 This review opens the door to the consideration of how mechanosensing and mechanotransduction can affect the nanomechanics of the NPC.
This review opens the door to the consideration of how mechanosensing and mechanotransduction can affect the nanomechanics of the NPC.
Abstract
Recent evidence suggests that mechanical deformation of the cell nucleus regulates the nuclear import of the transcriptional activators of genes involved in primary physiological cell responses such as stem cell differentiation. In addition, this nuclear mechanosensing response is de-regulated in pathological states, such as cancer and neurodegeneration. One hypothesis that could greatly advance the field is that the deformation of the nuclear envelope activates nuclear pore complexes through a direct mechanical link. The understanding of this possible mechanism for nuclear pore complex stretch-activation entails studying the mechanical connection of this complex to the nuclear envelope at the nanoscale. The nanomechanics of the nuclear pore complex is thus emerging as a novel research field, bridging nanoscience with nanotechnology. This review examines the frontier of research methodologies that are potentially useful for building a computational model of this interaction. This includes, for example, electron tomography to assess the geometrical features of the nuclear pore complex and nanoindentation to estimate its mechanical properties and that of the nuclear envelope. In order to summarize the state-of-the-art and perspectives in the field of NPC nanomechanics, this review covers highly interdisciplinary experimental and theoretical research methodologies pertaining to the fields of physics, chemistry, biology, materials and mechanics.
Insight, innovation, integration
We explore the literature supporting the hypothesis of a relation between transport through the nuclear pore complex (NPC) and mechanical forces that it experiences from the surroundings, providing insights into a possible mechanism of NPC stretch-activation. The frontier technology for nuclear pore complex characterization together with computational simulations would be a powerful tool to interpret research into a possible mechanism of nuclear pore stretch activation. We demonstrate integration of technology and biology regarding: (1) characterization techniques for the nanostructure of the NPC and the assembly of the nuclear envelope/lamina/NPC; (2) techniques to obtain the pore architecture and boundary conditions for numerical analysis; and (3) modelling techniques of the relationship between nucleocytoplasmic transport and the mechanical forces transmitted on the NPC.
Cell responses are based on biochemical signals, which enable structural internal changes such as cytoskeletal remodeling, contraction and stretching. The cell's ability to feel external stimuli and transform them into internal chemical reactions is known as mechanosensing and mechanotransduction, respectively.1–4 A recently published review5 provides a highly detailed history of scientific publications regarding the mechanical interaction of cells with their microenvironment under physiological conditions, mostly from an experimental/microstructural point of view. However, the work also reports on efforts linking the observed microstructural aspects of the cell with computational models to describe cell behavior.6–9 Similarly to the physiological case, the appearance of cellular pathological processes and diseases10–12 can also be linked to external stimuli when they have a negative impact on cell functions.13
In eukaryotic cells, nuclear pore complexes (NPCs) are the gates through which molecular exchange and genetic transport between the cytoplasm and the nucleus take place (Fig. 1). The NPC literally pierces the nuclear envelope (NE) of the nucleus allowing the exchange of molecules between the nucleus and the cytoplasm. The exchange of molecules and genetic information through the nuclear envelope, and how this affects cell differentiation, adaptability, and also disease, depend on how this trafficking takes place through the NPC. Both chemical and mechanical factors are involved in cell motility and remodeling, and such responses are led by the response of the NPC in regulating the transport of signaling molecules between the nucleus and the cytoplasm. The question as to how the NPC guides this molecular exchange is still unanswered. However, over the last few years researchers have made major breakthroughs in the fields of nanoscale imaging and mechanical characterization in terms of understanding the main geometrical and structural features of the NPC. On the other hand, the mechanical behavior of the NPC and the mechanisms by which this behavior can affect nucleocytoplasmic transport remain poorly understood. The new frontier techniques available today, in both experimental and computational fields, are potentially useful for building a computational model of the mechanical behavior of the NPC in response to nuclear envelope stretching.14–16 This review aims to highlight these advances and their potential application in order to unveil the nanomechanics of the NPC.
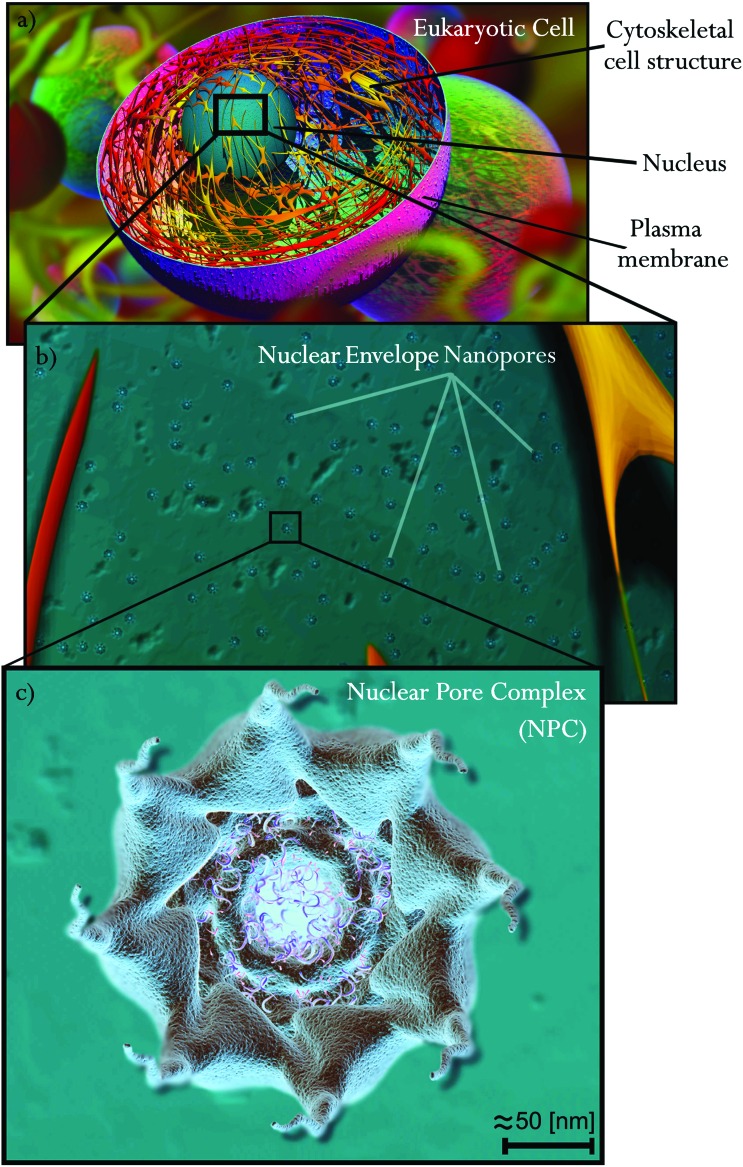
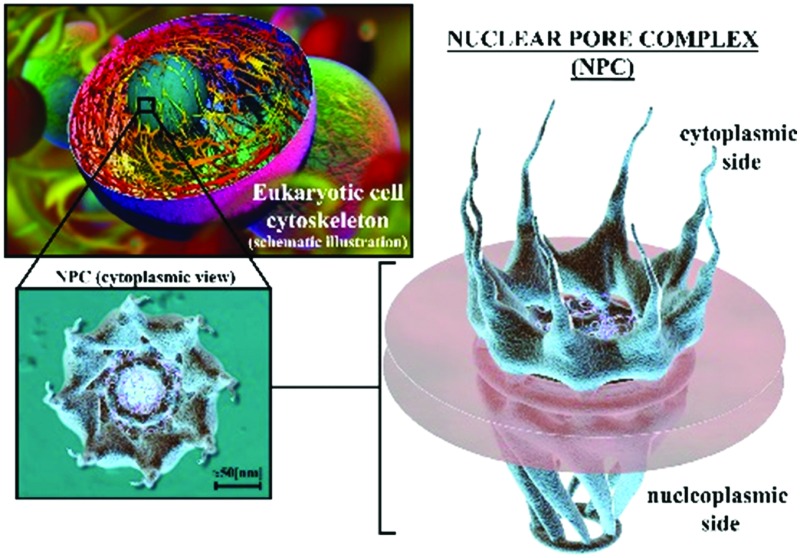
Fig. 1. 3D rendered illustration of a eukaryotic cell and its NPC distribution in the NE. (a) Cytoskeletal structure. (b) Distributed nanopores in the nuclear envelope, zoom-in of a eukaryotic cell, and a cytoplasmic side view. (c) Zoom-in of one of the 8-fold rotational symmetric nanopores; the nuclear pore complex, represented in an axial direction (cytoplasmic side).
The review is organized as follows. In Section 1, “Geometry and structure of the nuclear pore complex”, we focus on the current knowledge of the NPC architecture and geometry, and the most advanced imaging techniques used for accurately describing these features. Section 2, entitled “Chemo-mechanics of the nuclear pore complex”, describes the multiscale and multiphysics nature of the relationship between cell deformation and nucleocytoplasmic transport. Section 3, “Mechanical properties of the nuclear pore complex: modeling at multiple scales, from coarse-grained to atomistic scales”, describes the main efforts concentrated on characterizing the material properties of the NE–NPC assembly, the recent contributions to understanding the mechanical behavior of the NPC, and its role in the regulation of the molecular exchange through the NPC. At the end of Section 3 there is a summary of the main advances in the computer simulation of NPC mechanics. Section 4, “Conclusions and perspectives”, provides our concluding remarks and perspectives for studying the dependence of the NPC on mechanical stimuli, with an emphasis on the role that computational modeling will play in this regard.
1. Geometry and structure of the nuclear pore complex
This section summarizes the main advances regarding the mechanical dependency of the nucleoplasmic exchange of the NPC. Particular emphasis is placed on the mechanical factors affecting the transport of solutes, and on the visualization and computational techniques used to study this process. The term pore complex was introduced in ref. 17 to define the nucleocytoplasmic structure channel in animal cells visualized using an electron microscope (EM) and with an NPC diameter average of 100 nm and an inner-to-outer membrane separation between 20 and 30 nm. The molecular structure of the NPC and how this influences nucleocytoplasmic transport is a very active field of research. Nucleoporins that build the structure of the pores also participate in NPC activity in different ways depending above all on their location within the NPC structure. In this regard, important progress has been recently made in understanding the composite structure and functionality of the NPCs.18–20 These studies demonstrate similarities in the higher-order structure of the inner and outer rings of the NPC,20 but also show that the structure of the molecular machines within the NPC can undergo compositional arrangements as a function of cell types.18 A more recent study has performed a reconstruction of the composite structure for the entire NPC symmetric core using combined bottom-up and top-down approaches.21–23 The results of this study show a high degree of symmetry in the NPC structure, which provides the basis to explain how a limited vocabulary of proteins can generate such a large macromolecular structure. The thickness of a single NPC has been demonstrated to be less than 180 nm along its maximum length (perpendicular to the NE24–26). Therefore, visualization techniques at the nanoscale resolution are required in order to study how the structures of the nucleoporins and the NPC change under the effect of external chemo-mechanical factors.
Advances in imaging techniques have now made it possible to visualize the NPC structure at the nanoscale, in addition to providing important insights into nucleocytoplasmic transport. Recent studies have reported similar dimensions of the NPC structure, with a maximum external diameter of the nucleoporin–NE connector of 125 nm for the Dictyostelium discoideum nuclei, and 114 nm for the hNPC (human nuclear pore complex), respectively.24,27 These results are consistent with other studies.25 New imaging techniques and image acquisition protocols are constantly being developed in order to improve the accuracy of the image, thus leading to better characterization of the NPC structure, and nucleocytoplasmic cargo transport. In the reviews in ref. 28 and 29, atomic force microscopy/scanning force microscopy (AFM/SFM) are highlighted as useful techniques to analyze changes in the NPC shape in relation to different functional states. These reviews mention the work in ref. 30 that demonstrates the potential of using SFM to analyze structural changes in native NPCs and the central channel, according to different calcium-rated environments. Similarly,31 we can use AFM and energy-filtered transmission electron microscopy (EFTEM)25 on the cytoplasmic face of the NPC in an Xenopus laevis oocyte to analyze the occlusion of the free and passive diffusion of small and intermediate-sized molecules across the central channel under different calcium depletion conditions. One of the first studies on the characterization of the mechanical behavior of NPCs was performed in ref. 14. In this work, the authors use AFM to both develop NPC density maps and to measure the stiffness of the NPC. Furthermore, in ref. 91 the authors show the first experimental work that reveals FG Nup conformational dynamics inside the native NPC by high-speed atomic force microscopy. Field emission scanning electron microscopy (FE-SEM), a variety of electron microscopy, was used in ref. 32 to study the overall structure of NPCs, and they were able to identify its main parts.
Transmission electron microscopy (TEM) along with cryogenic samples (cryo-electron microscopy and tomography, Cryo-ET) has become a very useful tool to obtain ultra high-resolution images of the general structure of NPCs in their almost native environment. Using this technique,33 the authors showed electron micrographs of Xenopus NPCs with which it was possible to create a 3D reconstruction of the NPCs. Their results were very similar to the 3D reconstructions obtained using Cryo-ET enhanced with advanced image processing techniques.34 In this regard,34 the authors describe the NPC structure of Dictyostelium discoideum during cargo (labeled with gold nanoparticles) translocation along the channel. Cryo-ET has also been key in identifying different nucleoporins and structural arrangements of the NPCs in different states and cells.25,27,35,36 The advent of super-resolution fluorescence microscopy techniques such as direct stochastic optical reconstruction microscopy (dSTORM) has enabled the position of proteins of nuclear complexes in an isolated nucleus to be mapped. A combined SEM and dSTORM study was used to generate a super-resolution protein map of the NPCs in a Xenopus laevis oocyte NE.37 In addition, the work in ref. 38 shows an analysis of the structure of the nuclear pore complex scaffold by the use of stochastic super-resolution microscopy in combination with single-particle averaging. In addition to the image-based structural analysis techniques used for the study and visualization of pore complexes, single molecule fluorescence microscopy can be used to perform in vivo studies of the nucleocytoplasmic transport of cargos and smaller molecules by passive diffusion through a single pore.39–41
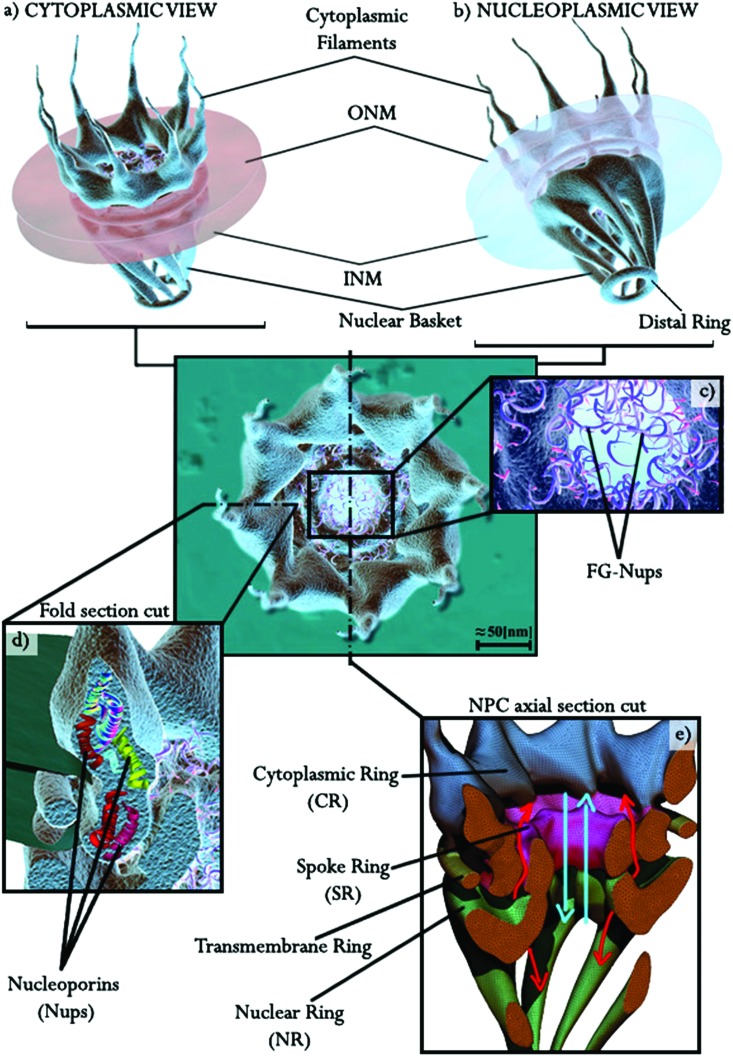
The modern visualization techniques and imaging protocols detailed above provide insights into the molecular structure of the NPCs. As previously mentioned, the NPC (Fig. 2) is a multiprotein structure embedded in the NE that connects the outer nuclear membrane with the inner nuclear membrane. It is mainly made up of three coaxial 8-fold rotational symmetric rings assembled together,24,25,33,42 see Fig. 1c for an axial view of the cytoplasmic side, and Fig. 2a and b for 3D illustrations of the NPC from the cytoplasmic and nucleoplasmic sides, respectively. This 8-fold rotational symmetry can present, in some cases, 9 or 10-fold variations which lead to changes in the dimensions of the NPC central channel and in NPC structural dimensions.43 Such modifications could be caused by local NE inhomogeneity as suggested in ref. 37. The three main rings of the NPC (see Fig. 2) are differentiated by their spatial location: (i) the spoke ring, (ii) the cytoplasmic ring, and (iii) the nuclear ring. The spoke ring serves as the junction/scaffold between the outer nuclear membrane and the inner nuclear membrane. Initial insights into the connection between the NE and the NPC are provided in ref. 33 and 35. A more recent work developed at the molecular resolution showed a more detailed nucleoporin connection between the NPC and the NE.27,44 The cytoplasmic ring, which is located on the cytoplasm side of the NPC, is connected directly to the spoke ring and also weakly connected to the outer nuclear membrane. The cytoplasmic ring works as the initial filter of the NPC for molecules and cargos with the help of the cytoplasmic filaments that serve as cargo receptors. The nuclear ring is submerged in the nucleoplasm and is directly connected with the spoke ring as in the case of the cytoplasmic ring. A specific structure of filaments of 10 nm diameter, known as the nuclear basket, grows from the nuclear ring and ends in a smaller ring called the distal ring.32 Around 30 different groups of nucleoporin repetitions can be found in the NPC.24,45 For detailed descriptions of the most important breakthroughs in the last few years, see reviews in ref. 44 and 46–50 and the papers in ref. 21, 27, 45, 51 and 52. These studies provide a thorough overview of the molecular arrangements of the most important Nups according to their main tasks, both structural and translocational. In particular, the review in ref. 47 gives a very detailed description of the NPC structure from the macro to the atomic level.
Fig. 2. The nuclear pore complex. (a) Cytoplasmic and (b) nucleoplasmic sides. (c) Phenylalanine–glycine (FG) nucleoporin filament illustration of the central channel. (d) A section cut of one symmetric fold and illustration of the nucleoporins (Nups) that form the NPC. (e) An axial/vertical section cut of the NPC. The cytoplasmic ring (CR) in blue is directly connected to the cytoplasmic filaments and the central ring/spoke ring (SR) is shown in purple, which connects to the nucleoplasmic ring (NR) shown in green with the nuclear basket filaments. The translocation paths are represented in light blue (the central channel) and red (two of the secondary channels for passive diffusion).
Differences in the structure and diameter of the transport channels are also reported in ref. 26. In this study, the authors show an ultra high-resolution 3D reconstruction of the X. laevis oocytes NPC with strong differences in the nucleoporin arrangement and structure due to transcription inhibition induction. In their work, X. laevis oocytes were treated with Actinomycin D leading above all to changes in the central channel ring (the internal part of the spoke ring) and the nuclear ring. These changes caused modifications in both the central channel diameter and the secondary channels, which the authors demonstrated to be around the central channel ring, and of a smaller diameter. There are thus still a number of unanswered questions regarding the NPC. For instance, the role of the central plug/transporter24 was highly unclear until 3D reconstructed Cryo-Electron Tomography (Cryo-ET) images demonstrated that the central plug/transporter is related to cargo translocation instead of a proper structural part belonging to the NPC spoke ring.34
2. Chemo-mechanics of the nuclear pore complex
The NE separates the cytoplasmic and nucleoplasmic environments and acts as the selective permeable membrane in the exchange of nanoconstituents and genetic information.53 Thus, the NE is of vital importance since such exchange selectivity regulates cell chemo-mechanical responses, adaptability, and transformations according to its particular cell function as part of a complex multicellular organism. Deformation of the plasma membrane of the eukaryotic cell, caused by cell contraction when interacting with the extracellular matrix, induces large deformations in the cell cytoskeleton (Fig. 1a), and the nuclear lamina to which the cytoskeleton is connected. This then deforms the bilayered nuclear envelope, connected to the nuclear lamina from the nuclear side.54,55 The layer of the nucleus in contact with the cytoplasm is known as the outer nuclear membrane, whereas the layer on the nuclear side is known as the inner nuclear membrane (Fig. 2a). This deformation mechanism suggests that the NE is subjected to large deformations due to external factors.56,57 Two recent studies have shown that the large nuclear deformation induced during migration may lead to a rupture of the nuclear envelope that provokes nucleocytoplasmic leakage and subsequent DNA damage leading to cell death.58,59
This perviousness of the NE is possible thanks to the presence of nanopores, called nuclear pore complexes (NPCs), distributed along the NE, which are responsible for selecting which molecules and cargos are allowed to pass through the NE60 (Fig. 1b and c). These NPCs (Fig. 1c and 2)28,41,46 are made up of a large number of proteins called nucleoporins45 (Fig. 2d). These nucleoporins form a structure that acts as: (i) a mechanical scaffold between the inner and outer nuclear membranes of the NE by joining both of them34,61 and (ii) a selective filter for the import–export transport of molecules between the nucleus and the cytoplasm.40 This transport is dependent on the size of the molecules to be exchanged through the NPC. The transport of small molecules normally occurs via passive diffusion, whereas larger molecules are actively transported cargos.39,62
The molecular exchange between the nucleus and cytoplasm, and the role of the NPC in nucleocytoplasmic transport, has gained considerable attention in the last decade. Along the NPC, there are two different types of channels where the molecular exchange takes place, see Fig. 2e: (i) the main central channel controlled by phenylalanine–glycine repeats nucleoporins (Fig. 2c) that “hook” both small molecules and cargos,34 and serves as the main permeability barrier, and (ii) a group of secondary channels around the central channel. The role of the permeability barrier played by the nucleoporins has been demonstrated in a number of recently published works.28,40,50,60,63 In this regard, the reviews in ref. 64 and 65 and the work in ref. 66 present a detailed description of the cargo transport (import–export of large molecules) through the NPC in association with phenylalanine–glycine–nucleoporins (FG–nucleoporins). The authors of ref. 40 proposed a brush-like mechanism as a cargo selective barrier of the central channel in which the density of phenyl–glycine–repeat nucleoporins acts as the barrier by reducing the effective space within the central pore. The strong influence of the FG-repeats, and therefore the permeability barrier of the central channel, is also studied in ref. 67 and 68. These studies also propose a model of translocation in which the FG-repeats are incorporated as a selectivity filter that restricts passage through the central channel. In ref. 51 the authors propose a Brownian affinity gating mechanism for this nucleocytoplasmic transport. The authors suggest that the diffusive movement of FG–nucleoporins may contribute to the selective binding of macromolecules. The calcium in the cytosol has also been found to play an important role. Stoffler et al. 30 demonstrated how the NPC of Xenopus oocytes opens or closes its distal ring on the nucleoplasmic side with the absence or presence of calcium in the cytosol. Similar calcium responses of NPCs were also found in the work outlined in ref. 31. The group of secondary channels with a smaller diameter around the central channel is believed to ensure the nucleocytoplasmic exchange of small molecules and ions via passive diffusion.26,35 A more detailed description is given in Section 1.
Consequently, it seems that cell adaptability, structural changes and thus normal and abnormal cell functionality are dependent on both internal and external chemical reactions as well as mechanical factors. Therefore, the cell response is highly dependent on the reaction of the NE and the NPC to these chemomechanical stimuli and how that affects the nucleocytoplasmic exchange of solutes.
3. Mechanical properties of the nuclear pore complex: modeling at multiple scales, from coarse-grained to atomistic scales
NPCs are multi-protein assemblies (linked together with the NE and nuclear fibrous lamina) which work as a selective barrier between the nucleus and the cytoplasm. Thus, as a structural assembly, it seems reasonable to hypothesize that the deformation of the NE due to mechanosensing could affect the structure of the NPC and therefore induce translocation of molecules.69 It is known that the NE is deformed due to cell response to external stimuli, i.e. forces corresponding to the instant when mechanotransduction takes place.70 Over the last two decades, there have been several breakthroughs in understanding the influence of chemical factors on the nucleocytoplasmic transport of molecules regulated by NPCs.21,26,33,34,42,44 However, the influence of mechanical factors on this molecular exchange through the nuclear membrane is still a frontier field.14,15,71
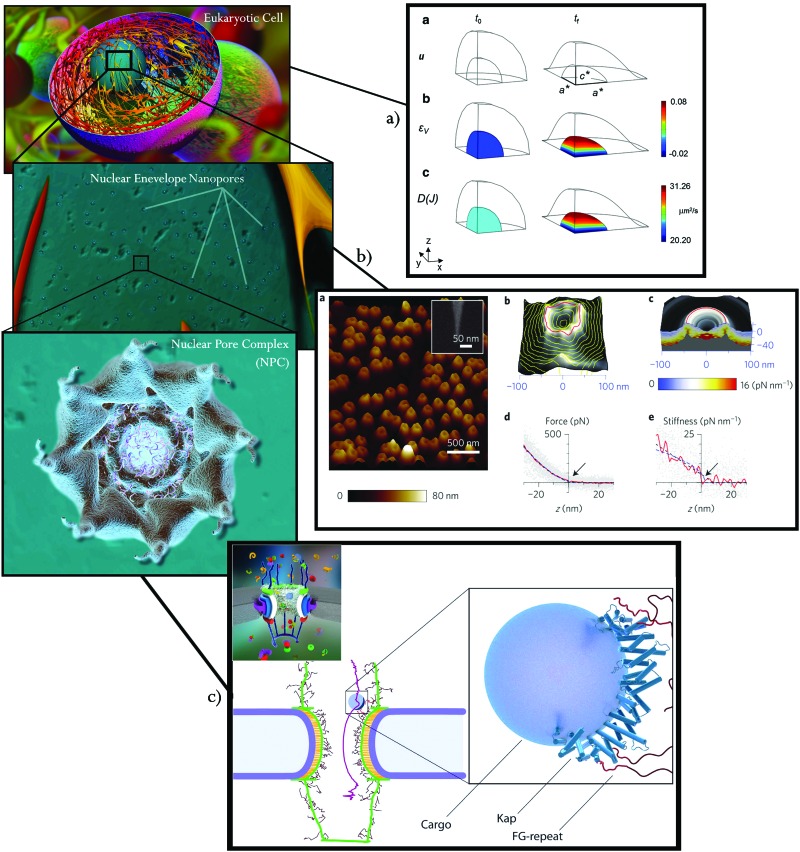
Mechanical testing has been demonstrated to be very useful for quantitative estimations of the nanomechanical properties of the pore and translocation information within the central channel as revealed in ref. 14. In this study, nanoscale stiffness topography analysis of the cytoplasmic side of the NPCs was performed using AFM nanoindentation, thus providing an estimate of the density of the NPC in the NE. For example, Fig. 3b, taken from the author's publication and corresponding to their Fig. 1, shows an image of the AFM results, from the cytoplasmic side, of an isolated nuclear envelope, the average force data measured and the stiffness calculated in the order of [pN nm–1].
Fig. 3. Published examples of experimental and numerical simulations of the mechanics of the NPC and the nucleocytoplasmic transport at different scales. (a) Finite element simulation of the passive diffusive flux from the cytoplasm to the nucleus as a function of nuclear deformation. The image was taken from ref. 70, licensed under CC BY 4.0 ; https://creativecommons.org/licences/by/4.0/ – corresponding to Fig. 6 in the scientific publication. (b) Tapping-mode AFM image of an isolated NE. The figure was taken from ref. 14, corresponding to Fig. 1 in the scientific publication. Adapted and reprinted by permission from Macmillan Publishers Ltd: [Nat. Nanotechnol.] (ref. 14), copyright (2015). (c) Numerical simulation of a cargo trajectory through an NPC. Numerical prediction of a 15 nm cargo trajectory in purple (left image) by its interaction with the FG-repeats (red), right image. The image was taken and slightly modified from ref. 71, licensed under CC BY 4.0 ; https://creativecommons.org/licences/by/4.0/ – corresponding to Fig. 1 and 7 in the scientific publication, both figures have been joined together into one single figure.
The study of the NPC mechanical/structural response also requires knowledge of its interaction with the NE/lamina assembly. In contrast to the many studies performed on the structure of NPCs and the biochemical signaling associated with NPC translocation, only a few studies have looked into the morphological changes of NPCs induced by the deformation of the NE/lamina assembly. The NE is considered to have a viscous fluid-like behavior. However, the NPCs are physically attached to the NE and the lamina network, which is directly connected to the cytoskeleton. Therefore, the stretching of the lamina by the cytoskeleton induces a deformation of the NE. In other words, the lamina gives solid–elastic material properties to the whole bilayer envelope assembly, as suggested in ref. 72, where the authors developed an AFM system to carry out force measurements through indentation on HeLa cells, in order to quantify the elastic modulus of the NE–lamina assembly. The estimated values were in the order of a few kilopascals to 10 KPa (as the authors mention), and lower values for the elastic modulus in the experiments carried out in the areas over the nucleus were found, and higher values in the peripherical areas of the cells. Aspiration experiments of the NE–NPC–lamina macrostructure73,74 suggested the existence of a mechanical coherence between the lamina network and the NPC nucleoporins at the nuclear ring level, and the solid-shear force resistance of the whole NE complex assembly.
It has been suggested that the mechanical response of the NPC affects the diffusion of cargos and smaller molecules through the NPC,71 and in particular that NPC stiffness and deformation can affect this process.15,75 However, this hypothesis is still under debate among the scientific community. The review in ref. 47 presents the state of the art regarding in silico studies of phenyl–glycine nucleoporins serving as the selective barrier of the NPC. In a series of papers, Moussavi-Baygi et al. 71,76 developed a numerical model to examine the mechanism of nucleocytoplasmic exchange. They proposed a coarse-grained model where the chemo-mechanical interactions at the atomic-scale are homogenized to the macromolecular scale to mimic the cargo transport interaction with phenyl–glycine nucleoporins. The models were validated against experimental data. Fig. 3c shows one of the images resulting from their work,71 which depicts the average path (in purple) of over 150 independent simulations of a 15 nm-sized cargo complex during the translocation process. The work in ref. 16 also shows the importance of nucleoporin-like charge regions with the use of a coarse-grained model. The molecular dynamics numerical method has been recently shown to be a potential tool to analyze and simulate these interactions at the atomistic scale as shown in ref. 77 and 78.
The energetic efficiency and stability of nucleocytoplasmic transport and NPC motion have also been analysed using static, modal, and transient analysis simulations, carried out on the 8-fold rotational symmetry of the NPC.15 The results suggest that this 8-fold distribution symmetry maximizes the bending stiffness of each fold, which is desirable for the normal functioning of the pore as the authors mention. Modal analysis simulations are useful when searching for more mechanically favorable modes of the deformation of complex protein structures, which have direct implications on selective cargo translocation.75,79 An agent-based model was also proposed in ref. 80 and 81 to analyze nucleocytoplasmic molecular diffusion through the NPC. The results suggest that that the affinity gradients in the nucleoporins directly affect the transport rate.
At a nuclear scale, the authors of ref. 70 attempt to numerically predict a strain-dependent passive diffusion of solutes between the nucleus and the cytoplasm (Fig. 3a). In their analysis, the whole nucleus is deformed and assumed as a permeable material in order to calculate the diffusive flux as a function of nuclear deformation. The results reinforce the hypothesis that under higher levels of strain of the nucleus, there is an increase in the flux of solutes through the NE, likely contributing to the nuclear import of mechanobiological transcription factors involved in stem cell differentiation.
4. Conclusions and perspectives
In this review we have briefly summarized the main aspects which, from a mechanistic perspective, play a significant role in the behavior and functioning of the NPC. We have described how the NPC is a highly complex macromolecular structure, which plays a vital role in regulating the nucleocytoplasmic exchange of constituents at the nanoscale and in gene expression. We have also described how many different studies have focused on analyzing NPC behavior, with strong insights and breakthroughs from a biochemical point of view.47,50
Finally, we have described how the mechanical modeling and simulation of NPC behaviour, which depends on nuclear stretching, is a highly challenging field which remains essentially unexplored. From a mechanobiological point of view, this is due to the highly complex nanostructure of the NPCs and the microstructural assembly of the NE–lamina–NPC. Another reason is that frontier techniques and protocols are needed to obtain experimental measurements and the quasi-realistic boundary conditions at the nanoscale that can be fed into a numerical analysis.
Today, the advent of novel ultra-high resolution imaging techniques, in addition to the continuous growth of computational power that permits the implementation of sophisticated visualization algorithms and complex numerical simulations of molecular/macromolecular structures, open the door to a whole new world of possibilities and questions. For example, how the mechanical strains that are transmitted to the NPC regulate pore activation, and thus the translocation of molecules? Several studies have started to look into these mechanisms,15,79 however there is still a long way to go before gaining a full understanding of this complex correlation. The mechanics of the NPC and its specific response to the deformation of the NE, due to the cell's mechanosensitive loop, may be key to understanding a plethora of vital processes such as cell remodeling, proliferation, mechanotransduction and cell adaptation. In addition, modeling the mechanical behavior of the NE and its failure mechanisms will help to explain the process of nuclear envelope rupture and repair during cell migration in highly confined spaces and depletion of nuclear lamins, a key mechanism behind cancer metastasis.58,59
Numerical modeling and computational simulations of the NPC structure will play a crucial role in understanding the behavior of cell mechanosensing and mechanotransduction in the future. However, current numerical techniques require a significantly large number of assumptions and simplifications in order to achieve physically significant results. In this regard, viscous fluid–solid interaction methods, highly nonlinear material behavior of the different components, and complex boundary conditions need to be formulated and applied. Furthermore, numerical models need to satisfy a number of requirements in terms of geometry, mechanical properties and boundary conditions, according to experimental observations in order to validate them, and to demonstrate the importance and viability of the in silico modeling of the NPCs. For example, in ref. 70 and 82 the authors measured different NE deformations in hundreds of differentially-spread cells. To control the nuclear shape, the authors of ref. 82 used a flat substrate with variable stiffness while the authors of ref. 70 used a 3D environment (i.e. a laser-printed “nichoid” microstructure), which allows the cells to interact with each other, as well as with the engineered extracellular matrix. Such biomimetic environments provide a close to in vivo mimicking of the nuclear deformations. These experimental studies lead the way towards a first approximation of the appropriate boundary conditions that should be used in simulations of the NPC–NE.
In addition, the use of very innovative numerical methods, such as particle-based methods,83 makes it possible to simulate the interaction of fluids/solids with different properties. This is possible by the discretization of the model as small moving particles and calculating their interaction based on the physical rules described by the user. An extensive review on the application of this technique in cells for tissue mechanics problems has recently been published.84 A multiscale finite element approach driven by the surface tension of fluids is presented in ref. 85. Here the authors show a Eulerian–Lagrangian finite element method based on a surface tension approach. This makes it possible to simulate the highly nonlinear deformation and merging of meshes as the authors show with an example of a water drop with different viscosities. Furthermore, the immersed finite element method that allows modeling of solid–fluid interactions is an attractive choice for the simulation of cell mechanics.86 Thin shell dynamic re-meshing models and textile-like mechanical behavior have also been presented for modeling highly nonlinear deformable structures.87–89 The authors present simulations of thin shells with different material properties such as textiles, plastics or metal under several stress conditions such as fractures or crumpling. Also, physics-based numerical computational algorithms that allow the recreation of the high deformations of solids, and non-linear material properties, are worth considering when highly complex interactions require realistic simulations.90 In the context of a NPC stretch-activation model, these methods could be used to better predict the interactions and responses of highly nonlinear materials and multiphase models, as in the case of a viscous-fluid material behavior of the nuclear envelope in interaction with stiffer and complex structures such as the nuclear pores in addition to a fibred anisotropic material like the lamina.
Despite the fact that these frontier modeling techniques are normally applied in very different fields of mechanobiology, all of them are based on well-established physical, mechanical and thermodynamic laws. Thus they have great potential for the prediction of NPC mechanics, responses, fluxes and the nucleocytoplasmic exchange of nanoconstituents.
Conflict of interest
The authors declare that they have no conflict of interest.
Acknowledgments
We would like to thank Dr Michele M. Nava's scientific advice and expertise in cell mechanotransduction. Politecnico di Milano supported Jose Felix Rodriguez Matas under the “Enrolment of International Faculty” program (grant no. RMJ5RIST01). This project received funding from the European Research Council (ERC) under the European Union's Horizon 2020 research and innovation programme (grant agreement no. 646990 – NICHOID). This publication only reflects the author's view and the Agency is not responsible for any use that may be made of the information it contains.
Biographies

Alberto García
Alberto García obtained his degree in Industrial Engineering (Mechanics) from the “Universidad de Oviedo” (Spain) and his PhD (European Mention) in Computational Mechanics (bioengineering) from the “Universidad de Zaragoza” (Spain). After working as a mechanical designer and as an analyst (freelance), he started as a postdoctoral researcher at “Dublin City University” (Ireland) in biomechanics and medical imaging (MRI and DTI). In 2015 he became a senior researcher at the Laboratory of Biological Structure Mechanics (LaBS) of the “Politecnico di Milano”, focusing his research on the nuclear pore complex. Recently he moved to the “Universitat Politècnica de Catalunya” as a lecturer and member of the Lacàn Group.

José F. Rodríguez Matas
José F. Rodríguez Matas received his BE degree in mechanical engineering from the Universidad Simón Bolívar, Caracas, Venezuela, in 1993, and his PhD in mechanical engineering from the University of Notre Dame, IN, in 1999. He is currently an associate professor of bioengineering in the Department of Chemistry, Materials and Chemical Engineering “Giulio Natta” at Politecnico di Milano. His research interests concern computational mechanics applied to the biomechanics of soft tissues and couple biophysical problems. He is particularly involved in nonlinear finite element applications, and the development of efficient computational tools for cardiac electrophysiology and couple problems.

Manuela T. Raimondi
Manuela T. Raimondi is an associate professor of bioengineering at Politecnico di Milano, where she teaches the course “technologies for regenerative medicine” to graduate students of the MS program in biomedical engineering. She is the founder and head of the Mechanobiology Lab and of the Interdepartmental Live Cell Imaging lab. She integrates multiphysics/multiscale computational modeling with advanced cell culture techniques, including synthetic stem cell niches and micromechanical bioreactors, for investigating basic stem cell mechanobiology. She is currently the Principal Investigator of an ERC-funded project entitled “Mechanobiology of nuclear import of transcription factors modelled within a bioengineered stem cell niche”.
References
- Kim D.-H., Chambliss A. B., Wirtz D. Soft Matter. 2013;9:5516–5523. doi: 10.1039/C3SM50798J. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D.-H., Khatau S. B., Feng Y., Walcott S., Sun S. X., Longmore G. D., Wirtz D. Sci. Rep. 2012;2:555. doi: 10.1038/srep00555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dahl K. N., Ribeiro A. J., Lammerding J. Circ. Res. 2008;102:1307–1318. doi: 10.1161/CIRCRESAHA.108.173989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dupont S., Morsut L., Aragona M., Enzo E., Giulitti S., Cordenonsi M., Zanconato F., Le Digabel J., Forcato M., Bicciato S. Nature. 2011;474:179–183. doi: 10.1038/nature10137. [DOI] [PubMed] [Google Scholar]
- Nava M. M., Raimondi M. T., Pietrabissa R. Biomech. Model. Mechanobiol. 2014;13:929–943. doi: 10.1007/s10237-014-0558-8. [DOI] [PubMed] [Google Scholar]
- Cañadas P., Wendling-Mansuy S., Isabey D. J. Biomech. Eng. 2006;128:487–495. doi: 10.1115/1.2205867. [DOI] [PubMed] [Google Scholar]
- Kardas D., Nackenhorst U., Balzani D. Biomech. Model. Mechanobiol. 2013;12:167–183. doi: 10.1007/s10237-012-0390-y. [DOI] [PubMed] [Google Scholar]
- Deshpande V. S., McMeeking R. M., Evans A. G. Proc. Natl. Acad. Sci. U. S. A. 2006;103:14015–14020. doi: 10.1073/pnas.0605837103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan J. L., Tien J., Pirone D. M., Gray D. S., Bhadriraju K., Chen C. S. Proc. Natl. Acad. Sci. U. S. A. 2003;100:1484–1489. doi: 10.1073/pnas.0235407100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Günther U., Schuppan D., Bauer M., Matthes H., Stallmach A., Schmitt-Gräff A., Riecken E.-O., Herbst H. Am. J. Pathol. 1999;155:493–503. doi: 10.1016/S0002-9440(10)65145-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gimbrone M. A., Topper J. N., Nagel T., Anderson K. R., Garcia-Cardeña G. Ann. N. Y. Acad. Sci. 2000;902:230–240. doi: 10.1111/j.1749-6632.2000.tb06318.x. [DOI] [PubMed] [Google Scholar]
- Paszek M. J., Zahir N., Johnson K. R., Lakins J. N., Rozenberg G. I., Gefen A., Reinhart-King C. A., Margulies S. S., Dembo M., Boettiger D. Cancer Cell. 2005;8:241–254. doi: 10.1016/j.ccr.2005.08.010. [DOI] [PubMed] [Google Scholar]
- Ingber D. Ann. Med. 2003;35:564–577. doi: 10.1080/07853890310016333. [DOI] [PubMed] [Google Scholar]
- Bestembayeva A., Kramer A., Labokha A. A., Osmanović D., Liashkovich I., Orlova E. V., Ford I. J., Charras G., Fassati A., Hoogenboom B. W. Nat. Nanotechnol. 2015;10:60–64. doi: 10.1038/nnano.2014.262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf C., Mofrad M. R. Biophys. J. 2008;95:2073–2085. doi: 10.1529/biophysj.108.130336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peyro M., Soheilypour M., Ghavami A., Mofrad M. R. PLoS One. 2015;10:e0143745. doi: 10.1371/journal.pone.0143745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson M. L. J. Biophys. Biochem. Cytol. 1959;6:147–156. doi: 10.1083/jcb.6.2.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ori A., Banterle N., Iskar M., Andrés-Pons A., Escher C., Bui H. K., Sparks L., Solis-Mezarino V., Rinner O., Bork P. Mol. Syst. Biol. 2013;9:648. doi: 10.1038/msb.2013.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Appen A., Kosinski J., Sparks L., Ori A., DiGuilio A. L., Vollmer B., Mackmull M.-T., Banterle N., Parca L., Kastritis P. Nature. 2015:140–143. doi: 10.1038/nature15381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosinski J., Mosalaganti S., von Appen A., Teimer R., DiGuilio A. L., Wan W., Bui K. H., Hagen W. J., Briggs J. A., Glavy J. S. Science. 2016;352:363–365. doi: 10.1126/science.aaf0643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuwe T., Correia A. R., Lin D. H., Paduch M., Lu V. T., Kossiakoff A. A., Hoelz A. Science. 2015;347:1148–1152. doi: 10.1126/science.aaa4136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuwe T., Bley C. J., Thierbach K., Petrovic S., Schilbach S., Mayo D. J., Perriches T., Rundlet E. J., Jeon Y. E., Collins L. N. Science. 2015;350:56–64. doi: 10.1126/science.aac9176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin D. H., Stuwe T., Schilbach S., Rundlet E. J., Perriches T., Mobbs G., Fan Y., Thierbach K., Huber F. M., Collins L. N. Science. 2016;352:aaf1015. doi: 10.1126/science.aaf1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck M., Förster F., Ecke M., Plitzko J. M., Melchior F., Gerisch G., Baumeister W., Medalia O. Science. 2004;306:1387–1390. doi: 10.1126/science.1104808. [DOI] [PubMed] [Google Scholar]
- Stoffler D., Feja B., Fahrenkrog B., Walz J., Typke D., Aebi U. J. Mol. Biol. 2003;328:119–130. doi: 10.1016/s0022-2836(03)00266-3. [DOI] [PubMed] [Google Scholar]
- Eibauer M., Pellanda M., Turgay Y., Dubrovsky A., Wild A., Medalia O. Nat. Commun. 2015;6:7532. doi: 10.1038/ncomms8532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bui K. H., von Appen A., DiGuilio A. L., Ori A., Sparks L., Mackmull M.-T., Bock T., Hagen W., Andrés-Pons A., Glavy J. S. Cell. 2013;155:1233–1243. doi: 10.1016/j.cell.2013.10.055. [DOI] [PubMed] [Google Scholar]
- Lim R. Y., Aebi U., Stoffler D. Chromosoma. 2006;115:15–26. doi: 10.1007/s00412-005-0037-1. [DOI] [PubMed] [Google Scholar]
- Stolz M., Stoffler D., Aebi U., Goldsbury C. J. Struct. Biol. 2000;131:171–180. doi: 10.1006/jsbi.2000.4301. [DOI] [PubMed] [Google Scholar]
- Stoffler D., Goldie K. N., Feja B., Aebi U. J. Mol. Biol. 1999;287:741–752. doi: 10.1006/jmbi.1999.2637. [DOI] [PubMed] [Google Scholar]
- Wang H., Clapham D. E. Biophys. J. 1999;77:241–247. doi: 10.1016/S0006-3495(99)76885-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldberg M. W., Allen T. D. J. Mol. Biol. 1996;257:848–865. doi: 10.1006/jmbi.1996.0206. [DOI] [PubMed] [Google Scholar]
- Akey C. W., Radermacher M. J. Cell Biol. 1993;122:1–19. doi: 10.1083/jcb.122.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck M., Lučić V., Förster F., Baumeister W., Medalia O. Nature. 2007;449:611–615. doi: 10.1038/nature06170. [DOI] [PubMed] [Google Scholar]
- Maimon T., Elad N., Dahan I., Medalia O. Structure. 2012;20:998–1006. doi: 10.1016/j.str.2012.03.025. [DOI] [PubMed] [Google Scholar]
- Rigort A., Bäuerlein F. J., Villa E., Eibauer M., Laugks T., Baumeister W., Plitzko J. M. Proc. Natl. Acad. Sci. U. S. A. 2012;109:4449–4454. doi: 10.1073/pnas.1201333109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Löschberger A., Franke C., Krohne G., van de Linde S., Sauer M. J. Cell Sci. 2014;127:4351–4355. doi: 10.1242/jcs.156620. [DOI] [PubMed] [Google Scholar]
- Szymborska A., de Marco A., Daigle N., Cordes V. C., Briggs J. A., Ellenberg J. Science. 2013;341:655–658. doi: 10.1126/science.1240672. [DOI] [PubMed] [Google Scholar]
- Kubitscheck U., Grünwald D., Hoekstra A., Rohleder D., Kues T., Siebrasse J. P., Peters R. J. Cell Biol. 2005;168:233–243. doi: 10.1083/jcb.200411005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tu L.-C., Fu G., Zilman A., Musser S. M. EMBO J. 2013;32:3220–3230. doi: 10.1038/emboj.2013.239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adams R. L., Wente S. R. Cell. 2013;152:1218–1221. doi: 10.1016/j.cell.2013.02.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akey C. W. J. Mol. Biol. 1995;248:273–293. doi: 10.1016/s0022-2836(95)80050-6. [DOI] [PubMed] [Google Scholar]
- Hinshaw J. E., Milligan R. A. J. Struct. Biol. 2003;141:259–268. doi: 10.1016/s1047-8477(02)00626-3. [DOI] [PubMed] [Google Scholar]
- Schwartz T. U. Curr. Opin. Struct. Biol. 2005;15:221–226. doi: 10.1016/j.sbi.2005.03.003. [DOI] [PubMed] [Google Scholar]
- Mi L., Goryaynov A., Lindquist A., Rexach M., Yang W. Sci. Rep. 2015;5:9372. doi: 10.1038/srep09372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suntharalingam M., Wente S. R. Dev. Cell. 2003;4:775–789. doi: 10.1016/s1534-5807(03)00162-x. [DOI] [PubMed] [Google Scholar]
- Lim R. Y., Ullman K. S., Fahrenkrog B. Int. Rev. Cell Mol. Biol. 2008;267:299–342. doi: 10.1016/S1937-6448(08)00632-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez-Martinez J., Rout M. P. Curr. Opin. Cell Biol. 2012;24:92–99. doi: 10.1016/j.ceb.2011.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bilokapic S., Schwartz T. U. Curr. Opin. Cell Biol. 2012;24:86–91. doi: 10.1016/j.ceb.2011.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck M., Glavy J. S. Nucleus. 2014;5:119–123. doi: 10.4161/nucl.28739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rout M. P., Aitchison J. D., Suprapto A., Hjertaas K., Zhao Y., Chait B. T. J. Cell Biol. 2000;148:635–652. doi: 10.1083/jcb.148.4.635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leksa N. C., Brohawn S. G., Schwartz T. U. Structure. 2009;17:1082–1091. doi: 10.1016/j.str.2009.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hetzer M. W. Cold Spring Harbor Perspect. Biol. 2010;2:a000539. doi: 10.1101/cshperspect.a000539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lombardi M. L., Jaalouk D. E., Shanahan C. M., Burke B., Roux K. J., Lammerding J. J. Biol. Chem. 2011;286:26743–26753. doi: 10.1074/jbc.M111.233700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osmanagic-Myers S., Dechat T., Foisner R. Genes Dev. 2015;29:225–237. doi: 10.1101/gad.255968.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang N., Tytell J. D., Ingber D. E. Nat. Rev. Mol. Cell Biol. 2009;10:75–82. doi: 10.1038/nrm2594. [DOI] [PubMed] [Google Scholar]
- Isermann P., Lammerding J. Curr. Biol. 2013;23:R1113–R1121. doi: 10.1016/j.cub.2013.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raab M., Gentili M., de Belly H., Thiam H., Vargas P., Jimenez A., Lautenschlaeger F., Voituriez R., Lennon-Duménil A., Manel N. Science. 2016;352:359–362. doi: 10.1126/science.aad7611. [DOI] [PubMed] [Google Scholar]
- Denais C. M., Gilbert R. M., Isermann P., McGregor A. L., te Lindert M., Weigelin B., Davidson P. M., Friedl P., Wolf K., Lammerding J. Science. 2016;352:353–358. doi: 10.1126/science.aad7297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran E. J., King M. C., Corbett A. H. Biochim. Biophys. Acta, Mol. Cell Res. 2014;1843:2784–2795. doi: 10.1016/j.bbamcr.2014.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devos D., Dokudovskaya S., Alber F., Williams R., Chait B. T., Sali A., Rout M. P. PLoS Biol. 2004;2:e380. doi: 10.1371/journal.pbio.0020380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hülsmann B. B., Labokha A. A., Görlich D. Cell. 2012;150:738–751. doi: 10.1016/j.cell.2012.07.019. [DOI] [PubMed] [Google Scholar]
- Labokha A. A., Gradmann S., Frey S., Hülsmann B. B., Urlaub H., Baldus M., Görlich D. EMBO J. 2013;32:204–218. doi: 10.1038/emboj.2012.302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jamali T., Jamali Y., Mehrbod M., Mofrad M. Int. Rev. Cell Mol. Biol. 2011;287:233–286. doi: 10.1016/B978-0-12-386043-9.00006-2. [DOI] [PubMed] [Google Scholar]
- Schmidt H. B., Görlich D. Trends Biochem. Sci. 2016;41:46–61. doi: 10.1016/j.tibs.2015.11.001. [DOI] [PubMed] [Google Scholar]
- Peyro M., Soheilypour M., Lee B., Mofrad M. Sci. Rep. 2015;5:1–14. doi: 10.1038/srep15795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters R. Traffic. 2005;6:421–427. doi: 10.1111/j.1600-0854.2005.00287.x. [DOI] [PubMed] [Google Scholar]
- Peters R. BioEssays. 2009;31:466–477. doi: 10.1002/bies.200800159. [DOI] [PubMed] [Google Scholar]
- Gupta S., Marcel N., Sarin A., Shivashankar G. PLoS One. 2012;7:e53031. doi: 10.1371/journal.pone.0053031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nava M. M., Fedele R., Raimondi M. T. Biomech. Model. Mechanobiol. 2015:1–11. doi: 10.1007/s10237-015-0737-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moussavi-Baygi R., Jamali Y., Karimi R., Mofrad M. R. PLoS Comput. Biol. 2011;7:e1002049. doi: 10.1371/journal.pcbi.1002049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yokokawa M., Takeyasu K., Yoshimura S. J. Microsc. 2008;232:82–90. doi: 10.1111/j.1365-2818.2008.02071.x. [DOI] [PubMed] [Google Scholar]
- Rowat A. C., Foster L., Nielsen M., Weiss M., Ipsen J. H. J. R. Soc., Interface. 2005;2:63–69. doi: 10.1098/rsif.2004.0022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowat A., Lammerding J., Ipsen J. H. Biophys. J. 2006;91:4649–4664. doi: 10.1529/biophysj.106.086454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lezon T. R., Sali A., Bahar I., Levitt M. PLoS Comput. Biol. 2009;5:e1000496. doi: 10.1371/journal.pcbi.1000496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moussavi-Baygi R., Jamali Y., Karimi R., Mofrad M. Biophys. J. 2011;100:1410–1419. doi: 10.1016/j.bpj.2011.01.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao C. L., Mahboobi S. H., Moussavi-Baygi R., Mofrad M. R. PLoS One. 2014;9:e93709. doi: 10.1371/journal.pone.0093709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahboobi S. H., Javanpour A. A., Mofrad M. R. PLoS One. 2015;10:e0112969. doi: 10.1371/journal.pone.0112969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D.-N., Nguyen C.-T., Bathe M. J. Struct. Biol. 2011;173:261–270. doi: 10.1016/j.jsb.2010.09.015. [DOI] [PubMed] [Google Scholar]
- Azimi M., Mofrad M. R. PLoS One. 2013;8:e81741. doi: 10.1371/journal.pone.0081741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azimi M., Bulat E., Weis K., Mofrad M. R. Mol. Biol. Cell. 2014;25:3643–3653. doi: 10.1091/mbc.E14-06-1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Discher D. E., Janmey P., Wang Y.-l. Science. 2005;310:1139–1143. doi: 10.1126/science.1116995. [DOI] [PubMed] [Google Scholar]
- Premžoe S., Tasdizen T., Bigler J., Lefohn A. and Whitaker R. T., Computer Graphics Forum, 2003, pp. 401–410. [Google Scholar]
- Van Liedekerke P., Palm M., Jagiella N., Drasdo D. Computational Particle Mechanics. 2015;2:401–444. [Google Scholar]
- Thürey N., Wojtan C., Gross M. and Turk G., ACM Transactions on Graphics (TOG), 2010, p. 48. [Google Scholar]
- Rüberg T., Aznar J. M. G. Advanced Modeling and Simulation in Engineering Sciences. 2016;3:1. doi: 10.1186/s40323-016-0077-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narain R., Samii A. and O'Brien J. F., ACM Transactions on Graphics (TOG), 2012, vol. 31, p. 152. [Google Scholar]
- Narain R., Pfaff T. and O'Brien J. F., ACM Transactions on Graphics (TOG), 2013, vol. 32, p. 51. [Google Scholar]
- Pfaff T., Narain R., de Joya J. M. and O'Brien J. F., ACM Transactions on Graphics (TOG), 2014, vol. 33, p. 110. [Google Scholar]
- Irving G., Schroeder C. and Fedkiw R., ACM Transactions on Graphics (TOG), 2007, p. 13. [Google Scholar]
- Sakiyama Y., Mazur A., Kapinos L. E., Lim R. Y. H. Nat. Nanotechnol. 2016;11:719–723. doi: 10.1038/nnano.2016.62. [DOI] [PubMed] [Google Scholar]