Abstract
Our research group demonstrated, in a precedent study, the prognostic power of the 3p microsatellites alterations (MAs) detectable in exhaled breath condensate (EBC) in NSCLC patients. The analysis of genetic markers in the EBC might have precious clinical and economic consequences when inserted in diagnostic and follow up programs for lung cancer.
The aim of this study was to evaluate the prognostic value of a new panel of MAs in the EBC of patients with NSCLC.
We enrolled 45 NSCLC patients during a period of 36 months and the follow-up period was 156 weeks. We analyzed MAs for eight markers in EBC samples: D3S2338, D3S1266, D3S1300, D3S1304, D3S1289, D5S2094, D3S1313, and AFMa305ye1.
Our study showed that the presence of more than 2 simultaneous MAs reduces outcome in NSCLC patients. The new panel of eight microsatellites markers proposed in EBC samples could have a potential clinical role in assessing survival in lung cancer patients.
Keywords: lung cancer, exhaled breath condensate, microsatellite, prognosis.
Introduction
In a previous study 1 we analysed the prognostic power of genetic alterations of a panel of 5 microsatellites located at the 3p chromosome in the exhaled breath condensate (EBC) of 61 lung cancer patients. The impact on patients' outcome of microsatellite alterations selected resulted in great clinical importance and remarkably influenced by the number of microsatellite alterations (MAs) involved. The interest for genetic alterations as MAs on tumour suppressor genes have recently aroused a lot of interest 2-4.
Microsatellite instability and loss of heterozygosity represents molecular disorders acquired by the cell during neoplastic transformation 5. Several MAs at chromosome 3, 9, 17 have been associated to lung cancer 6-11. Recently MAs that activated the FHIT gene function have been also validated as biomarkers of risk for progressive disease in patients belonging to the multicentre European study for the Early detection of lung cancer (EUELC) 10. The reliability increases when these alterations are detectable in samples that are completely non invasive to collect such as the exhaled breath condensate (EBC). Some years ago, our group demonstrated the high sensitivity and specificity of this sample for the study of somatic alterations reporting a complete overlap of MAs at 3 p in the EBC and paired lung tissue of non small cell lung cancer (NSCLC) patients 7.
The analysis of genetic markers in the EBC might have precious clinical and economic consequences when inserted in diagnostic and follow up programs for lung cancer patients who had difficulty undergoing further exams after those routine for diagnosis and staging of the disease. The complete non invasiveness and simplicity of the EBC collection allows it to be well accepted by patients. Samples are obtained simply by breathing for ten minutes in the condenser and thus providing important genetic information coming from the epithelial cells of airways 12. This knowledge, that at the moment, is only used for research goals, could in the near future, change the diagnostic and therapeutic approach in lung cancer.
Considering the potentiality of the study of a larger panel of MAs in the EBC of lung cancer patients which would give doctors and patients the prognostic tools in order to make clinical and life decisions, we tested other 3 MAs to add to the previous panel of MAs already investigated for their outcome value in lung cancer. We selected the microsatellite D5S2094 located near the XRCC4 locus involved in DNA repair mechanisms, the microsatellite D3S1313 located near the FHIT gene and largely known for its involvement in lung cancerogenesis and microsatellite AFMa305ye1 located in the gene coding for IGF1 receptor. Our aim was to analyse the prognostic value of this new panel of microsatellites in the EBC of patients with NSCLC diagnosis.
Materials and method
Patients
During a 36-month period from January 2011 to January 2013, 45 consecutive patients with NSCLC (29 men, 65±5years) were enrolled. All subjects received a histological diagnosis of NSCLC at the Department of Thoracic Surgery, La Madonnina Hospital, Bari and at the Department of Respiratory Diseases of the Foggia University Hospital.
Written informed consent was obtained from all subjects upon approval of the study by the Ethics Committees of the two hospitals. All the patients were enrolled in the study for exhaled and plasmatic microsatellite analysis immediately after histology, and prior to undergoing any forms of anti-cancer therapy whatsoever, including primary surgery.
The patients underwent standard diagnostic and staging procedures. The diagnosis of NSCLC was made either by bronchoscopic biopsy or by transthoracic needle aspiration. Twenty-six subjects received a diagnosis of squamous carcinoma, while the remaining patients were diagnosed with adenocarcinoma. Overall, 13 NSCLC patients were classified as stage I cases, stage II in 10 cases, stage III in 9 cases and stage IV in 13 cases.
Patients with other primitive tumours or with other cardiorespiratory diseases, except for chronic obstructive pulmonary diseases, were excluded from the study.
All the patients underwent EBC and whole blood (WB) collection at enrolment.
Six patients were further excluded because they were lost in follow-up. As a result, a total of 39 patients were enrolled in this survival study.
EBC and WB collection
The EBC and WB were collected as previously described 1.
Microsatellite analysis
DNA was extracted from WB using a QIAamp DNA Mini Kit (Qiagen, Italy), according to the “blood and body fluid protocol”; DNA was then eluted in 100µl of sterile bidistilled water and stored at-20 ◦C. The whole EBC was used for PCR amplification as previously described 6. All the samples of the EBC DNA samples had a positive β-actin gene fragment. Both EBC and WB DNAs were amplified by fluorescent PCR. The analysis of microsatellite alterations was performed using eight polymorphic microsatellite markers on chromosome 3p: 3p24.2 (D3S2338), 3p23 (D3S1266), 3p14.2 (D3S1300, FHIT locus), 3p25-26 (D3S1304), 3p21 (D3S1289), 5q14,2 (D5S2094), 3p14.2 (D3S1313), 15q26.3 (AFMa305ye1) as previously described 6; Genescan TM2.1 software (Applied Biosystems, Foster City, USA) was used for genotyping. This sensitive and quantitative technique allowed allele ratio estimation by measuring the peak height of both alleles 6. Only cases showing heterozygosity were considered as informative and retained for further analysis. Loss of heterozygosis (LOH) and the presence of allele shifts indicating genomic instability in EBC-DNA were recorded and compared with the WB-DNA profile. Each genotype result was confirmed by at least two independent experiments.
Statistical analysis
The Kaplan-Meier method was used to estimate the probability of survival as a function of time, and survival differences were analyzed using the log rank test. The level of significance was set at p <0.05.
Results
The follow-up period was 3 years for the 39 patients that finally were enrolled in this survival study. Patients' survival was calculated as the interval between the date of diagnosis and date of death or date of the last follow-up visit and accounted for the most important clinical endpoint. All patients enrolled in the study, who expired due to lung cancer, were not stratified for other causes of death.
Microsatellite alterations in the DNA from exhaled breath condensate and blood: Survival analysis
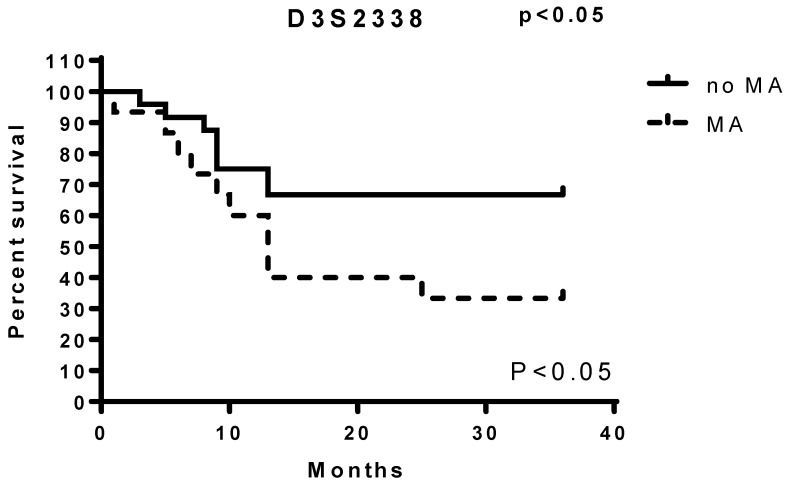
By analysing microsatellite alterations at each of the loci studied we found that the MAs at D3S2338 were those more significantly associated with unfavourable prognostic value (Figure 1). The results of this analysis indicate that the Ma at this locus is an independent variable that decreases survival in patients with lung carcinoma. While only the MA at AFMa305ye1 locus was observed to not have an impact on survival (Table 1), all MAs at other explored loci showed to have a prognostic significance as their presence was observed in patients with more reduced survival (Figure 1).
Figure 1.
Survival differences by MAs analysis at each of the loci studied in EBC/DNA D3S2338.
Table 1.
Survival analysis (X2 and p value for log rank test) for microsatellite alterations at each locus.
| Long rank test | |||
|---|---|---|---|
| X2 | p | ||
| D3S1266 | 0.5016 | 0.4788 | |
| AFMa305ye1 | 0.0001 | 0.9916 | |
| D3S1289 | 0.3479 | 0.5553 | |
| D3S1300 | 0.3324 | 0.5642 | |
| D3S1313 | 0.6228 | 0.4278 | |
| D5S2094 | 1.561 | 0.2115 | |
| D3S2338 | 3.728 | 0.043 | |
| D3S1304 | 1.258 | 0.2621 |
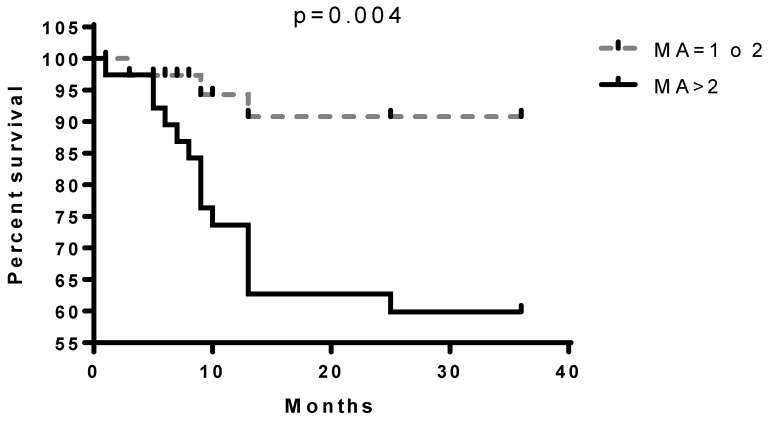
The contemporary existence of more than two MAs (LOH or MI) in loci analysed resulted associated with a reduced survival of patients (Fig. 2) compared to the absence of MAs or the presence of a single one.
Figure 2.
Survival differences in the presence of more than two MAs in all loci.
Discussion
With this study we tested a new panel of microsatellites located in tumour suppressor genes and observed that the presence of more than 2 simultaneous MAs in patients greatly reduces their survival. The microsatellite D3S2338 (Fig. 2) has the strongest correlation with patients survival even though D3S1266, D3S1304, D3S1289, D3S1300, D3S1313, D3S2094 MAs showed to be related to a low outcome. Since lung cancer is a polygenic disease that requires an accumulation of genetic alterations, the singular MA doesn't give us sufficient information that can modify our approach toward the disease. On the other hand, by increasing the panel of mutated genes that could be analysed surely will give us more opportunities to better explore, understand and treat lung cancer and will allow a more accurate prediction of tumour behaviour and patients' outcome in NSCLC 13. For this reason, our group, for several years now, has believed in this new genetic approach to the tumour, looking for a panel of genetic alterations involved in lung cancer that might reach clinical value. In 2009 we focused on 5 MAs at 3p, that, when accumulated showed a strong correlation with patients' survival. With this study we intended to give a further contribution to this field adding other new MAs to the panel of microsatellite markers on chromosome 3p with prognostic significance for lung cancer. The results of this study also confirmed that to have more MAs on the panel explored is associated with poorer prognosis and led us to support the idea that the accumulation of genetic alterations accelerates the cancerogenesis and therefore the progression of the disease leading to an unfavourable prognosis.
The analysis of the panel proposed in the EBC supports the potential role of this non invasive analysis in assessing survival in lung cancer patients although ours is just an exiguous contribution in the identification of most genes involved in this disease.
References
- 1.Carpagnano GE, Spanevello A, Carpagnano F. et al. Prognostic value of exhaled microsatellite alterations at 3p in NSCLC patients. Lung Cancer. 2009;64(3):334–40. doi: 10.1016/j.lungcan.2008.09.004. [DOI] [PubMed] [Google Scholar]
- 2.Fong KM, Zimmerman PV, Smith PJ. Correlation of loss of heterozygosity at 11p with tumour progression and survival in non-small cell lung cancer. Genes Chromosomes Cancer. 1994;10(3):183–9. doi: 10.1002/gcc.2870100306. [DOI] [PubMed] [Google Scholar]
- 3.Fong K, Zimmermal PV, Smith PJ. Tumor progression and loss of heterozygosity at 5q and 18q in non small cell lung cancer. Cancer Res. 1995;55:220–3. [PubMed] [Google Scholar]
- 4.Tseng RC, Chang JW, Hsien FJ. et al. Genomewide loss of heterozygosity and its clinical association in non small cell lung cancer. Int J Cancer. 2005;117(2):241–7. doi: 10.1002/ijc.21178. [DOI] [PubMed] [Google Scholar]
- 5.Shen C, Wang X, Tian L. et al. Microsatellite alteration in multiple primary lung cancer. J Thorac Dis. 2014;6(10):1499–505. doi: 10.3978/j.issn.2072-1439.2014.09.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carpagnano GE, Foschino-Barbaro MP, Mulé G. et al. 3p microsatellite alterations in exhaled breath condensate from patients with non-small cell lung cancer. Am J Respir Crit Care Med. 2005;172(6):738–44. doi: 10.1164/rccm.200503-439OC. [DOI] [PubMed] [Google Scholar]
- 7.Carpagnano GE, Foschino-Barbaro MP, Spanevello A. et al. 3p microsatellite signature in exhaled breath condensate and tumor tissue of patients with lung cancer. Am J Respir Crit Care Med. 2008;177(3):337–41. doi: 10.1164/rccm.200707-1136OC. [DOI] [PubMed] [Google Scholar]
- 8.Carpagnano GE, Palladino GP, Gramiccioni C. et al. Exhaled ERCC-1 and ERCC-2 microsatellite alterations in NSCLC patients. LungCancer. 2010;68(2):305–7. doi: 10.1016/j.lungcan.2010.01.020. [DOI] [PubMed] [Google Scholar]
- 9.Lee HJ, Jang SJ, Chun SM. et al. Comparison of genetic and epigenetic alterations at 11 tumor suppressor loci in pulmonary sclerosing hemangioma and adenocarcinoma. Exp Lung Res. 2011;37(6):344–53. doi: 10.3109/01902148.2011.577267. [DOI] [PubMed] [Google Scholar]
- 10.Verri C, Roz L, Conte D. et al. EUELC Consortium. Fragile histidine triad gene inactivation in lung cancer: the European Early Lung Cancer project m. J Respir Crit Care Med. 2009;179(5):396–401. doi: 10.1164/rccm.200807-1153OC. [DOI] [PubMed] [Google Scholar]
- 11.Ludovini V, Pistola L, Gregorc V. et al. Plasma DNA, microsatellite alterations, and p53 tumor mutations are associated with disease-free survival in radically resected non-small cell lung cancer patients: a study of the Perugia multidisciplinary team for thoracic oncology. J Thorac Oncol. 2008;3(4):365–73. doi: 10.1097/JTO.0b013e318168c7d0. [DOI] [PubMed] [Google Scholar]
- 12.Powell CA. Waiting to exhale. Am J Respir Crit Care Med. 2008;177:246–7. doi: 10.1164/rccm.200710-1604ED. [DOI] [PubMed] [Google Scholar]
- 13.Zhou X, Kemp BL, Khuri FR. et al. Prognostic implication of microsatellite alteration profiles in early stage non small cell lung cancer. Clin Cancer Res. 2008;6:559–65. [PubMed] [Google Scholar]