Da Silva and Jantsch discuss work by Lawrence et al. demonstrating a link between the nucleus and cytoplasm for DNA repair.
Abstract
Assaults to our DNA take place at a high frequency and are incompatible with life. In this issue, Lawrence et al. (2016. J. Cell Biol. https://doi.org/10.1083/jcb.201604112) demonstrate that a novel complex links the nucleus with cytoplasmic microtubules for the promotion of DNA repair by homologous recombination.
The importance of DNA damage repair pathways was recognized by the award of the Nobel Prize in Chemistry in 2015 to Tomas Lindahl, Paul Modrich, and Aziz Sancar, and is reflected in the many genetic syndromes associated with mutations in repair components. Homologous recombination (HR) and nonhomologous end joining (NHEJ) are the two major DNA repair pathways that mend lesions occurring on DNA double strand breaks (DSBs). HR relies on homologous sequences for repair, it is mainly used after S-phase in mitotic cells, and is the predominant repair pathway during meiosis. In contrast, NHEJ does not require DNA homology because it simply religates the broken extremities flanking a DNA break, resulting in intrinsic mutagenic potential.
During meiosis, HR promotes the formation of physical connections (chiasmata) between parental chromosomes that are necessary for the faithful chromosome segregation into gametes. HR is also the major repair pathway for the products of genotoxic insults, such as interstrand cross-links (ICLs) that form via alkylating agents (i.e., nitrogen mustard) or the intercalating chemotherapeutic drug cisplatin. These agents distort DNA double strands, which blocks replication forks, causing replication arrest and cell death. Repair of these toxic intermediates is therefore crucial for cell survival. The NHEJ pathway is largely used for repairing DSBs before DNA replication. In Caenorhabditis elegans, NHEJ is normally kept inactive during meiosis, although the regulatory mechanisms that act in germ cells are largely unknown. Blocking the HR pathway and/or excess genotoxic stress can activate the NHEJ repair pathway during gametogenesis, leading to the formation of toxic mutations and chromosome translocations (Adamo et al., 2010). Therefore, meiotic cells use mechanisms to preferentially use HR over NHEJ. In this issue, Lawrence et al. shed light on a novel player involved in the error-free repair of DNA damage by HR. This study is remarkable because it provides evidence of DNA damage sensitivity on an organismal level with the analysis performed in a true tissue. Via the use of specific well-characterized alleles, this study lends support to the importance of linkages between the cytoplasm and the nucleus in the DNA repair process.
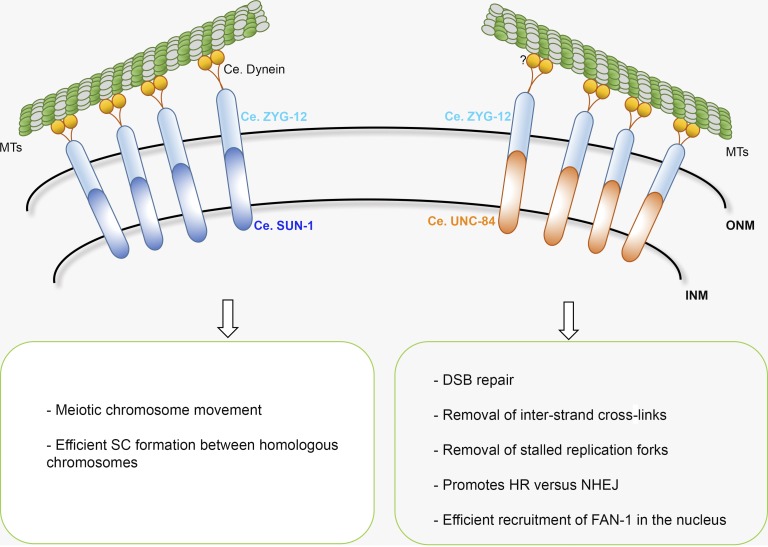
The formation of a physical connection between the cytoplasm and nucleus is crucial for numerous biological processes, including nuclear migration and positioning, centrosome (or spindle pole body) attachment to the nucleus, telomere anchorage to the nuclear periphery, DNA repair, mechanotransduction, and the transmission of cytoplasmic forces during meiotic chromosome movement (Kim et al., 2015). This physical connection is mediated by binding of protein partners known as LINC (linker of nucleoskeleton and cytoskeleton) complexes in the perinuclear space that form a bridge connecting the two membranes (Chang et al., 2015). LINC complexes comprise members of two conserved protein families, SUN (Sad1 and UNC-84) and KASH (Klarsicht, ANC-1, and SYNE/nesprin-1 homology) domain proteins, located in the inner and outer membranes of the nuclear envelope. The N termini of SUN-domain proteins localize to the nucleus, where they can interact with both chromatin and the lamina. In contrast, the cytoplasmic domains of KASH domain proteins can bind to cytoskeletal and motor proteins (Fig. 1).
Figure 1.
Functional diversification of different LINC complexes in the C. elegans germline. The SUN-1–ZYG-12 LINC complex drives chromosome movement and establishes synapsis between homologous chromosomes in meiosis, whereas the UNC-84–ZYG-12 LINC complex is involved in DNA repair. By binding to the ZYG-12 KASH protein, both SUN-1 and UNC-84 can span the nuclear membranes to establish a connection between the nucleus and cytoplasm. Through motor proteins, the LINC complex is connected to microtubules. The dynein motor protein drives meiotic chromosome movement; less is known about the motors involved in the DNA repair process. INM, inner nuclear membrane; MTs, microtubules; ONM, outer nuclear membrane; SC, synaptonemal complex.
After DNA damage, enhanced movement of a specific DNA locus has been frequently observed, although numerous studies have reported the opposite or a general increase in DNA mobility (Dion and Gasser, 2013). The increased mobility of broken DNA ends might accelerate the repair process or may help to sequester them within a specialized compartment, such as the nuclear periphery. Several previous studies support one or the other of these possibilities. DNA mobility has been shown to depend on 53BP1, which is important for DNA repair pathway selection, the LINC proteins SUN1/2, microtubules, nesprins, and kinesins (Lottersberger et al., 2015). Interestingly, Lottersberger et al. (2015) demonstrated that DNA movement promotes, instead of prevents, aberrant chromosome fusion by NHEJ. However, they argued that the enhanced mobility of broken DNA ends might benefit repair by bringing the correct ends into close proximity or by aborting ectopic recombination intermediates. The directed transport of persistent DNA breaks to the nuclear periphery is proposed to promote their correct repair (Nagai et al., 2008; Khadaroo et al., 2009; Oza et al., 2009; Horigome et al., 2014; Su et al., 2015). In particular, in Drosophila melanogaster this mechanism depends on SUN-domain proteins and nuclear pores and promotes the repair of complicated heterochromatic lesions: after transportation to the nuclear periphery, the strand exchange protein RAD-51 was efficiently loaded onto the heterochromatic break (Ryu et al., 2015). The combined knockout of SUN1 and SUN2 in tissue culture cells elicits DNA damage in addition to impaired DNA damage signaling (Lei et al., 2012). Lei et al. (2012) also demonstrated that both SUN1 and SUN2 interact with the DNA-dependent kinase Ku70 and Ku80 proteins. However, it is unclear how the interaction of repair factors with a SUN domain protein can influence DNA damage. A study of Schizosaccaromyces pombe reported the interesting observation that damaged DNA colocalizes with the SUN/KASH module Sad1/Kms1 (Swartz et al., 2014). As Kms1 does not have a role in vegetative growth, mutants do not display cell division defects, which would confound the analysis of DNA repair. It appears that Kms1 mutants, as well as mutants with microtubule defects, are less efficient at repairing DNA breaks via homology-directed repair. It is thus likely that chromosome mobility would reinforce the use of a certain repair pathway. The results presented in Lawrence et al. (2016) neither support a model where the LINC complex contributes to DNA repair by sequestering a break to the nuclear periphery nor a model where the LINC complex has a role in DNA damage signaling. Rather, their data are in agreement with a model where motion-driven repair favors HR over NHEJ. It remains to be shown whether this involves direct interaction of the LINC complex with the NHEJ machinery.
SUN proteins expressed in the C. elegans germline include matefin/SUN-1, which has a major role in meiotic chromosome end–led mobility and centrosome attachment in embryos (Woglar and Jantsch, 2014) and is thus an essential gene, and UNC-84, which has no apparent role in chromosome segregation under unchallenged conditions. The expression of UNC-84 in germcells makes the germline an excellent system for investigating the role of this protein in maintaining chromosome stability. In worms, related paralogs have often differentiated to such an extent that they serve specific, nonoverlapping functions, allowing mutant phenotypes to be precisely defined. Germ cells undergo mitotic divisions before engaging in meiosis, enabling DNA repair pathways to be analyzed in both mitotic and meiotic cells within the gonad. Moreover, the spatiotemporal organization of meiocytes during extended G2 stage within this tissue allows the progress of meiotic recombination (i.e., through the induction and repair of DSBs) to be studied at specific stages of gametogenesis. High-resolution microscopy combined with genetics provides an opportunity for the detailed analysis of chromosomal abnormalities and aberrant DNA repair outcomes, making the C. elegans germline an extremely powerful platform for studying meiosis.
Lawrence et al. (2016) took advantage of the C. elegans germline to examine how SUN proteins function in DNA repair, providing evidence that UNC-84, an inner nuclear membrane SUN-domain protein involved in nuclear migration and anchoring in the soma (reviewed in Chang et al., 2015), promotes HR-dependent repair of both meiotic and ectopically induced DSBs by inhibiting NHEJ at DNA damage sites. unc-84 mutants were sensitive to ICL damage caused by cisplatin or nitrogen mustard, with surviving progeny displaying developmental defects that were also seen in mutants with ectopically activated NHEJ (Adamo et al., 2010). Lawrence et al. (2016) found that the replication A protein coated and stabilized single-stranded DNA at early time points after release from genotoxic insult and was replaced with the RAD-51 recombinase in later stages of HR. Under most conditions examined (i.e., hydroxyurea replication blockage, irradiation, or cisplatin challenge), RAD-51 loading was markedly delayed, but this could be suppressed by blocking NHEJ, as shown in the unc-84; cku-70/cku-80 double mutants. Similarly, endogenous DSBs induced during meiosis had delayed RAD-51 loading. Moreover, UNC-84 localization in the gonad responded to genotoxic insult, whereas its enrichment in nuclei undergoing repair also promoted the membrane enrichment of the outer nuclear membrane KASH protein ZYG-12 (Fig. 1).
Using unc-84 alleles with different point mutations in the N-terminal nuclear domain or the SUN domain necessary for interacting with the KASH partner protein, Lawrence et al. (2016) elegantly showed that the SUN–KASH interaction is necessary for unc-84–dependent HR promotion. Consistent with this finding, UNC-84 could interact with the ZYG-12 KASH partner in vitro, and zyg-12 depletion and microtubule poisoning induced phenotypes similar to those of unc-84 mutants, such as ICL hypersensitivity and reduced RAD-51 loading at early time points after cisplatin exposure. However, unlike in unc-84 mutant worms, loss of NHEJ in zyg-12 mutants exacerbated embryonic lethality upon cisplatin exposure, suggesting that in the absence of ZYG-12 NHEJ is required to some extent for DNA repair. Therefore, ZYG-12 might have a more complex role in DNA repair. In addition, Lawrence et al. (2016) observed persistent RAD-51 accumulation, suggesting that the LINC complex contributes to DSB processing after HR promotion.
Mutants in the Fanconi Anemia (FA) DNA repair pathway were previously shown to display ICL hypersensitivity that could be rescued by blocking NHEJ (Adamo et al., 2010). Similarly, Lawrence et al. (2016) showed that unc-84; cku-70/cku-80 worms have improved viability upon cisplatin treatment compared with unc-84 single mutants. The possibility of a role for UNC-84 in regulating faithful DNA repair upon ICL exposure was strengthened by their finding that both UNC-84 and the FA core protein FCD-2/FANCD2 are essential for nuclear enrichment of the FAN-1 nuclease, which operates in the FA pathway. Interestingly, unc-84–dependent FAN-1 recruitment did not depend on interaction between UNC-84 and its KASH partner, suggesting that UNC-84 contributes to DNA repair in multiple ways. Finally, the authors extrapolated their findings with the demonstration that SUN1 knockdown in human cells rendered them hypersensitive to ICL agents in a NHEJ-dependent manner.
The study by Lawrence et al. (2016) provides extremely important insights into the regulation of DNA repair by showing in vivo that promotion of HR, the most reliable and effective DNA repair pathway, requires communication between the nuclear and cytosolic compartments. Further experiments are required to assess whether UNC-84 acts directly at the site of DNA breaks as a signal transducer or instead acts as a force provider through ZYG-12 and microtubules to establish DNA contacts favoring repair. The nuclear accumulation of FAN-1 independent of the nuclear membrane bridge function supports a model in which UNC-84 contributes to repair at several levels by promoting both the HR pathway and FAN-1 processing of recombination intermediates. Further study of the UNC-84 protein interactome will improve our understanding of the series of events necessary to block unscheduled NHEJ repair and activate the machinery required for ICL repair.
Acknowledgments
We apologize to those colleagues whose work could not be cited due to length restrictions.
Research in the Jantsch laboratory is funded by the Austrian Science Fund (SFB-F34 and P28464). Nicola Silva is funded by an Interdisciplinary Cancer Research fellowship (INDICAR), which has received funding from the Mahlke-Obermann Stiftung and the European Union Seventh Framework Programme for research, technological development, and demonstration under grant agreement number 609431.
The authors declare no competing financial interests.
References
- Adamo A., Collis S.J., Adelman C.A., Silva N., Horejsi Z., Ward J.D., Martinez-Perez E., Boulton S.J., and La Volpe A.. 2010. Preventing nonhomologous end joining suppresses DNA repair defects of Fanconi anemia. Mol. Cell. 39:25–35. 10.1016/j.molcel.2010.06.026 [DOI] [PubMed] [Google Scholar]
- Chang W., Worman H.J., and Gundersen G.G.. 2015. Accessorizing and anchoring the LINC complex for multifunctionality. J. Cell Biol. 208:11–22. 10.1083/jcb.201409047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dion V., and Gasser S.M.. 2013. Chromatin movement in the maintenance of genome stability. Cell. 152:1355–1364. 10.1016/j.cell.2013.02.010 [DOI] [PubMed] [Google Scholar]
- Horigome C., Oma Y., Konishi T., Schmid R., Marcomini I., Hauer M.H., Dion V., Harata M., and Gasser S.M.. 2014. SWR1 and INO80 chromatin remodelers contribute to DNA double-strand break perinuclear anchorage site choice. Mol. Cell. 55:626–639. 10.1016/j.molcel.2014.06.027 [DOI] [PubMed] [Google Scholar]
- Khadaroo B., Teixeira M.T., Luciano P., Eckert-Boulet N., Germann S.M., Simon M.N., Gallina I., Abdallah P., Gilson E., Géli V., and Lisby M.. 2009. The DNA damage response at eroded telomeres and tethering to the nuclear pore complex. Nat. Cell Biol. 11:980–987. 10.1038/ncb1910 [DOI] [PubMed] [Google Scholar]
- Kim D.I., Birendra K.C., and Roux K.J.. 2015. Making the LINC: SUN and KASH protein interactions. Biol. Chem. 396:295–310. 10.1515/hsz-2014-0267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence K., Tapley E.C., Cruz V.E., Li Q., Aung K., Hart K.C., Schwartz T.U., Starr D.A., and Engebrecht J.. 2016. LINC complexes promote homologous recombination in part through inhibition of nonhomologous end joining. J. Cell Biol. 10.1083/jcb.201604112 [DOI] [PMC free article] [PubMed]
- Lei K., Zhu X., Xu R., Shao C., Xu T., Zhuang Y., and Han M.. 2012. Inner nuclear envelope proteins SUN1 and SUN2 play a prominent role in the DNA damage response. Curr. Biol. 22:1609–1615. 10.1016/j.cub.2012.06.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lottersberger F., Karssemeijer R.A., Dimitrova N., and de Lange T.. 2015. 53BP1 and the LINC complex promote microtubule-dependent DSB mobility and DNA repair. Cell. 163:880–893. 10.1016/j.cell.2015.09.057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagai S., Dubrana K., Tsai-Pflugfelder M., Davidson M.B., Roberts T.M., Brown G.W., Varela E., Hediger F., Gasser S.M., and Krogan N.J.. 2008. Functional targeting of DNA damage to a nuclear pore-associated SUMO-dependent ubiquitin ligase. Science. 322:597–602. 10.1126/science.1162790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oza P., Jaspersen S.L., Miele A., Dekker J., and Peterson C.L.. 2009. Mechanisms that regulate localization of a DNA double-strand break to the nuclear periphery. Genes Dev. 23:912–927. 10.1101/gad.1782209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryu T., Spatola B., Delabaere L., Bowlin K., Hopp H., Kunitake R., Karpen G.H., and Chiolo I.. 2015. Heterochromatic breaks move to the nuclear periphery to continue recombinational repair. Nat. Cell Biol. 17:1401–1411. 10.1038/ncb3258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su X.A., Dion V., Gasser S.M., and Freudenreich C.H.. 2015. Regulation of recombination at yeast nuclear pores controls repair and triplet repeat stability. Genes Dev. 29:1006–1017. 10.1101/gad.256404.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swartz R.K., Rodriguez E.C., and King M.C.. 2014. A role for nuclear envelope-bridging complexes in homology-directed repair. Mol. Biol. Cell. 25:2461–2471. 10.1091/mbc.E13-10-0569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woglar A., and Jantsch V.. 2014. Chromosome movement in meiosis I prophase of Caenorhabditis elegans. Chromosoma. 123:15–24. 10.1007/s00412-013-0436-7 [DOI] [PMC free article] [PubMed] [Google Scholar]