Abstract
Cancer cells transformation into a normal state or into a cancer cell population which is less tumorigenic than the initial one is a challenge that has been discussed during last decades and it is still far to be solved. Due to the highly heterogeneous nature of cancer cells, such transformation involves many genetic and epigenetic factors which are specific for each type of tumor. Different methods of cancer cells reprogramming have been established and can represent a possibility to obtain less tumorigenic or even normal cells. These methods are quite complex, thus a simple and efficient method of reprogramming is still required. As soon as induced pluripotent stem cells (iPSC) technology, which allowed to reprogram terminally differentiated cells into embryonic stem cells (ESC)-like, was developed, the method strongly attracted the attention of researches, opening new perspectives for stem cell (SC) personalized therapies and offering a powerful in vitro model for drug screening. This technology is also used to reprogram cancer cells, thus providing a modern platform to study cancer-related genes and the interaction between these genes and the cell environment before and after reprogramming, in order to elucidate the mechanisms of cancer initiation and progression. The present review summarizes recent advances on cancer cells reprogramming using iPSC technology and shows the progress achieved in such field.
Keywords: Reprogramming, Yamanaka`s factors, Cancer cells, Induced pluripotent stem cells.
Introduction
The term pluripotency refers to the ability of a stem cell (SC) to differentiate into all derivatives of the three germ layers: endoderm, mesoderm or ectoderm. Such SC population should express in vitro markers of pluripotent cells, form cystic embryoid bodies, which are in vitro three-dimensional model of early embryo, and produce teratomas in vivo. When reintroduced into early embryos, pluripotent cells are able to contribute partially or completely to the new organism development and to transmit pluripotent cells phenotype to the next generation 1. Nevertheless, SC potency varies from the pluripotent state such as embryonic stem (ES) cells to incompletely or partially potent state as adult stem cells (ASC). ASC may produce differentiated cells derived from the three germ layers, however may not exhibit all aforementioned characteristics of pluripotent SC 2,3.
A huge effort has been done in many research centers around the world in order to develop, test and implement protocols which could be used in medicine. Such protocols could be therapeutically effective to treat or even cure many human diseases, such as diabetes, Parkinson and others, including different injuries that are today considered incurable 4,5. The growing quest for an ideal cell with a high differentiation potential in the target tissue has raised a vast number of questions and several problems that must be faced by scientists, such as immunohistocompatibility between donor and recipient, as well as, bioethics issues.
In 2006, Takahashi and colleagues 6 suggested a manner to solve the issue of bioethics, immunohistocompatibility and pluripotency, using a protocol quite ingenious. The group reprogrammed somatic cells turning them similar to pluripotent ESC-like using retroviral vectors containing genes expressed during early developmental stage. These reprogrammed SC were called induced pluripotent stem cells (iPSC).
Cancer research also acquired a new turn due to iPSC technology. The reprogramming of cancer cells is an interest approach to study cancer-related genes and the interaction between these genes and cell environment before and after reprogramming, in order to elucidate the mechanisms of different stages of cancer development. Additionally, reprogramming cancer cells is one of the ways for discovering novel cancer treatments. Cancer cells may also be reverted into an immature state and, therefore, be able to differentiate into derivatives of the three germ layers. Moreover, using iPSC technology is possible to transform cancer cells into highly immunogenic tumor antigen-presenting dendritic cells, which represent a promising approach for cancer immunotherapy 7,8. While the reprogramming of normal somatic cells is abundantly highlighted in scientific literature 9,10,11,12,13, reprogramming of cancer cells received less attention 14,15,16. After a short introduction in reprogramming technology of somatic cells, the present review will be focused on cancer cells reprogramming using iPSC technologies.
iPSC Technology
As soon as pluripotent cells were discovered, many scientists have addressed their studies to better understand the molecular mechanisms of pluripotency. They demonstrated that pluripotent cells express a unique set of transcription factors (TF), which do not serve only as markers, but also are functionally important for the pluripotency maintenance 17. Takahashi and Yamanaka 6 proposed the use of such TF for the induction of a pluripotent state in somatic cells. More than 20 TF were tested individually or in combination by the group to induce pluripotency in terminally differentiated somatic cells and, finally, the main candidates were selected: octamer 4 (Oct4), SRY box-containing gene 2 (Sox2), Kruppel-like factor 4 (Klf4) and the oncogene c-Myc. These factors together (OSKM) were named as Yamanaka´s factors and they were crucial for the generation of iPSC from mouse embryonic fibroblasts (MEF). Yamanaka´s factors were also introduced into adult tail-tip fibroblasts derived from C57/BL6-129 mice of different ages and they were also able to reverse these cells into a pluripotent state. Later, the same method of reprogramming was successfully tested in human somatic cells 9.
In spite of being an innovative method, iPSC methodology had low efficiency of reprogramming, once the best efficiency reported in scientific literature for human fibroblasts was 0.1%, which turns the method laborious and time consuming 18. Different groups looked for improving reprogramming technology and many efforts have been done aiming at: (1) increasing the efficiency of reprogramming; (2) obtaining a pluripotent SC population capable of differentiating into the three germ layers-derived cell types in vitro and in vivo and (3) eliminating the use of viral plasmid transfection. In order to achieve such purposes, different somatic cells, such as: cord blood 19, peripheral blood T e B lymphocytes 20,21, keratinocytes 22, pancreatic β cells 23, amnion-derived cells 24, adipose SC 25, neural SC 26, astrocytes 27, dental pulp SC 28 and others have been reprogrammed. All these cell types have been reported to generate iPSC but also showed variable reprogramming efficiencies and kinetics 19,28. Besides OSKM, other key TF have been considered for reprogramming, such as Nanog and Lin-28, which also were efficient for iPSC generation 29.
Numerous studies have shown that reprogramming TF can be introduced in cells by using adenovirus or lentivirus. However, the use of retrovirus remains the most used method since higher efficiency levels were obtained. Recently, non-integrating reprogramming methods have been developed to deliver factors into the cells in a safer manner, than using viral methods. The most widely techniques used for generating integration-free human iPSC are: Sendai-virus (SeV), episomal (Epi) and mRNA 30. Interesting that these three non-integrating techniques were used in parallel with two integrating (retro- and lentiviral vectors) methods to compare their reprogramming efficiencies, which were: mRNA = 2.7%, SeV = 0.077%, Epi = 0.013%, Lenti/Retro = 0.27%. All methods produced good-quality iPSC, however significant differences were registered in aneuploidy rates, reprogramming efficiencies, reliability and time consuming 30.
Cancer and Reprogramming
The idea of cancer cells reprogramming is not new and other methods, such as nuclear reprogramming of somatic cells by the injection of tumor cells - embryonic carcinoma into normal blastocyst 31, by in vitro hybridization of cancer cells with ESCs 32 and somatic cell nuclear transfer (SCNT) technique, which implants an enucleated oocyte in a donor nucleus from a cancer cell 33 were already used to suppress the tumorigenic phenotype 34. Several cancer cell lines have already been reprogrammed using one of the aforementioned methods. It was possible to reprogram embryonic carcinoma cells into an almost normal state by transferring a tumor cell nucleus to an enucleated mouse oocyte, taking an advantage from the embryonic microenvironment. Resulting blastocysts showed the ability to develop, but they had the same tumorigenic potential as the donor cells 35. Other study used SCNT technology to reprogram melanoma cells. In this study, ESC-like were produced and were able, when re-introduced into recipient early embryo, to complete the normal development and to produce healthy offspring 35. Recent reports provided additional evidences that the malignant phenotype of cancer cells could be suppressed in embryonic niche, accompanied by alternative expression of miRNA and by epigenetic regulation, such as DNA methylation 36,14. Taken together, these data confirm the importance of genetic changes in the tumor development and raise the possibility that in certain tumor types, epigenetic changes may play a predominant role. Although epigenetic changes contribute to tumorigenesis, it is still poorly understood how it occurs and also whether it is reversible 37.
The classical view of carcinogenesis mechanisms has considered the tissue de- differentiation during the malignant process 38. The most modern version is based on the hypothesis that cancer stem cells (CSC) arise from SC of a primitive tissue or from a specific population of progenitor cells that can assume self-renewal and unlimited growth properties 39. The iPSC technology is based on the reprogramming of somatic cells into ESC-like by ectopic expression of different TF. When this reprogramming occurs, epigenetics markers are also re-established. All of these premises lead to the conclusion that the iPSC technology can be useful for cancer cells reprogramming, which in some cases may lose or achieve a less tumorigenic state. However, these reprogrammed cells vary in reprogramming status at a large extent and, in turn, may accomplish a partial or a complete pluripotent state.
Cancer Cell Lines Reprogramming Using iPSC Technologies
Different cancer cell lines were used in reprogramming experiments 40,41. One important study employed R545-melanoma cell line, which is trisomic for chromosomes 8 and 11 and conditionally express the oncogene H-Ras 42. In H-Ras transgenic animals, which also carry deletion of the ink4a/Arf (tumor suppressor locus), RAS transgene is activated specifically in melanocytes after the administration of doxycycline, resulting in melanoma formation 42. R545-melanoma cell line was a smart choice to test reprogramming by TF, once the same group had already showed reprogramming of R545-genome by SCNT 35. R545-melanoma cells were infected by lentiviral vectors expressing Oct4, Klf4 and c-Myc (OKM). ESC-like were obtained after 14 days of reprogramming and further analysis demonstrated that viral gene expression was silenced following reprogramming. Demethylation of Oct4 and Nanog promoters also occurred and R545-derived iPSC were able to form teratomas, as well as to give rise to chimeras.
Carette and co-workers 43 reprogrammed KBM7 cell line previously obtained from a carrier of a chronic myelogenous leukemia (CML). KBM7 cell line possesses BCR-ABL mutation and shows karyotypic instability in culture 44. Retrovirus containing OSKM were used to infect KBM7 cells. ESC-like colonies appeared at day 21 after reprogramming and expressed the pluripotent cell markers Oct4, Nanog and Sox2, while the expression of hematopoietic markers CD43 and CD45 was lost. The expression levels of Nanog and Sox2 was similar to human ESC. After reprogramming, the percentage of methylated cytosine-phosphate-guanosine (CpG) sites of Oct4 and Nanog promoter regions was significantly lower than in KBM7 cell line and was comparable to human ESC. Also, KBM7-iPSC still presented abnormal karyotype and maintained the expression of BCR-ABL oncogene. The authors tested weather the removal of one or more TF would be crucial for KBM7 cells reprogramming. Interesting that removal of c-Myc induced cell death, while the exclusion of Oct4, Sox2 or Klf4 from reprogramming mixture decreased iPSC phenotype and colonies formation. These incompletely reprogrammed colonies also maintained the expression of CD43, typical of parental cell line. ESC-like colonies of KBM7-iPSC were able to differentiate in vitro into hematopoietic-like cells that were positive for CD34, CD43 and CD45 and into neuronal-like cells. Also, these ESC-like colonies of KBM7-iPSC formed teratomas in NOD-SCID mice. In contrast to original KBM7 cells, which are imatinib-sensitive (drug used to treat different types of leukemia), reprogrammed cells and their non-hematopoietic derivatives were shown to be completely resistant to this drug 43. This study raises interesting questions: (i) Does reprogramming method with removal of c-Myc may be used as a tool to combat CML? (ii) Does KBM7-iPSC may be used as an in vitro model to study imatinib-sensitivity?
Other authors 45 used eight different gastrointestinal cancer cell (GCC) lines to obtain iPSC-like cells. Several retroviral and lentiviral plasmids containing OSKM were tested and the selected plasmid was introduced into cancer cells by using lipofectamine; iPSC-like colonies were formed at day 31 after reprogramming. Such GCC-derived induced pluripotent cancer cells (GCC-iPCC) expressed endogenous Oct3/4, Sox2, Klf4, c-Myc and oncogenes, such as BCL2 and KRAS, besides the tumor suppressor genes TP53, P16, PTEN, FHIT and RB1. Nanog expression increased significantly after reprogramming and achieved an expression level comparable to the pluripotent teratocarcinoma cell line used as positive control. Similar to KBM7-iPSC, these GCC-iPCC showed epigenetic modifications which occurred in CpG sites in the Nanog promoter, confirming the immature status of this gene in comparison to its status in parental cell line and in in vitro differentiated GCC-iPCC derivatives. These reprogrammed cells also showed the capacity to form embryoid-like bodies, which adhered to plastic, producing attached cells named by authors as PostiPC cells. PostiPC cells were capable to differentiate in vitro into derivatives of the three germ layers. Tumorigenic properties of PostiPC cells in compassion to parental GCC were tested in NOD/SCID mice. The results revealed the reduction of tumorigenesis in “spontaneously differentiated” PostiPC cells. However, this study did not provide any information about teratoma formation or tumorigenic potential of GCC-iPCC. Further, the authors used fluoropyrimidine 5-fluorouracil (5-FU) in parental GCC and PostiPC cells and demonstrated that PostiPC cells were significantly more sensitive to this drug when compared to the parental cell line, suggesting that PostiPC cells could be more sensitive to therapeutic agents 45.
Osteosarcoma cell lines (Saos-2, MG-63, G-292 and U-2 OS) were also reprogrammed into a pluripotent state using OSKM retroviral transduction method. After reprogramming, these cells showed morphology resembling ESC colonies and expressed alkaline phosphatase, besides pluripotent markers: Oct4, SSEA4, TRA-1-60 and TRA-1-81. Reprogrammed sarcoma cells showed capacity to differentiate into adipocytes and osteocytes. They showed variable responses of reprogramming in respect of efficiency and long term culture effect 46.
Different from aforementioned cell lines, human skin cancer cells were reprogrammed using mir-302 microRNA (miRNA), which is expressed in slow-growing human ESC. The mir-302 - transfected cells expressed markers of ESC and showed a highly demethylated genome. These mirPS cells were able to differentiate into neuron-, chondrocyte-, fibroblast-, and spermatogonia-like primordial cells. The use of intronic mir-302 transfection represents a new and promise tool for the generation of pluripotent stem cells derived from cancerous cells 47.
Table 1 summarizes current knowledge which used iPSC technology to harvest cancer-derived reprogrammed cells. All features mentioned in the table refer to a characteristic necessary to classify a pluripotent cell. Considering the efficiency of reprogramming methods or the combination of TF used during the process, there is no completely reprogrammed population of cancer cells.
Table 1.
Studies relating cancer-derived cells reprogrammed by iPSC technology.
| CANCER CELL LINES | Karyotype | Reprogramming methods | Viral gene expression | Epigenetic Modification | In vitro Differentiation capacity | Teratomas/ Tumors formation | Chimeras | Drug sensitivity | Authors |
|---|---|---|---|---|---|---|---|---|---|
| Mice Melanoma R545 cell line Ras-induction | Trisomy Chromosomes 8 and 11 | Lentiviral OKM1 | Silenced | Demethylation Oct4 and Nanog Promoters | Unknown | Yes | Yes | No tumors in the absence of DOX2 | Utikal et al., 2009 |
| Human Leukemia KBM7 CML | Tetraployd, chromossomes 9 and 22 Ph(+) | a. Retrovirus OSKM3 | Unknown | Partly demethylated Oct4 and Nanog Incomplete | Neuronal- and hemat - like cells | Yes | Not applied | Non-hemat. Derivatives are imatinib resistant - Cell type specific drug sensitivity | Carette et al., 2010 |
| b. Retrovirus OSK4 Incomplete Reprogramming | Unknown | Unknown | __ | ||||||
| Human gastrointestinal cancer cells | Abnormal | Retrovirus and Lentivirus + Lipofectamine + OSKM | Silenced | Demethylation of Nanog promoter; Histone modification | Derivatives of three germ line | GCC5 - Tumor GCC - iPSC Unknown PotsiPC cells - Tumor | Not applied | PotsiPC cells -more sensitive to 5-FU6 and Differentiation Inducing drugs | Miyoshi et al., 2010 |
| Human gastrointestinal cancer cells | Abnormal | Retrovirus and Lentivirus + Lipofectamine + OSKM | Long term culturing - down regulation of endogenous OCT4, SOX2, KLF4 and c-MYC up regulation and high tumorigenic ability | Nagai et al., 2010 | |||||
| Human Osteosarcoma and liposarcoma | Abnormal | Lentivirus OSKM + Nanog +Lin28 |
Silenced | Unknown | Ectoderm and endoderm derivatives; Mesoderm with less efficiency | Tumors less aggressive than parental line. No tumor after injection into mice of differentiated Cancer derived iPC7 cells |
Not applied | Unknown | Zhang et al., 2013 |
| Human lung adenocarcinoma A549 epithelial cell line | Abnormal | Lentivirus + OSLN8 + hypoxia | Unknown | Partly demethylated Oct4 | Unknown | Highly aggressive tumors | Not applied | Unknown | Mathieu et al., 2011 |
| HCT116 colorectal cancer cells |
Abnormal | Lentivirus + OSLN + hypoxia | Unknown | Unknown | Unknown | Reduced Tumor Formation | Not applied | Unknown | Hoshino et al., 2012 |
| HCT116 colorectal cancer cells + TR53-deficient |
Abnormal | Lentivirus + OSLN + hypoxia | Unknown | Unknown | Unknown | Highly aggressive tumors | Not applied | Unknown | Hoshino et al., 2012 |
| Increased efficiency | |||||||||
| Solid primary human cancer - pancreatic ductal adenocarcinoma (PDAC) | Aberrant karyotype ~20 chromosomal aberrations |
Lentiviral (dox)- regulated vector + OSKM |
Unknown | Demethylation Oct4 and Nanog Promoters | Unknown | Yes Restricted Mostly endodermal | Not applied | Unknown | Kim et al., 2013 |
| Generates pancreatic intraepithelial neoplasia | |||||||||
Abbreviations: 1Oct4, Klf4 and c-Myc; 2Doxiciclin; 3Oct4, Sox2, Klf4 and c-Myc; 4Oct4, Sox2 and c-Myc; 5Gastrointestinal cancer cell; 65-fluorouracil; 7induced pluripotent cancer; 8Oct4, Sox2, Lin-28 and Nanog.
Cancer Cell Lines Reprogramming Using iPSC Technologies and Hypoxia
It is of common knowledge that hypoxia helps to maintain undifferentiated state of normal cells and CSC, as well as it also effects proliferation and cell-fate commitment 48,49. In order to test the effect of hypoxia on cancer cells reprogramming, human lung adenocarcinoma A549 epithelial cell line was infected first by retrovirus expressing hypoxia-inducible factor (HIF) and next by lentivirus expressing Oct4, Sox2, Nanog and Lin28 (OSLN). ESC-like were collected at day 12 after reprogramming and the expression levels of endogenous Nanog and Oct4 in A549 iPS-like cells were lower than in normal human ESC, used as control. The promoter of Oct4 was only partially unmethylated, thus suggesting that these cells were not fully reprogrammed. The tumorigenic capacity of these partially reprogrammed A549 iPSC-like colonies was assessed in vivo. The colonies were injected into the femoral muscle of immune compromised mice and the cells rapidly produced highly aggressive tumors 50. The results obtained by Mathieu and colleagues 50 suggest that hypoxia targets are crucial for maintaining the stemness in malignant cells.
Hypoxia was also used for reprogramming HCT116 colorectal cancer cells and mutant TP53-deficient HCT116 cells, which showed increased iPCC generation efficiency. Additionally, both of these cells showed reduced proliferation, invasive and tumorigenic capacities after reprogramming. Transplantation of iPCC derived from TP53-deficient HCT116 cells into NOD/SCID mice resulted in more rapidly tumor formation, when compared to the same cells without the mutation 51 (Table 1).
Reprogramming of Solid Primary Human Cancer
According to current knowledge, only one study reported the reprogramming of solid primary human cancer - pancreatic ductal adenocarcinoma (PDAC) - using iPSC technology. Usually, PDAC is detected at advanced stage thus having a poor prognosis to the patient, with less than 5% survival rate. PDAC is initiated by a mutation at KRAS (proto-oncogene that encodes an ~21 kDa small GTPase), which is present in more than 90% of PDAC cases 52. There are no in vitro models in scientific literature which allow study early stages of PDAC. Therefore, reprogramming of PDAC cells may be a tool for studying the progress of the disease. Kim and colleagues 53 obtained biopsies from nine patients and isolated epithelial cancer cells. These cells were infected with doxycycline (dox)-regulated lentiviral vector expressing OSKM, while the margin cells of the biopsies were used as control. Four ESC-like lines from nine tumors were obtained. They harbored the same KRAS G12D mutation observed in the initial tumor epithelial population, besides possesses aberrant karyotype (~20 chromosomal aberrations). Reprogrammed cancer cells expressed pluripotent cell makers, such as: Oct4 and Nanog (original tumors were negative for such markers). Demethylation was observed in several sites of Nanog and Oct4 promoters of reprogrammed cancer cells, which showed similar demethylation pattern as human ESC. Reprogrammed cancer cells formed embryoid bodies in vitro and teratomas in vivo. These teratomas generated mostly endodermal structures in contrast to human ESC used as control, which produced mainly neuronal cell lines. Only one reprogrammed PDAC cell line, when injected into immunodeficient mice, generated pancreatic intraepithelial neoplasia (PanIN), precursor of PDAC. The cells isolated from PanIN-like structure secreted proteins that are expressed in human PDAC during cancer progression. The authors used iPSC technology and succeeded to generate partially reprogrammed cells from PDAC and also provided a human cell model of early pancreatic cancer 53 (Table 1).
Reprogramming of Cancer Cells into a Less Tumorigenic State and Derivation of Potentially Malignant Cells from iPSC
Cells reprogramming may have different objectives and one of them consist on the establishment of in vitro models to study the mechanisms of cancer cells transformation into a less tumorigenic state, as well as the transformation of normal cells into a malignant state. Both of these models, especially in combination, are of great interest once it may help to understand the mechanisms of malignant transformation and answer the question of how normal cells may become cancer cells. Although Utikal and co-authors 41 already demonstrated the reprogramming of melanoma cells into almost typical iPSC, they did not show if this state is reversible. Also, it was not clear whether normal iPSC in a tumor microenvironment will have a risk of malignant transformation. This is important as the main issue of iPSC transplantation into patients is about safety concerns, once these cells tend to form teratomas. Recently, it has been shown that mouse iPSC cultured in conditioned media by cancer cell lines showed tumorigenic capacity 54,55. After transplantation into nude mice, all iPSC lines showed formation of malignant tumors and one iPSC line enhanced angiogenesis formation. This work demonstrated that iPSC is an interesting model for the study of malignant cell transformation and also alerts us about possible malignant transformation of normal iPSC and their derivatives.
Li-Fraumeni syndrome (LFS) is a rare autosomal dominant disease, related to mutations at the TP53 gene and characterized by the occurrence of multiple tumors in the same person. Lee and collaborators 56 obtained iPSC from skin fibroblasts of LFS patient with osteosarcoma. Mesenchymal stem cells (MSC) derived from these iPSC upon differentiation were able to differentiate into osteoblasts, which in turn, generated osteosarcoma after their injection into nude mice. This study demonstrated that malignant transformation depends, at least in this case, on cell specification, once iPSC-derived MSC were not able to form any type of tumor. It is still interesting to derive iPSC from primary tumor cells of LFS patients. The generation of such pairs of cell lines obtained from normal and malignant cells of the same patient is a model of interest for studying the mechanisms of tumor transformation, tumor progression and the reversion to normal or malignant state.
Why do We Want to Reprogram Cancer Cells?
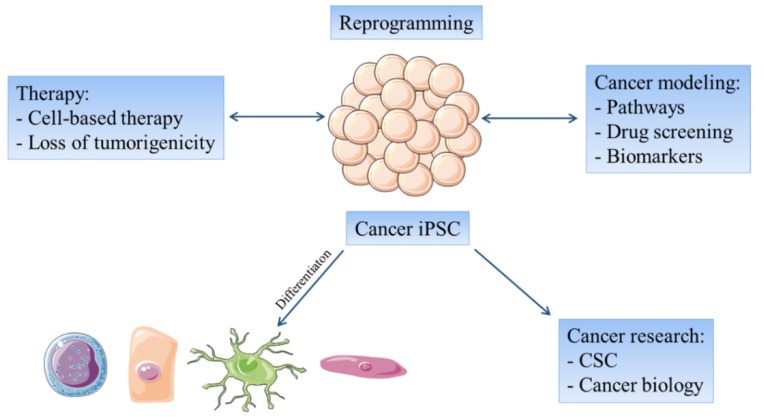
The reprogramming of cancer cells have several basic aims: (1) to explore the possibility to normalize in vivo the malignant phenotype of such cells, as an alternative to conventional therapeutic protocols; (2) to yield a larger CSC population, which would be available for experimental manipulation and for exploration of their biological properties to better understand resistant tumors and reduce relapses; (3) to produce less tumorigenic iPSC derived from cancer cells that may be differentiated into a specific cell type e.g. dendritic cells or cytotoxic T cells, in order to prepare specific cancer vaccines; (4) to use cancer-derived iPSC for pharmacological screenings 57; (5) to use reprogrammed cancer cells to create novel therapeutic targets against CSC by combining, for example, small non-coding RNAs with efficient drug delivery systems 58; and (6) to create a powerful tool for distinguishing epigenetic and genetic alterations that occur during tumor development and progression 40 (Figure 1).
Figure 1.
The reprogramming of cancer cells focuses on possible therapeutic use of iPSC-like cancer cells and at cancer remodeling. Cancer- derived iPSC lines can be differentiated into different cell types in order to investigate the features of cancer progression and drug screening or to develop cell-based therapies. On the other hand undifferentiated cancer-derived iPSC lines may be useful for further cancer research.
Several factors can enhance the efficiency of iPSC generation, such as cell cycle checkpoints mediated by the cyclin-dependent kinase inhibitor family. The transient inhibition of these proteins may significantly improve iPSC generation 33,59, although the ability of the resultant cells to become non tumorigenic is not completely understood. During reprogramming, cells increase their intolerance to different types of DNA damage. The mechanisms which occur in cancer cells and then undergo an incomplete form of reprogramming, such as the presence of CSC that may increase the heterogeneity of a cancer cell population, remain unclear 58. It has been proposed that two types of SC coexist in normal and in cancer microenvironment and that these cells populations are transient and are regulated by epigenetic controls 60,61. Emerging evidence indicates that quiescent and active SC subpopulations that are in lower metabolic and proliferative states, respectively, may coexist in several tissues 61. Endogenous expression levels of TF during early embryogenesis and in pluripotent cells could be relevant for tumor cell malignancy and transformation 62.
Conclusions
The investigation of cellular reprogramming and pluripotency encompasses several important decades in the recognition of similarity between normal pluripotent cells and tumor cells. Cellular mechanisms involved in normal pluripotent SC differentiation and in abnormal growth and differentiation of CSC in the neoplasia were also addressed 40,63.
Recent studies demonstrate that Yamanaka`s method 6 can be successfully used for reprogramming cancer cell. Similar to normal cells, cancer cells also showed variable reprogramming efficiencies and kinetics. They respond differentially to each reprogram method when removing or adding TF in the reprogramming vector. It seems that in contrast to normal cells, which after reprogramming and in vitro long-term cultivation tend to preserve their pluripotent properties, cancer reprogrammed cells demonstrate down regulation of pluripotent genes and up regulation of the oncogene c-Myc.
These cells also showed epigenetic modifications following reprogramming. Until now, the majority of cancer cells used for reprogramming experiments is from cancer cell lines, which are already highly instable and frequently do not reflect the true scenario of cancer cells populations, when compared to the model of primary cells isolated from tumors. Therefore, the number of studies using primary cancer cells should be increased in order to provide more adequate models for drug screening.
The small number of studies described in this present review already demonstrates the usefulness of this model for cancer studies. Herein, we show that although great advances on reprogramming efficiency of cancer cells have been made, the heterogeneity of such cells remains to be investigated as a novel therapeutic approach. The most striking conclusion is that cancer cells reprogramming results in the ability of the cancer cells to re-engage and terminally execute normal cellular differentiation pathways with consequent reduction of tumorigenic properties 42.
Acknowledgments
Funding: This work was supported by grant 2010/51051-6 from FAPESP (Sao Paulo Research Foundation). The funder had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Authorship
DC and LM: writing the first draft of manuscript, figure and table production, references organization, and manuscript revision.
AP: participation in the manuscript writing and organization.
IK: research design, writing of the manuscript and critical revision for important intellectual content.
References
- 1.Thomson J, Itskovitz-Eldor J, Shapiro S, Waknitz M, Swiergiel J, Marshall V. et al. Embryonic stem cell lines derived from human blastocysts. Science. 1998;282:1145–1147. doi: 10.1126/science.282.5391.1145. [DOI] [PubMed] [Google Scholar]
- 2.Eisenberg LM, Eisenberg C.A. Stem cell plasticity, cell fusion, and transdifferentiation, Birth Defects Res. Part C - Embryo Today Rev. 2003;69:209–218. doi: 10.1002/bdrc.10017. [DOI] [PubMed] [Google Scholar]
- 3.Onichtchouk DV, Voronina AS. Regulation of Zygotic Genome and Cellular Pluripotency. Biochemistry. 2015;80:1723–1733. doi: 10.1134/S0006297915130088. [DOI] [PubMed] [Google Scholar]
- 4.Accomasso L, Gallina C, Turinetto V, Giachino C. Stem Cell Tracking with Nanoparticles for Regenerative Medicine Purposes. An Overview. 2015;2016:1–23. doi: 10.1155/2016/7920358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li K, Kong Y, Zhang M, Xie F, Liu P, Xu S. Differentiation of pluripotent stem cells for regenerative medicine. Biochem. Biophys. Res. Commun. 2016;471:4–7. doi: 10.1016/j.bbrc.2016.01.182. [DOI] [PubMed] [Google Scholar]
- 6.Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–76. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 7.Lin FK-Y, Chui Y-L. Generation of Induced Pluripotent Stem Cells from Mouse Cancer Cells. Cancer Biother. Radiopharm. 2012;27:694–700. doi: 10.1089/cbr.2012.1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sachamitr P, Hackett S, Fairchild PJ. Induced pluripotent stem cells: Challenges and opportunities for cancer immunotherapy. Front. Immunol. 2014;5:1–9. doi: 10.3389/fimmu.2014.00176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K. et al. Induction of Pluripotent Stem Cells from Adult Human Fibroblasts by Defined Factors. Cell. 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 10.Yu J, Vodyanik MA, Smuga-Otto K, Antosiewicz-Bourget J, Frane J.L, Tian S. et al. Induced pluripotent stem cell lines derived from human somatic cells. Science. 2007;318:1917–20. doi: 10.1126/science.1151526. [DOI] [PubMed] [Google Scholar]
- 11.Zhou Q, Brown J, Kanarek A, Rajagopal J, Melton DA. In vivo reprogramming of adult pancreatic exocrine cells to β-cells. Nature. 2008;455:627–632. doi: 10.1038/nature07314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kaji K, Norrby K, Paca A, Mileikovsky M, Mohseni P, Woltjen K. Virus-free induction of pluripotency and subsequent excision of reprogramming factors. Nature. 2009;458:771–5. doi: 10.1038/nature07864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Okita K, Matsumura Y, Sato Y, Okada A, Morizane A, Okamoto S. et al. A more efficient method to generate integration-free human iPS cells. Nat. Methods. 2011;8:409–412. doi: 10.1038/nmeth.1591. [DOI] [PubMed] [Google Scholar]
- 14.Kasai T, Chen L, Mizutani A, Kudoh T, Murakami H, Fu L. et al. Cancer stem cells converted from pluripotent stem cells and the cancerous niche. J. Stem Cells Regen. Med. 2014;10:2–7. doi: 10.46582/jsrm.1001002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Volinia S, Nuovo G, Drusco A, Costinean S, Abujarour R, Desponts C. et al. Pluripotent stem cell miRNAs and metastasis in invasive breast cancer. J. Natl. Cancer Inst. 2014;106:1–8. doi: 10.1093/jnci/dju324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Curry E.L, Moad M, Robson CN, Heer R. Using induced pluripotent stem cells as a tool for modelling carcinogenesis. World J. Stem Cells. 2015;7:461–9. doi: 10.4252/wjsc.v7.i2.461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen L, Daley GQ. Molecular basis of pluripotency. Hum. Mol. Genet. 2008;17:23–27. doi: 10.1093/hmg/ddn050. [DOI] [PubMed] [Google Scholar]
- 18.Huangfu D, Osafune K, Maehr R, Guo W, Eijkelenboom A, Chen S. et al. Induction of pluripotent stem cells from primary human fibroblasts with only Oct4 and Sox2. Nat. Biotechnol. 2008;26:1269–1275. doi: 10.1038/nbt.1502. [DOI] [PubMed] [Google Scholar]
- 19.Haase A, Olmer R, Schwanke K, Wunderlich S, Merkert S, Hess C. et al. Generation of Induced Pluripotent Stem Cells from Human Cord Blood. Cell Stem Cell. 2009;5:434–441. doi: 10.1016/j.stem.2009.08.021. [DOI] [PubMed] [Google Scholar]
- 20.Hanna J, Markoulaki S, Schorderet P, Carey BW, Beard C, Wernig M. et al. Direct Reprogramming of Terminally Differentiated Mature B Lymphocytes to Pluripotency. Cell. 2008;133:250–264. doi: 10.1016/j.cell.2008.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Brown ME, Rondon E, Rajesh D, Mack A, Lewis R, Feng X. et al. Derivation of induced pluripotent stem cells from human peripheral blood T lymphocytes. PLoS One. 2010;5:1–9. doi: 10.1371/journal.pone.0011373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Aasen T, Raya A, Barrero MJ, Garreta E, Consiglio A, Gonzalez F. et al. Efficient and rapid generation of induced pluripotent stem cells from human keratinocytes. Nat. Biotechnol. 2008;26:1276–1284. doi: 10.1038/nbt.1503. [DOI] [PubMed] [Google Scholar]
- 23.Stadtfeld M, Brennand K, Hochedlinger K. Reprogramming of Pancreatic ß Cells into Induced Pluripotent Stem Cells. Curr Biol. 2008;18:1–11. doi: 10.1016/j.cub.2008.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhao HX, Li Y, Jin HF, Xie L, Liu C, Jiang F. et al. Rapid and efficient reprogramming of human amnion-derived cells into pluripotency by three factors OCT4/SOX2/NANOG. Differentiation. 2010;80:123–129. doi: 10.1016/j.diff.2010.03.002. [DOI] [PubMed] [Google Scholar]
- 25.Sun N, Panetta NJ, Gupta DM, Wilson KD, Lee A, Jia F. et al. Feeder-free derivation of induced pluripotent stem cells from adult human adipose stem cells. Proc. Natl. Acad. Sci. U. S. A. 2009;106:15720–5. doi: 10.1073/pnas.0908450106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kim JB, Zaehres H, Araúzo-Bravo MJ, Schöler HR. Generation of induced pluripotent stem cells from neural stem cells. Nat. Protoc. 2009;4:1464–70. doi: 10.1038/nprot.2009.173. [DOI] [PubMed] [Google Scholar]
- 27.Ruiz S, Brennand K, Panopoulos AD, Herrerías A, Gage FH, Izpisua-Belmonte JC. High-efficient generation of induced pluripotent stem cells from human astrocytes. PLoS One. 2010;5:1–9. doi: 10.1371/journal.pone.0015526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Beltrão-Braga PCB, Pignatari GC, Maiorka PC, Oliveira NAJ, Lizier NF, Wenceslau CV. et al. Feeder-free derivation of induced pluripotent stem cells from human immature dental pulp stem cells. Cell Transplant. 2011;20:1707–19. doi: 10.3727/096368911X566235. [DOI] [PubMed] [Google Scholar]
- 29.Yan X, Qin H, Qu C, Tuan RS, Shi S, Huang GT-J. iPS Cells Reprogrammed From Human Mesenchymal-Like Stem/Progenitor Cells of Dental Tissue Origin. Stem Cells Dev. 2010;19:469–480. doi: 10.1089/scd.2009.0314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schlaeger TM, Daheron L, Brickler TR, Entwisle S, Chan K, Cianci A. et al. A comparison of non-integrating reprogramming methods. Nat. Biotechnol. 2015;33:58–63. doi: 10.1038/nbt.3070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Papaioannou VE. Ontogeny, pathology, oncology. Int. J. Dev. Biol. 1993;37:33–37. [PubMed] [Google Scholar]
- 32.Tada M, Takahama Y, Abe K, Nakatsuji N, Tada T. Nuclear reprogramming of somatic cells by in vitro hybridization with ES cells. Curr. Biol. 2001;11:1553–1558. doi: 10.1016/s0960-9822(01)00459-6. [DOI] [PubMed] [Google Scholar]
- 33.Marión RM, Strati K, Li H, Murga M, Blanco R, Ortega S. et al. A p53-mediated DNA damage response limits reprogramming to ensure iPS cell genomic integrity. Nature. 2009;460:1149–53. doi: 10.1038/nature08287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gurdon JB, Wilmut I. Nuclear transfer to eggs and oocytes. Cold Spring Harb. Perspect. Biol. 2011;3:1–14. doi: 10.1101/cshperspect.a002659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hochedlinger K, Blelloch R, Brennan C, Yamada Y, Kim M, Chin L. et al. Reprogramming of a melanoma genome by nuclear transplantation. GENES Dev. 2004;18:1875–1885. doi: 10.1101/gad.1213504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Postovit L-M, Margaryan NV, Seftor EA, Kirschmann DA, Lipavsky A, Wheaton WW. et al. Human embryonic stem cell microenvironment suppresses the tumorigenic phenotype of aggressive cancer cells. Proc. Natl. Acad. Sci. U. S. A. 2008;105:4329–34. doi: 10.1073/pnas.0800467105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Muñoz P, Iliou MS, Esteller M. Epigenetic alterations involved in cancer stem cell reprogramming. Mol. Oncol. 2012;6:620–636. doi: 10.1016/j.molonc.2012.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–74. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 39.Kreso A, Dick JE. Evolution of the cancer stem cell model. Cell Stem Cell. 2014;14:275–91. doi: 10.1016/j.stem.2014.02.006. [DOI] [PubMed] [Google Scholar]
- 40.Utikal J, Maherali N, Kulalert W, Hochedlinger K. Sox2 is dispensable for the reprogramming of melanocytes and melanoma cells into induced pluripotent stem cells. J. Cell Sci. 2009;122:3502–10. doi: 10.1242/jcs.054783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang X, Cruz FD, Terry M, Remotti F, Matushansky I. Terminal differentiation and loss of tumorigenicity of human cancers via pluripotency-based reprogramming. Oncogene. 2013;32:2249–60. doi: 10.1038/onc.2012.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chin L, Tam A, Pomerantz J, Wong M, Holash J, Bardeesy N. et al. Essential role for oncogenic Ras in tumour maintenance. Nature. 1999;400:468–472. doi: 10.1038/22788. [DOI] [PubMed] [Google Scholar]
- 43.Carette J.E, Pruszak J, Varadarajan M, Blomen VA, Gokhale S, Camargo FD. et al. Generation of iPSCs from cultured human malignant cells. Blood. 2010;115:4039–4042. doi: 10.1182/blood-2009-07-231845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kotecki M, Reddy PS, Cochran BH. Isolation and characterization of a near-haploid human cell line. Exp. Cell Res. 1999;252:273–280. doi: 10.1006/excr.1999.4656. [DOI] [PubMed] [Google Scholar]
- 45.Miyoshi N, Ishii H, Nagai K, Hoshino H, Mimori K, Tanaka F. et al. Defined factors induce reprogramming of gastrointestinal cancer cells. Proc. Natl. Acad. Sci. U. S. A. 2010;107:40–5. doi: 10.1073/pnas.0912407107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Choong PF, Teh HX, Teoh HK, Ong HK, Choo KB, Sugii S. et al. Heterogeneity of osteosarcoma cell lines led to variable responses in reprogramming. Int. J. Med. Sci. 2014;11:1154–1160. doi: 10.7150/ijms.8281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lin S-L, Chang DC, Chang-Lin S, Lin C-H, Wu DTS, Chen DT. et al. Mir-302 reprograms human skin cancer cells into a pluripotent ES-cell-like state. RNA. 2008;14:2115–24. doi: 10.1261/rna.1162708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Li Z, Bao S, Wu Q, Wang H, Eyler C, Sathornsumetee S. et al. Hypoxia-Inducible Factors Regulate Tumorigenic Capacity of Glioma Stem Cells. Cancer Cell. 2009;15:501–513. doi: 10.1016/j.ccr.2009.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mohyeldin A, Garzón-Muvdi T, Quiñones-Hinojosa A. Oxygen in stem cell biology: A critical component of the stem cell niche. Cell Stem Cell. 2010;7:150–161. doi: 10.1016/j.stem.2010.07.007. [DOI] [PubMed] [Google Scholar]
- 50.Mathieu J, Zhang Z, Zhou W, Wang AJ, Heddleston JM, Pinna CMA. et al. HIF induces human embryonic stem cell markers in cancer cells. Cancer Res. 2011;71:4640–4652. doi: 10.1158/0008-5472.CAN-10-3320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hoshino H, Nagano H, Haraguchi N, Nishikawa S, Tomokuni A, Kano Y. et al. Hypoxia and TP53 deficiency for induced pluripotent stem cell-like properties in gastrointestinal cancer. Int. J. Oncol. 2012;40:1423–1430. doi: 10.3892/ijo.2012.1346. [DOI] [PubMed] [Google Scholar]
- 52.Eser S, Schnieke A, Schneider G, Saur D. Oncogenic KRAS signalling in pancreatic cancer. Br. J. Cancer. 2014;111:1–6. doi: 10.1038/bjc.2014.215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kim J, Hoffman JP, Alpaugh RK, Rhimm AD, Reichert M, Stanger BZ. et al. An iPSC Line from Human Pancreatic Ductal Adenocarcinoma Undergoes Early to Invasive Stages of Pancreatic Cancer Progression. Cell Rep. 2013;3:2088–2099. doi: 10.1016/j.celrep.2013.05.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chen L, Kasai T, Li Y, Sugii Y, Jin G, Okada M. et al. A model of cancer stem cells derived from mouse induced pluripotent stem cells. PLoS One. 2012;7:1–9. doi: 10.1371/journal.pone.0033544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yan T, Mizutani A, Chen L, Takaki M, Hiramoto Y, Matsuda S. et al. Characterization of Cancer Stem-Like Cells Derived from Mouse Induced Pluripotent Stem Cells Transformed by Tumor-Derived Extracellular Vesicles. Journal of Cancer. 2014;5:572–584. doi: 10.7150/jca.8865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lee DF, Su J, Kim HS, Chang B, Papatsenko D, Zhao R. et al. Modeling familial cancer with induced pluripotent stem cells. Cell. 2015;161:240–254. doi: 10.1016/j.cell.2015.02.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Liras A. Future research and therapeutic applications of human stem cells: general, regulatory, and bioethical aspects. J. Transl. Med. 2010;8:131. doi: 10.1186/1479-5876-8-131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Dewi D, Ishii H, Haraguchi N, Nishikawa S, Kano Y, Fukusumi T. et al. Reprogramming of gastrointestinal cancer cells. Cancer Sci. 2012;103:393–399. doi: 10.1111/j.1349-7006.2011.02184.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Utikal J, Polo JM, Stadtfeld M, Maherali N, Kulalert W, Walsh RM. et al. Immortalization eliminates a roadblock during cellular reprogramming into iPS cells. Nature. 2009;460:1145–8. doi: 10.1038/nature08285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Haraguchi N, Ishii H, Mimori K, Tanaka F, Ohkuma M, Kim HM. et al. CD13 is a therapeutic target in human liver cancer stem cells. J Clin Invest. 2010;120:3326–39. doi: 10.1172/JCI42550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Li L, Clevers H. Coexistence of quiescent and active adult stem cells in mammals. Science. 2010;327:542–5. doi: 10.1126/science.1180794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ben-Porath I, Thomson MW, Carey VJ, Ge R, Bell GW, Regev A. et al. An embryonic stem cell-like gene expression signature in poorly differentiated aggressive human tumors. Nat. Genet. 2008;40:499–507. doi: 10.1038/ng.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tung PY, Knoepfler PS. Epigenetic mechanisms of tumorigenicity manifesting in stem cells. Oncogene. 2015;34:2288–2296. doi: 10.1038/onc.2014.172. [DOI] [PMC free article] [PubMed] [Google Scholar]