomes are fully closed. Perhaps most surprisingly, the main defect Nguyen et al. (2016) observed in cells knocked out for all ATG8 members was not at the level of autophagosome biogenesis or during the selective encapsulation of mitochondria, but at the autophagosome–lysosome fusion step (Fig. 1). To determine the cause of this fusion defect, the authors analyzed the recruitment of the adaptor protein PLEKHM1 to autophagosomes by fluorescence microscopy. PLEKHM1 was previously shown to bind to autophagosomes via LC3/GABARAPs (McEwan et al., 2015), and the researchers found that PLEKHM1 failed to localize to autophagosomes when all LC3/GABARAPs are knocked out. Therefore, they speculated that its absence was responsible for the autophagosome–lysosome fusion defect. Among the LC3/GABARAPs, analysis of cells deficient for the LC3 subfamily only showed that LC3 proteins do not appear to have any essential functions during PINK/Parkin-dependent mitophagy and starvation-induced autophagy, based on analyses of the degradation of mitochondrial DNA, CoxII, and p62 as well as the mtKeima assay, which measures autophagosome–lysosome fusion. In contrast, the depletion of all GABARAPs resulted in a marked decrease in autophagic activity, demonstrating that the LC3s cannot fully compensate for the loss of GABARAPs. Consistently, PLEKHM1 was still recruited to autophagosomes lacking LC3s, but much less so in GABARAP-depleted cells.
Although the results of Nguyen et al. (2016) show that the LC3/GABARAPs are not essential for autophagosome formation under the conditions tested, they do seem to play some role during this process because autophagosomes in cells knocked out for all ATG8 proteins formed at a lower rate and were smaller. This result indicates that ATG8 proteins function during isolation membrane expansion, for example, by acting as fusion or tethering factors, as proposed previously (Weidberg et al., 2011), or by recruiting and activating other ATG proteins, as presented earlier (Kraft et al., 2012; Joachim et al., 2015). Consistent with the findings by Nguyen et al. (2016), a recent study showed that blocking LC3/GABARAP conjugation to PE by knocking out ATG3, ATG5, or ATG7 slows down isolation membrane elongation and closure but does not abolish autophagosome formation (Tsuboyama et al., 2016). However, in contrast to Nguyen et al. (2016), Tsuboyama et al. (2016) report that the degradation of the inner autophagosomal membrane rather than the fusion of autophagosomes with lysosomes is blocked in cells that are deficient in LC3/GABARAP–PE conjugation. This difference may arise from the fact that Nguyen et al. (2016) knocked out LC3/GABARAPs, whereas Tsuboyama et al. (2016) knocked out ATG3 and ATG5 proteins, which are required for ATG8 conjugation to PE. Thus, LC3/GABARAP proteins could have conjugation-independent functions and/or ATG3 and ATG5 could have functions independent of LC3/GABARAP conjugation. More generally, the function of LC3/GABARAPs may be less intimately linked to the formation of double membrane structures (i.e., autophagosomes) than often assumed, as it becomes increasingly clear that the conjugation of these proteins to the membrane also occurs during processes that are not directly related to autophagosome formation. For instance, LC3/GABARAP conjugation has been shown to occur at phagosomes, which are plasma membrane–derived single-membrane structures (Sanjuan et al., 2007) and LC3C was shown to play a role during COPII-mediated ER–Golgi transport (Stadel et al., 2015), two processes that do not involve autophagosomes. Collectively, the current evidence strongly suggests that there is an LC3/GABARAP-independent mechanism for the formation of isolation membranes and autophagosomes that is rendered more robust by LC3/GABARAP proteins. Consistently, ATG5- and ATG7-independent, Golgi-related forms of autophagosome formation have been reported (Nishida et al., 2009). The results by Nguyen et al. (2016) lastly indicate that ATG8 proteins are not essential for autophagosome sealing, suggesting that other factors may mediate this membrane remodeling event.
Another surprising finding in the work of Nguyen et al. (2016) is that mitochondria are apparently still selectively engulfed by autophagosomes as assessed by electron microscopy and protease protection assays, even when all LC3/GABARAPs are missing. So far, it was widely assumed that the selective characteristic of autophagic processes was mediated by cargo receptors (e.g., optineurin, NDP52, and p62) that bind the cargo and link it to the isolation membrane via LIR-dependent interactions with membrane-localized LC3/GABARAPs. The results by Nguyen et al. (2016) suggest that there are likely other factors or mechanisms that link the cargo to the membrane in the absence of ATG8 proteins. Nguyen et al. (2016) used PINK/Parkin-dependent mitophagy induced by oligomycin/antimycin A treatment. Under these conditions, isolation membrane formation is initiated locally at the mitochondrial surface. However, these treatments also lead to inactivation of mTORC1; therefore, the mechanism of membrane expansion may be analogous to that triggered by starvation, which also requires mTORC1 inactivation (Lazarou et al., 2015). Under these conditions, the recruitment of cargo receptors and their binding to LC3/GABARAPs to link the cargo to the nascent autophagosome may not be required. It will be interesting to see if other less dramatic forms of selective autophagy also do not require LC3/GABARAPs proteins and to reanalyze the role of specific LC3/GABARAPs such as LC3C during other, specialized forms of selective autophagy, such as xenophagy (von Muhlinen et al., 2012). Overall, the work of Nguyen et al. (2016) suggests an important shift in the classical view of the essential function of ATG8 family proteins in autophagosome formation. More work will be needed to dissect the mechanisms and determinants of autophagosome biogenesis and the specific contributions of ATG8 family proteins in all types of autophagy.
Acknowledgments
The author declares no competing financial interests.
References
- Ichimura Y., Kirisako T., Takao T., Satomi Y., Shimonishi Y., Ishihara N., Mizushima N., Tanida I., Kominami E., Ohsumi M., et al. 2000. A ubiquitin-like system mediates protein lipidation. Nature. 408:488–492. 10.1038/35044114 [DOI] [PubMed] [Google Scholar]
- Joachim J., Jefferies H.B., Razi M., Frith D., Snijders A.P., Chakravarty P., Judith D., and Tooze S.A.. 2015. Activation of ULK kinase and autophagy by GABARAP trafficking from the centrosome is regulated by WAC and GM130. Mol. Cell. 60:899–913. 10.1016/j.molcel.2015.11.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirisako T., Baba M., Ishihara N., Miyazawa K., Ohsumi M., Yoshimori T., Noda T., and Ohsumi Y.. 1999. Formation process of autophagosome is traced with Apg8/Aut7p in yeast. J. Cell Biol. 147:435–446. 10.1083/jcb.147.2.435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraft C., Kijanska M., Kalie E., Siergiejuk E., Lee S.S., Semplicio G., Stoffel I., Brezovich A., Verma M., Hansmann I., et al. 2012. Binding of the Atg1/ULK1 kinase to the ubiquitin-like protein Atg8 regulates autophagy. EMBO J. 31:3691–3703. 10.1038/emboj.2012.225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazarou M., Sliter D.A., Kane L.A., Sarraf S.A., Wang C., Burman J.L., Sideris D.P., Fogel A.I., and Youle R.J.. 2015. The ubiquitin kinase PINK1 recruits autophagy receptors to induce mitophagy. Nature. 524:309–314. 10.1038/nature14893 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McEwan D.G., Popovic D., Gubas A., Terawaki S., Suzuki H., Stadel D., Coxon F.P., Miranda de Stegmann D., Bhogaraju S., Maddi K., et al. 2015. PLEKHM1 regulates autophagosome-lysosome fusion through HOPS complex and LC3/GABARAP proteins. Mol. Cell. 57:39–54. 10.1016/j.molcel.2014.11.006 [DOI] [PubMed] [Google Scholar]
- Nguyen T.N., Padman B.S., Usher J., Oorschot V., Ramm G., and Lazarou M.. 2016. Atg8 family LC3/GABARAP proteins are crucial for autophagosome–lysosome fusion but not autophagosome formation during PINK1/Parkin mitophagy and starvation. J. Cell Biol.:jcb.201607039 10.1083/jcb.201607039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishida Y., Arakawa S., Fujitani K., Yamaguchi H., Mizuta T., Kanaseki T., Komatsu M., Otsu K., Tsujimoto Y., and Shimizu S.. 2009. Discovery of Atg5/Atg7-independent alternative macroautophagy. Nature. 461:654–658. 10.1038/nature08455 [DOI] [PubMed] [Google Scholar]
- Sanjuan M.A., Dillon C.P., Tait S.W., Moshiach S., Dorsey F., Connell S., Komatsu M., Tanaka K., Cleveland J.L., Withoff S., and Green D.R.. 2007. Toll-like receptor signalling in macrophages links the autophagy pathway to phagocytosis. Nature. 450:1253–1257. 10.1038/nature06421 [DOI] [PubMed] [Google Scholar]
- Sou Y.S., Waguri S., Iwata J., Ueno T., Fujimura T., Hara T., Sawada N., Yamada A., Mizushima N., Uchiyama Y., et al. 2008. The Atg8 conjugation system is indispensable for proper development of autophagic isolation membranes in mice. Mol. Biol. Cell. 19:4762–4775. 10.1091/mbc.E08-03-0309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stadel D., Millarte V., Tillmann K.D., Huber J., Tamin-Yecheskel B.C., Akutsu M., Demishtein A., Ben-Zeev B., Anikster Y., Perez F., et al. 2015. TECPR2 cooperates with LC3C to regulate COPII-dependent ER export. Mol. Cell. 60:89–104. 10.1016/j.molcel.2015.09.010 [DOI] [PubMed] [Google Scholar]
- Stolz A., Ernst A., and Dikic I.. 2014. Cargo recognition and trafficking in selective autophagy. Nat. Cell Biol. 16:495–501. 10.1038/ncb2979 [DOI] [PubMed] [Google Scholar]
- Tsuboyama K., Koyama-Honda I., Sakamaki Y., Koike M., Morishita H., and Mizushima N.. 2016. The ATG conjugation systems are important for degradation of the inner autophagosomal membrane. Science. 10.1126/science.aaf6136 [DOI] [PubMed] [Google Scholar]
- von Muhlinen N., Akutsu M., Ravenhill B.J., Foeglein Á., Bloor S., Rutherford T.J., Freund S.M., Komander D., and Randow F.. 2012. LC3C, bound selectively by a noncanonical LIR motif in NDP52, is required for antibacterial autophagy. Mol. Cell. 48:329–342. 10.1016/j.molcel.2012.08.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weidberg H., Shpilka T., Shvets E., Abada A., Shimron F., and Elazar Z.. 2011. LC3 and GATE-16 N termini mediate membrane fusion processes required for autophagosome biogenesis. Dev. Cell. 20:444–454. 10.1016/j.devcel.2011.02.006 [DOI] [PubMed] [Google Scholar]
- Zaffagnini G., and Martens S.. 2016. Mechanisms of selective autophagy. J. Mol. Biol. 428:1714–1724. 10.1016/j.jmb.2016.02.004 [DOI] [PMC free article] [PubMed] [Google Scholar]