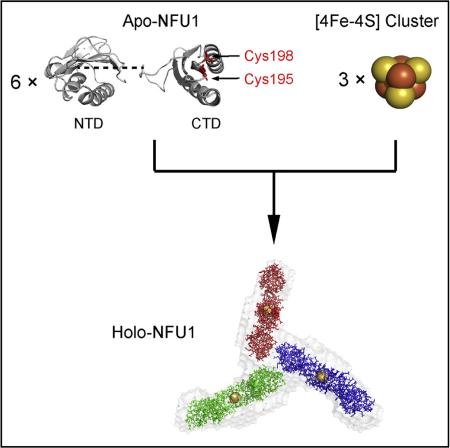
SUMMARY
Human mitochondrial NFU1 functions in the maturation of iron-sulfur proteins, and NFU1 deficiency is associated with a fatal mitochondrial disease. We determined three-dimensional structures of the N-and C-terminal domains of human NFU1 by nuclear magnetic resonance spectroscopy and used these structures along with small-angle X-ray scattering (SAXS) data to derive structural models for full-length monomeric apo-NFU1, dimeric apo-NFU1 (an artifact of intermolecular disulfide bond formation), and holo-NFUI (the [4Fe-4S] cluster-containing form of the protein). Apo-NFU1 contains two cysteine residues in its C-terminal domain, and two apo-NFU1 subunits coordinate one [4Fe-4S] cluster to form a cluster-linked dimer. Holo-NFU1 consists of a complex of three of these dimers as shown by molecular weight estimates from SAXS and size-exclusion chromatography. The SAXS-derived structural model indicates that one N-terminal region from each of the three dimers forms a tripartite interface. The activity of the holo-NFU1 preparation was verified by demonstrating its ability to activate apo-aconitase.
Graphical abstract
INTRODUCTION
Fe-S clusters are among the most ancient, yet versatile, inorganic protein co-factors (Beinert, 2000). The most common Fe-S clusters are [2Fe-2S] and [4Fe-4S] clusters, which are essential players in a variety of biological processes (Crack et al., 2012; Johnson et al., 2005; Lill, 2009). Despite their simple chemical composition, the biosynthesis of Fe-S clusters is a complex and strictly regulated process involving multiple protein components. The iron-sulfur cluster (ISC) system found in human mitochondria apparently arose from the bacterial ISC system as the result of an endosymbiotic event (Lill and Mühlenhoff, 2005, 2006). Genetic and functional defects in many of these proteins are associated with neurodegenerative, hematological, and metabolic diseases, underscoring the important role of the ISC system in human health (Maio and Rouault, 2015; Stehling et al., 2014).
NFU1 is a component of the human mitochondrial ISC system whose function is still under debate. Human NFU1 comprises two domains: an N-terminal domain (NTD) and a C-terminal domain (CTD), which contains the “CXXC” motif involved in cluster binding (Figure S1). NFU1 was initially thought to be an alternative scaffold to ISCU because it forms a [4Fe-4S] cluster when incubated with free iron and sulfide (Tong et al., 2003). Recent studies of patients afflicted by the fatal mitochondrial disease, multiple mitochondrial dysfunctions syndrome 1 (MMDS1), have shed light on the function of NFU1 (Cameron et al., 2011; Navarro-Sastre et al., 2011). The majority of patients who died before the age of 15 months carried a homozygous missense mutation on NFU1 (G193C) (Figure S1), which led to impaired NFU1 expression or function. The biochemical phenotype suggested that NFU1 is required for a subset of [4Fe-4S] proteins, including subunits of respiratory complexes I and II, and the radical S-adenosyl-methionine protein, lipoic acid synthase. NFU1 was thus characterized as a late-acting factor in the human mitochondrial ISC system that transfers [4Fe-4S] clusters to specific protein targets and facilitates their maturation. Despite its physiological importance, no three-dimensional (3D) structures have been reported for NFU1 in either its apoor holo-form.
We report here the combined use of nuclear magnetic resonance (NMR) spectroscopy and small-angle X-ray scattering (SAXS) to determine structural models for human mitochondrial NFU1. NMR spectroscopy was used to determine 3D structural models for the isolated NTD and CTD of NFU1. The structures of the domains were combined with SAXS data from a full-length construct of NFU1 in its apo- and holo-forms to derive structural models for these proteins. The holo-NFU1 preparation whose structure was investigated was demonstrated to be functionally active in the transfer of a [4Fe-4S] cluster to an acceptor protein, apo-aconitase.
RESULTS
Solution NMR Structures of the NTD and CTD Domains of Human NFU1
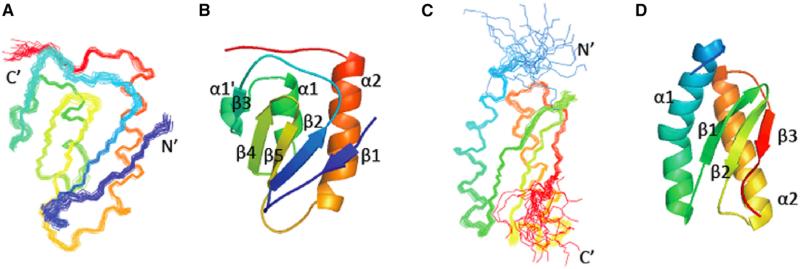
High-quality solution NMR structures were determined for both NFU1 NTD (UniProt Q9UMS0 residues 59–155) and NFU1 CTD (UniProt residues 162–247) (Figure 1 and Table 1). The NTD has a βββαββα fold with an additional short α turn α1′ between strand β3 and helix α1; the CTD has a αββαβ fold with a kink in the middle of helix α1. The β strands form an antiparallel β sheet (β1↓β2↑β5↓β4↑β3↓ for NTD and β1↑β2δβ3↑ for CTD), and the two helices pack on one side of the β sheet to form a two-layer sandwich topology.
Figure 1. Representations of the NMR Solution Structures of the Two Domains of NFU1.
(A–D) Superimposed conformers representing the solution structures of the (A) N-terminal domain (NTD) and (C) C-terminal domain (CTD). Ribbon diagrams representing the (B) NTD and (D) CTD. The residues in each domain are colored from blue at the N terminus to red at the C terminus.
Table 1.
NMR Structure Quality Statistics for NFU1 NTD and CTD Domains
| NTD | CTD | |
|---|---|---|
| NMR Distance and Dihedral Restraintsa | ||
| Distance restraints | ||
| Total NOE | 2,799 | 2,857 |
| Intra-residue | 507 | 556 |
| Inter-residue | ||
| Sequential (|i – j| = 1) | 676 | 701 |
| Medium-range (|i – j| % 4) | 493 | 711 |
| Long-range (|i – j| ≥ 5) | 1,123 | 889 |
| Intermolecular | ||
| Hydrogen bond | 50 | 46 |
| Total dihedral angle restraints | 112 | 120 |
| phi | 56 | 60 |
| psi | 56 | 60 |
| Total RDCs | ||
| Alignment medium 1b | 60 | 46 |
| Alignment medium 2b | 61 | 49 |
| Structure Statistics | ||
| Violations | ||
| RMSD of distance violation/restraintc (Å) | 0.01 | 0.01 |
| RMSD of dihedral angle violation/restraint (°) | 0.67 | 0.63 |
| Max distance restraint violationc (Å) | 0.43 | 0.34 |
| Max dihedral angle violation (°) | 7.30 | 6.7 |
| RMSD to representative structured (Å) | ||
| Heavy atoms | 0.57 ± 0.07 | 0.48 ± 0.05 |
| Backbone (N, Cα, C′) atoms | 1.15 ± 0.08 | 0.85 ± 0.07 |
| RPF scores | ||
| Recall | 0.988 | 0.959 |
| Precision | 0.969 | 0.975 |
| F measure | 0.979 | 0.967 |
| DP scores | 0.929 | 0.893 |
| Structure quality scores (raw/Z scoree) | ||
| Procheck G factor (phi/psi only) | −0.09/−0.04 | −0.05/0.12 |
| Procheck G factor (all dihedral angles) | −0.05/−0.30 | 0.04/0.24 |
| Verify3D | 0.40/−0.96 | 0.35/−1.77 |
| ProsaII (−ve) | 0.80/0.62 | 0.57/−0.33 |
| MolProbity clashscoref | 8.76/0.02 | 12.59/−0.63 |
| Ramachandran plot summary (%)f | ||
| Most favored regions | 97.4 | 98.7 |
| Allowed regions | 2.6 | 1.3 |
| Disallowed regions | 0 | 0 |
Analyzed for the 20 lowest energy refined NMR structures of each target, using PDBStat (Tejero et al., 2013), PSVS 1.4 (Bhattacharya et al., 2007), and RPF (Huang et al., 2005) software.
PEG and phage were used as alignment media 1 and 2.
Calculated by using sum over r−6.
Calculated from 20 refined structures for residues in well-defined regions that have sum of phi and psi order parameters S(phi) + S(psi) >1.8. NTD, 12–58, 61–67, 70–106; CTD, 17–48, 51–58, 64–93. RMSD values were calculated by MolMol (Koradi et al., 1996) relative to the representative structure (mediod of the ensemble).
With respect to mean and SD for a set of 252 X-ray structures with sequence lengths <500, resolution ≤ 1.80 Å, R factor ≤ 0.25 and R-free ≤ 0.28; a more positive Z score value indicates a “better-quality” score.
Statistics from MolProbity (Chen et al., 2010).
Size-Exclusion Chromatography Studies of Full-Length Apo-NFU1
The full-length NFU1 sample consisted of residues 16–254 of the 254-residue UniProt sequence. Because residues 1–9 are considered to be the transit peptide, which is cleaved off, the sequence approximated that of the mature protein. We denote this protein simply as NFU1. Under oxidizing conditions, apo-NFU1 showed both monomeric and dimeric species by size-exclusion chromatography (SEC) (Figure S2A). Non-reducing SDS-PAGE of the SEC peak corresponding to the dimer revealed bands corresponding to monomeric and dimeric apo-NFU1 (Figure S2B), whereas reducing SDS-PAGE (Figure S2C) of all fractions showed a band from monomeric apo-NFU1 alone. The results suggest that apo-NFU1 is largely monomeric in solution but can form a dimer that is partly disulfide-linked under oxidizing conditions. Similar dimer formation bridged by a disulfide bond was observed in Arabidopsis thaliana (At) CnfU, a plant Nfu-type protein (Yabe et al., 2008).
SAXS Studies of Apo-NFU1 Reveal Low-Resolution Models for Its Monomer and Dimer States
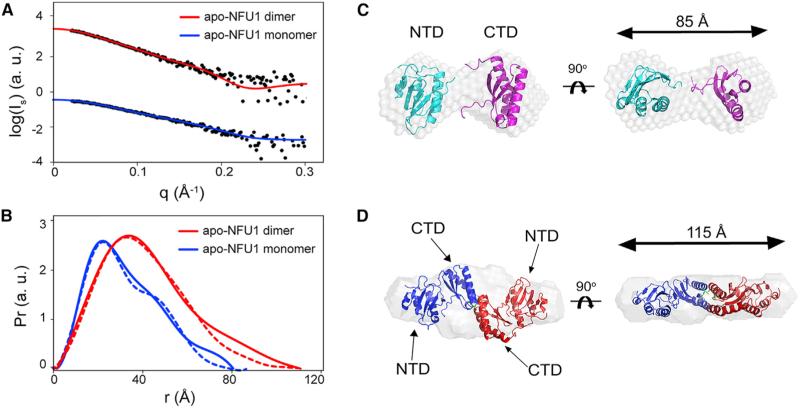
Monomeric apo-NFU1 was prepared in an anaerobic chamber to prevent disulfide bond formation, and dimeric apo-NFU1 was prepared in air and isolated by SEC. The radius of gyration (Rg) of the monomer and dimer were determined to be 25.1 and 33.2 Å, respectively; and the maximum end-to-end distances (Dmax) of the monomer and dimer were 85 and 115 Å, respectively (Figure 2 and Table 2). The molecular masses of the apo-NFU1 monomer and dimer determined by the Vc approach (Rambo and Tainer, 2013) were 28 and 57 kDa, respectively, consistent with theoretical values (Table 2). DAMMIF software (Franke and Svergun, 2009) was used to reconstruct ab initio structural models. Monomeric apo-NFU1 adopts a dumbbell-shaped structure with the NTD and CTD connected by a flexible linker (Figure 2C). We used SASREF software (Petoukhov and Svergun, 2005) to generate molecular models of apo-NFU1 consistent with the SAXS data. The solution NMR structures of the individual domains were used in the rigid body modeling simulation, which generated a model that superimposed well against the ab initio shape envelope model and also yielded a good fit to the experimental data with χ2 = 0.962 (Figures 2A–2C). The ab initio structural model also resembles the crystal structure of At CnfU (Yabe et al., 2008). The ab initio model of the disulfide-linked apo-NFU1 dimer derived from SAXS data revealed an envelope with P2 symmetry (Figure 2D) similar to the X-ray structure of the At CnfU dimer (Yabe et al., 2008). The model resulting from rigid body simulation fit the experimental SAXS data well (χ2 = 1.06), and it superimposed closely with the ab initio dummy atom model (Figures 2A and 2D).
Figure 2. SAXS Data from Apo-NFU1 Monomer and Dimer.
(A) Experimental SAXS data (black circles) collected for apo-NFU1 monomer (blue) and dimer (red) overlaid with computed scattering curves obtained from rigid body modeling.
(B) Pairwise distance distribution functions (Pr) derived from experimental SAXS data (solid lines) for apo-NFU1 dimer (red) and apo-NFU1 monomer (blue) compared with those derived from structures derived from rigid body modeling (dashed lines).
(C) Rigid body reconstructed model of monomeric apo-NFU1 superimposed on the ab initio dummy atom model reconstructed from experimental SAXS data by using the DAMMIF program from the ATSAS software suite.
(D) The rigid body reconstructed model of dimeric apo-NFU1 superimposed on the ab initio bead model reconstructed from experimental SAXS data by using the DAMMIF program.
Table 2.
Parameters Derived from SAXS and Comparison with Molecular Weight from the Chemical Structure
| Monomeric apo-NFU1 | Dimeric apo-NFU1 | Holo-NFU1 | |
|---|---|---|---|
| Radius of gyration from SAXS, Rg (Å) | 25.1 ± 0.4 | 33.2 ± 0.5 | 51.7 ± 1.2 |
| Maximum dimension from SAXS, Dmax (Å) | 82 ± 2 | 115 ± 4 | 180 ±8 |
| Molecular weight from SAXS determined by Vc (kDa) | 28 ±2 | 57 ±5 | 182 ± 15 |
| Molecular weight from chemical structure (kDa) | 27.0 | 54.0 | 164.112 |
Holo-NFU1 Is Oligomeric
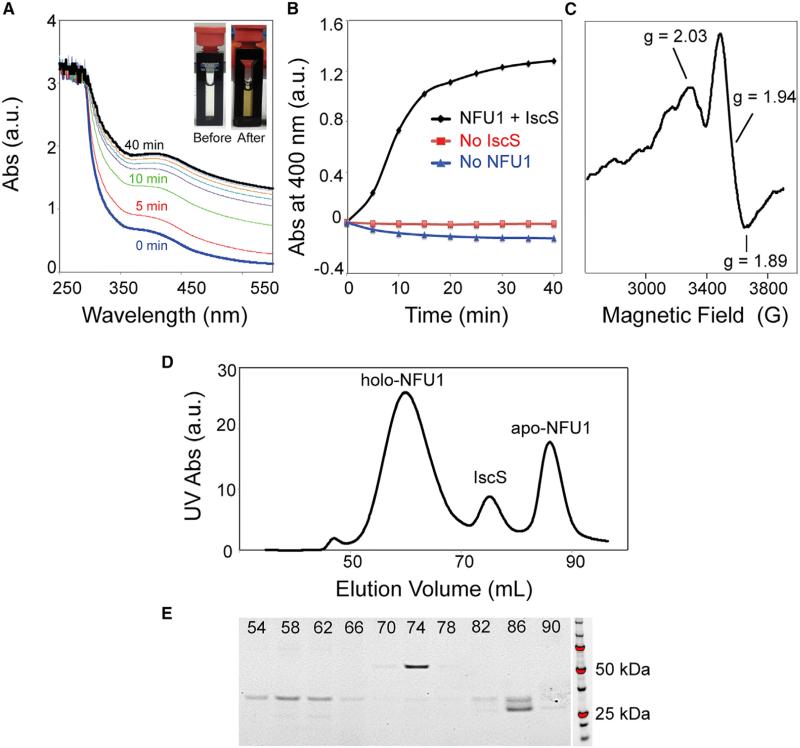
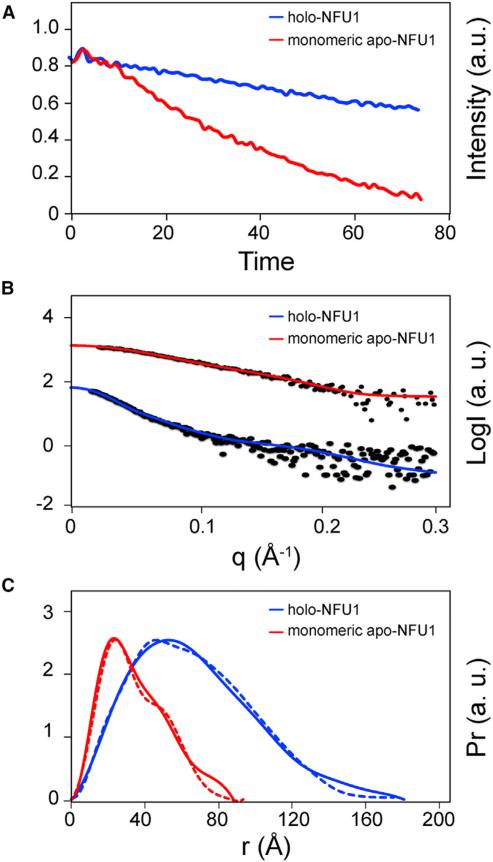
It has been shown that NFU1 can assemble a [4Fe-4S] cluster in vitro from free inorganic iron and sulfide (Tong et al., 2003). We used instead an in vitro reaction catalyzed by a 1/50 equivalent of Escherichia coli cysteine desulfurase (IscS), with Fe2(NH4)2(SO4)2 as the source of iron, L-cysteine as the source of sulfur, and DTT as the reductant. Cluster assembly was followed by optical (Figures 3A and 3B) and electron paramagnetic resonance (EPR) (Figure 3C) spectroscopies. The product of the cluster assembly reaction was subjected to SEC analysis on a Superdex 200 10/300 GL column installed in the anaerobic chamber. The SEC profile exhibited three peaks (Figure 3D), the identities of which were assayed by UV spectroscopy and SDS-PAGE (Figure 3E). The elution volume of holo-NFU1 corresponded to a homo-oligomer of about 160 kDa. The leading and training edges of the peak assigned to holo-NFU1 were re-chromatographed by SEC and found to exhibit single peaks at the same elution volume as the major peak itself (Figure S3). Data from sedimentation equilibrium were consistent with a hexamer (Figure S4). NMR diffusion experiments showed that the diffusion rate of holo-NFU1 (0.594 ± 0.011 × 10 −6 cm2/s) is much slower than that of monomeric apo-NFU1 (0.957 ± 0.021 × 10 −6 cm2/s) (Figure 4A); these diffusion rates correspond to 146 and 25.7 kDa for holo- and apo-NFU1, respectively (He and Niemeyer, 2003). SAXS data yielded a Rg = 51.7 and 25.1 Å and a Dmax = 180 and 82 Å, respectively, for holo- and monomeric apo-NFU1 (Figures 4B and 4C and Table 2). SAXS data by the Vc approach (Rambo and Tainer, 2013) yielded molecular weights of 182 and 28 kDa for holo- and apo-NFU1. The peptide chain of NFU1 contains only two cysteine residues (C195 and C198), both located in the CTD, and the cysteines from two subunits are expected to ligate one [4Fe-4S] cluster to form a dimer linked by the CTDs. Thus, these results suggest that holo-NFU1 exists as a trimer of dimers each containing a [4Fe-4S] cluster (164 kDa).
Figure 3. Production and Analysis of Holo-NFU1 in an Anaerobic Chamber.
(A) UV-visible absorption spectra of NFU1 during Fe-S cluster assembly. Spectra were taken at 5 min intervals. Inset, photograph of the cuvette before (left) and after (right) the Fe-S cluster reconstitution reaction.
(B) Time course of Fe-S cluster assembly followed at 400 nm. Control experiments carried out in the absence of IscS or NFU1 are shown in red and blue, respectively.
(C) X-band EPR spectrum of Fe-S cluster reconstituted on NFU1 reduced by one equivalent of dithionite. The g values are indicated.
(D) Size-exclusion chromatography (SEC) of the product of cluster assembly on NFU1.
(E) SDS-PAGE of SEC elution fractions of (D). The elution volume of each fraction is indicated.
Figure 4. Comparison of Monomeric Apo-NFU1 and Holo-NFU1 Produced by the Cluster Assembly Reaction.
(A) NMR diffusion results of monomeric apo-NFU1 (red) and holo-NFU1 (blue). The calculated diffusion rates for apo- and holo-NFU1 are 0.95 ± 0.021 × 10 −6 cm2/s and 0.59 ± 0.011 × 10 −6 cm2/s, respectively.
(B) Experimental SAXS data (black circles) collected for holo-NFU1 (blue) overlaid with the scattering curve calculated from rigid body modeling (see Figure 5A). For comparison, the experimental SAXS data for monomeric apo-NFU1 (red) are overlaid with the scattering curve calculated from rigid body modeling (see Figure 2A).
(C) Pairwise distance distribution function (Pr) derived from experimental SAXS data (solid line) for holo-NFU1 (blue) compared with that derived from the rigid body structural model (dashed line) (see Figure 5D). For comparison, Pr derived from experimental SAXS data (solid line) for monomeric apo-NFU1 (red) is compared with that derived from the rigid body structural model (dashed line) (See Figure 2B).
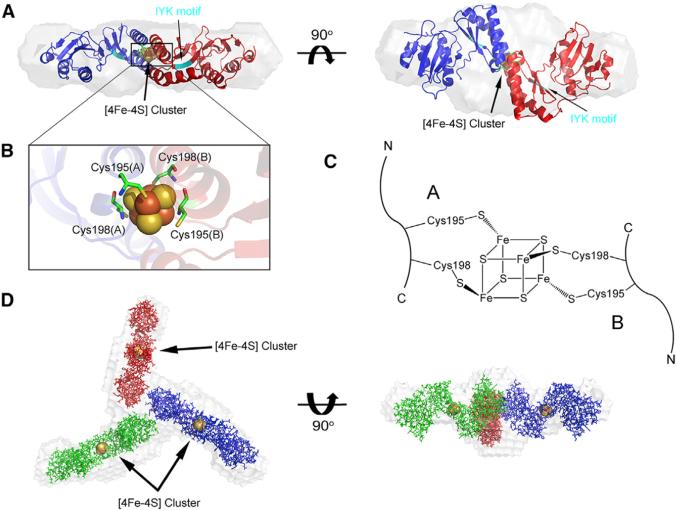
A structural model for holo-NFU1 was constructed from the combined NMR and SAXS data by use of DAMMIF software. As a first step, the [4Fe-4S] cluster was fitted into the SAXS density of dimeric apo-NFU1 (Figures 5A and 5B). Of the two possible ligation configurations, only that shown in Figure 5C fit the SAXS density. This dimeric structure was then fitted into the SAXS density for holo-NFU1, yielding a highly symmetrical (P3 symmetry) “Y” shaped structure (Figure 5D). The rigid body modeling construct agreed reasonably well with experimental SAXS data with χ2 = 1.16 (Figures 4B and 4C). Given that the CTDs are involved in Fe-S cluster binding, the tripartite interface appears to be formed by residues in the NTD.
Figure 5. Modeling of the [4Fe-4S] Cluster into the SAXS Density for holo-NFU1.
(A) Rigid body reconstructed model of the [4Fe-4S] cluster-containing dimer of NFU1 superimposed onto the ab initio dummy atom model reconstructed from the SAXS data for dimeric apo-NFU1 by using the DAMMIF program. The location of the IYF sequence motif in each subunit of the dimer is indicated in cyan, and the cyan arrow points to this sequence in one subunit.
(B) Expansion of the region of the [4Fe-4S] cluster.
(C) Configuration of cluster ligation consistent with the SAXS results. The alternative ligation pattern (i.e., that obtained by swapping of the positions of Cys195 and Cys198 in one of the chains) did not fit the dummy atom model.
(D) The rigid body reconstructed model of the trimer of cluster-containing dimers of NFU1 superimposed onto the ab initio dummy atom model reconstructed from the SAXS data for holo-NFU1 by using the DAMMIF program. The residue numbering is that of the protein construct; add 15 for that of the UniProt sequence.
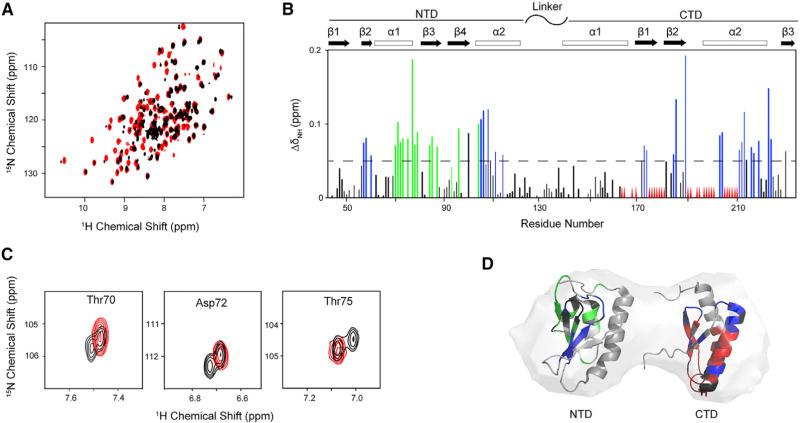
Comparison of 1H-15N TROSY-heteronuclear single-quantum correlation (HSQC) spectra of [U-15N]-apo-NFU1 (Figure 6A, red) and purified [U-15N]-holo-NFU1 (Figure 6A, black) revealed that Fe-S cluster assembly led to large chemical shift perturbations and significant line broadening. Control experiments showed that the perturbations were not a result of added of Fe2+ or cysteine desulfurase (data not shown). Careful analysis revealed that most of the NFU1 residues exhibiting large chemical shift perturbations (ΔδN-H > 0.05 ppm) are localized on the four β strands and helix α1 of the NTD (Figure 6B). Interestingly, in the NMR spectrum of holo-NFU1 a few NTD residues (T68-F73, T75, A77-A79, L84, A85, Q87, G93, and S97; numbered according to the construct—add 15 for UniProt numbering) exhibited two sets of peaks with equal peak volume (Figures 6A and 6B). Most of these residues are located in the α1 helix, and several in the β2 and β3 sheets (Figures 6B and 6D, residues colored green). Signals from three of these residues are shown as examples in Figure 6C. One set of the doubled peaks has chemical shifts similar to those of apo-NFU1, and the other set exhibits chemical shift perturbations. We assign the first set of peaks to the free NTDs and the second to the NTDs that form the tripartite interface suggested by the SAXS model for holo-NFU1. Most of the peaks exhibiting severe line broadening correspond to residues located in the CTD. This is consistent with paramagnetic relaxation enhancement of CTD residues close to the Fe-S cluster. Chemical shift perturbations of other CTD residues may be explained by conformational changes resulting from cluster binding.
Figure 6. Effect of [4Fe-4S] Cluster Formation on the NMR Spectrum of NFU1.
(A) Overlay of 1H-15N TROSY-HSQC spectra of apo-[U-15N]-NFU1 (red) and holo-[U-15N]-NFU1 after cluster assembly (black).
(B) Chemical shift (CS) perturbation of 1H-15N signals (ΔδNH) of [U-15N]-NFU1 resulting from Fe-S cluster assembly. Most of the NFU1 residues exhibiting large chemical shift changes upon cluster formation (ΔδN-H > 0.05 ppm) are localized on the four β strands and helix a1 of the NTD: S56–I60, G62, T70–F74, T76, A78–R81, A85–Q87, S96–K108, and D114. The green lines denote the residues that have two sets of peaks in the spectrum of holo-[U-15N]-NFU1 (T68-F73, T75, A77-A79, L84, A85, Q87, G93, and S96), and the red triangles denote residues whose peaks were broadened out beyond detection. CTD residues with peaks exhibiting major chemical shift changes: R168, Q171, D173, G181, D184, G185, L189, I202, L213–I217, E219, and V223). CTD residues with peaks exhibiting severe line broadening include V170, G175, D176, I178, K180, I186, K190, L191, S194, T196–C198, S201, and L205–Ile209. Chemical shift changes below the dashed line are not considered significant.
(C) Examples of residues that exhibit one peak in the NMR spectrum of apo-[U-15N]-NFU1 (red) but two sets of peaks in the NMR spectrum of holo-[U-15N]-NFU1 (black).
(D) Mapping of CS perturbation results of (B) onto the NMR structures of NTD and CTD NFU1 overplayed with the SAXS model. Color code: gray, not significantly affected (ΔδNH < 0.05 ppm); blue, significantly shifted (ΔδNH ≥ 0.05 ppm); red, broadened beyond detection; black, not assigned or overlapped. The numbering shown here is that of the protein construct itself; add 15 to yield the UniProt residue number for NFU1.
Holo-NFU1 Transfers [4Fe-4S] to Apo-Aconitase Leading to Its Activation
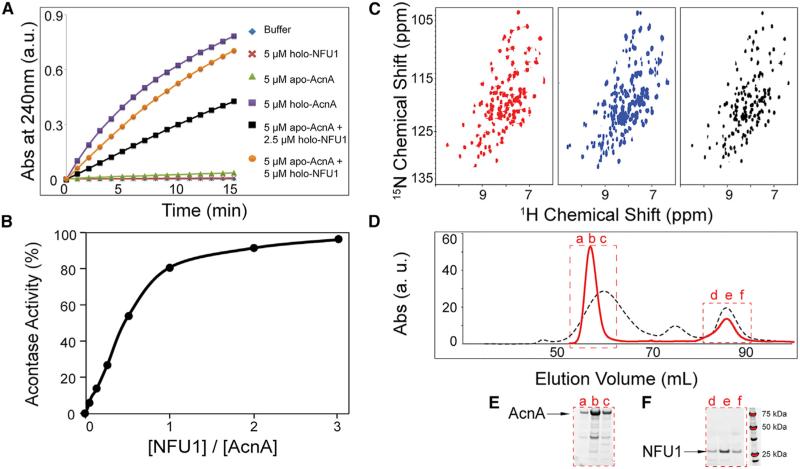
As a test of the functional activity of the holo-NFU1 preparation, we tested whether it was competent in cluster transfer to an acceptor protein, apo-aconitase. Holo-aconitase (AcnA) is a 78 kDa protein containing one [4Fe-4S] cluster that catalyzes the conversion of citrate to isocitrate via cis-aconitate in the tricarboxylic acid cycle (Beinert et al., 1996). The porcine mitochondrial AcnA used in this study shares 98% sequence identity with human mitochondrial AcnA. We found that neither holo-NFU1 nor apo-AcnA alone had AcnA activity, but that apo-AcnA became activated upon the addition of holo-NFU1 (Figure 7A). The results of these experiments showed progressive activation of apo-AcnA with maximal activity obtained with the amount of holo-NFU1 containing ~2 NFU1 chains (Figure 7B).
Figure 7. Transfer of [4Fe-4S] clusters from holo-NFU1 to apo-aconitase.
Oligomeric holo-NFU1 transfers Fe-S cluster to aconitase (AcnA) and activates AcnA activity, and NFU1 returns to its monomeric apo-form after Fe-S cluster transfer to AcnA.
(A) Time course of AcnA activation by various amounts of holo-NFU1, as followed by absorbance at 240 nm.
(B) AcnA activity as a function of the amount of added holo-NFU1 indicated in terms of the number of NFU1 chains.
(C) Left panel, 1H-15N TROSY-HSQC spectrum of [U-15N]-apo-NFU1; middle panel, 1H-15N TROSY-HSQC spectrum of [U-15N]-[4Fe-4S]-NFU1 after Fe-S cluster assembly; right panel, 1H-15N TROSY-HSQC spectrum of [U-15N]-[4Fe-4S]-NFU1 mixed with one equivalent of apo-AcnA.
(D) SEC profiles of NFU1 after Fe-S cluster assembly (black) and NFU1+AcnA after [4Fe-4S] cluster transfer from holo-NFU1 to apo-AcnA (red).
(E and F) SDS-PAGE of the SEC elution fractions denoted in (D).
We next used NMR to follow the cluster transfer reaction. The product of [4Fe-4S] cluster assembly on [U-15N]-NFU1 was mixed with equimolar (in terms of cluster sites) apo-AcnA in the anaerobic chamber and incubated for 2 hr. The 1H-15N TROSY-HSQC spectrum of the product (Figure 7C, right panel) closely resembled that of apo-[U-15N]-NFU1 (Figure 7C, left panel) and was distinct from that of holo-[U-15N]-NFU1 (Figure 7C, middle panel), confirming that the [4Fe-4S] cluster on NFU1 had been transferred and that the resulting apo-NFU1 was monomeric. The same product from the transfer was then assayed by anaerobic SEC. Two major peaks appeared in the SEC profile: as analyzed by SDS-PAGE and UV spectroscopy, the one eluting at ~57 mL contained holo-AcnA and the one eluting at ~87 mL contained apo-NFU1 (Figure 7D, red solid curve, and Figures 7E and 7F). Comparison with the SEC profile of the cluster assembly product (Figure 7D, black dashed curve) confirmed that oligomeric holo-NFU1 had dissociated into monomeric apo-NFU1 following cluster transfer.
DISCUSSION
The structures of the isolated human NFU1 NTD (PDB: 2LTM) and CTD (PDB: 2M5O) reported here are similar, respectively, to those of the Saccharomyces cerevisiae Nfu1 NTD (PDB: 2LTL) determined by the Northeast Structural Genomics Consortium and the mouse Nfu1 CTD (PDB: 1VEH), determined by the RIKEN Structural Genomics Initiative (Figure S5). The two NTD structures (PDB: 2LTM and 2LTL), with 33% sequence identity, were determined by G.L., G.T.M., and coworkers. These NTD structures are quite similar, with a Cα backbone root-mean-square deviation (RMSD) of 1.5 Å. The two CTD structures (PDB: 2M5O and 1VEH) have 91% sequence identity and a backbone Cα RMSD of 2.1 Å. Most of the structural differences between PDB: 2M5O and 1VEH are localized in the less-well-defined C-terminal backbone segment.
Our SAXS study indicates that human apo-NFU1 adopts a dumbbell shape with the NTD and CTD connected by a flexible linker (Figure 2A). The linker (G132–E150) is predicted by PONDR software (Li et al., 1999) to be highly disordered. Structural models generated by protein structure prediction software I-TASSER (Yang et al., 2015) agree well with our SAXS data with the flexible linker region G132–G150 connecting the CTD and NTD. A similar dumbbell-shaped structure with two domains connected by a flexible linker has been found in the crystal structure of At Cnfu (PDB: 2Z51), a plant Nfu-type protein (Yabe et al., 2008). Dimeric apo-NFU1, which is converted to monomers by the reductant DTT, appears to be the result of intermolecular disulfide bond formation. Notably, the crystal structure of At Cnfu shows a covalent dimer linked by a disulfide bridge (Yabe et al., 2008).
Although both [2Fe-2S] and [4Fe-4S] forms of Nfu2 from At Nfu2 have been reported (Gao et al., 2013), our results are consistent with previous findings that human holo-NFU1 assembles [4Fe-4S] clusters (Tong et al., 2003) and is involved in maturing proteins that contain [4Fe-4S] clusters (Cameron et al., 2011; Navarro-Sastre et al., 2011). We propose on the basis of the SAXS model (Figure 5D) that holo-NFU1 consists of a trimer of dimers. The cluster is bound by the two conserved cysteine residues in the CTDs of two apo-NFU1 subunits forming a dimer (Figures 5B and 5C). The NTDs of three of these dimers form a trimeric interface. The α1 helix of the NTD likely provides the major interface for trimer formation as many residues show two sets of peaks on the NMR spectrum of holo-NFU1. The results underscore the importance of both domains in forming holo-NFU1. A sequence alignment of mitochondrial NFU1 sequences (Figure S1) shows regions of high sequence identity in the NTD as well as the CTD. The CTD in general appears to be more conserved than NTD in mitochondrial NFU1 proteins; however, the NTD residues that are involved in the trimer formation are relatively more conserved especially among high eukaryotes (Figure S1). Because the disulfide-linked dimer of apo-NFU1 showed no evidence for aggregation (Figure 2), the propensity to trimerize appears to be a consequence of [4Fe-4S] cluster binding. NFU1 likely obtains its Fe-S clusters from an upstream protein (e.g., ISCU) through a cluster transfer reaction assisted by mitochondrial Hsp70-type chaperone mtHSP70 and J-type co-chaperone HSC20. The “IYK” tripeptide motif on the β1 strand (Figure 5A, residues colored cyan) of the CTD potentially is involved in interacting with HSC20 to guide the cluster transfer reaction as has been shown with other Fe-S proteins (Rouault, 2015).
Our in vitro results demonstrate that holo-NFU1 can transfer [4Fe-4S] to apo-AcnA to activate the enzyme. Upon cluster transfer, the holo-NFU1 oligomer converts to monomeric apo-NFU1. Although it was shown that patients with NFU1 defects had normal mitochondrial aconitase levels (Navarro-Sastre et al., 2011), the same study showed that yeast cells with NFU1 depletion have significantly decreased AcnA activity. In addition, two Nfu-type holo-proteins, E. coli NfuA and Staphylococcus aureus Nfu, have been shown to activate AcnA, and losses of NfuA or Nfu in both cases led to largely compromised AcnA activities in vivo (Bandyopadhyay et al., 2008; Mashruwala et al., 2015). Future studies are needed to address these discrepancies. Nonetheless, it is very likely that NFU1 specializes in the maturation of certain [4Fe-4S] proteins and requires other protein partners to be specifically targeted to proteins such as apo-lipoic acid synthase (LIAS). Possible candidates include BOLA3 and IBA57 (Cameron et al., 2011; Sheftel et al., 2012), the defects of which also cause multiple mitochondrial disorder syndromes (MMDS2 and MMDS3) with similar clinical and biochemical phenotypes as NFU1 deficiency (MMDS1) (Maio et al., 2014; Stehling et al., 2014).
The bundling of three [4Fe-4S] clusters in a single aggregate, may offer an efficient mechanism for cluster storage. In the 3D structure of holo-NFU1 as a trimer of dimers (Figure 5D) all three [4Fe-4S] clusters appear to be equally accessible to apo-protein targets. Thus, mitochondrial apo-proteins that require more than one [4Fe-4S] cluster, for example ABCE1, LIAS, NFUFS1, and NDUFS8 (Andreini et al., 2016), may be able to harvest them in a coordinated fashion.
EXPERIMENTAL PROCEDURES
Proteins and Buffers
Samples of NFU1 NTD (UniProt: Q9UMS0 residues 59–155 with N-terminal tag MGHHHHHHSH), NFU1 CTD (UniProt residues 162–247 with N-terminal tag MGHHHHHHSHM), and NFU1(59–254) containing both domains (UniProt residues 59–254 with N-terminal tag MGSSHHHHHHSSGLVPRGSH) used for studies carried out at Rutgers University were expressed and purified by standard protocols (Acton et al., 2011). The NTD and CTD each exhibited a single-site polymorphism (F118S for NTD, E168G for CTD) relative to the NFU1 sequence in UniProt. Non-isotope-labeled domains were expressed using auto-induction medium (Studier, 2005). [U-15N, 5%-13C]-, [U-15N, U-13C]-, and [U-2H, U-15N, U-13C]-enriched proteins were expressed using MJ9 minimal medium (Jansson et al., 1996). [U-15N, 5%-13C]-labeled proteins were generated for stereo-specific assignments of isopropyl methyl groups of valines and leucines (Neri et al., 1989) and for residual dipolar coupling (RDC) measurements (Tjandra et al., 1996). Samples were validated by MALDI-TOF mass spectrometry, and were >98% homogeneous based on analytical SDS-PAGE. For structural studies, the purified protein concentration was 0.4–1 mM in a buffer at pH 7.5 containing 5 mM DTT, 100 mM NaCl, 10 mM Tris-HCl 0.02% NaN3, and 5%–10% 2H2O. Some data were also obtained in a solution containing 5 mM DTT and 5% acetonitrile (pH 7.5).
IscS was prepared by a published method (Kim et al., 2012). The full-length NFU1 samples studied by NMR and SAXS at the University of Wisconsin-Madison consisted of residues 16–254 of the 254-residue UniProt sequence and had no extra residues as a tag. The NFU1 protein samples, both at natural abundance and labeled with 15N, were produced recombinantly from E. coli cells by standard protocols (Markley et al., 2009).
The HND buffer used in functional studies consisted of 50 mM HEPES (pH 7.8), 150 mM NaCl, and 5 mM DTT clarified by passage through a 0.2 μm filter (Millipore). For anaerobic experiments, the buffer was thoroughly degassed and equilibrated for at least 12 hr in an anaerobic chamber (Coy Laboratory) filled with 95% N2 and 5% H2. An O2 detector was installed inside the anaerobic chamber to ensure O2 < 1 ppm.
Production of Holo-NFU1
Holo-NFU1 was produced in the anaerobic chamber. The reaction mixture in HND buffer contained 100 μM Fe2(NH4)2(SO4)2, 2 μM E. coli cysteine desulfurase IscS, and 1 mM DTT. The reaction was initiated by adding 300 μM L-cysteine to bring the final volume to 1 mL. The reaction was carried out at 25° C in a 10 mm pathlength quartz cuvette sealed with a rubber septum. Spectra were collected on a UV-1700 UV-visible spectrophotometer (Shimadzu) equipped with a temperature-control utility. UVProbe 2.21 software (Shimadzu) was used to collect and analyze the data. To isolate holo-NFU1 protein for the NMR and cluster transfer studies, the product of the cluster assembly reaction mixture was loaded onto a Superdex 200 10/300 GL SEC column (GE Healthcare) installed inside the anaerobic chamber, and the eluted fraction containing holo-NFU1 was collected.
NMR Data Collection and Analysis
All NMR spectra were recorded at 25° C using cryogenic NMR probes. Triple resonance NMR data, simultaneous 3D 15N/13Caliphatic/13Caromatic-edited nu clear Overhauser effect spectroscopy (NOESY) (Shen et al., 2005) (mixing time: 100 ms) and 3D 13C-edited aromatic NOESY (mixing time: 100 ms) spectra were acquired on a Bruker Avance 800 MHz spectrometer. Two-dimensional constant-time 1H-13C HSQC spectra of [U-15N, 5%13C]-enriched samples were recorded with 28 and 42 ms constant-time delays on a Varian Inova 600 MHz spectrometer in order to obtain stereo-specific assignments for valines and leucines (Neri et al., 1989). 15N-1H RDCs were collected with either interleaved HSQC-TROSY or J-modulation sequences using samples aligned with phage or polyethylene-glycol-alkyl bicelles (Eletsky et al., 2012; Tjandra et al., 1996). NMR data were processed using the NMRPipe program (Delaglio et al., 1995) and analyzed using the XEASY program (Bartels et al., 1995). Spectra were referenced to external DSS.
Rotational correlation times (τc) were computed from one-dimensional (1D) 15N T1 and T2 relaxation data at 25° C (Aramini et al., 2011). τc was then calculated from the 15N T1/T2 ratio using the following approximation (Fushman et al., 1994; Kay et al., 1989):
| (Equation 1) |
where νN is the resonance frequency of 15N in Hz. Finally, values of τc were plotted against protein molecular weight and compared with data for known monomeric proteins. The τc results for the NTD and CTD (in disulfide-reduced state) domains demonstrate that under the conditions of these NMR measurements both domains are monomeric (Figure S6). For NFU1(59–254), the protein construct containing both domains, however, τc = 10.3 ns corresponded to ~17 kDa, a value lower than the expected molecular weight of 24 kDa, suggesting some dynamic flexibility between the two domains.
Chemical shift perturbations ΔδHN (absolute value) were calculated using Equation 2,
| (Equation 2) |
where ΔδH and ΔδN are the chemical shift changes in the 1H and 15N dimensions, respectively.
NMR Resonance Assignments
Sequence-specific backbone resonance assignments for NFU1 NTD, NFU1 CTD, and full-length NFU1 were determined in a largely automated fashion with the AUTOASSIGN program (Liu et al., 2005; Moseley et al., 2001). Simultaneous 3D 15N/13Caliphatic/13Caromatic-NOESY and CCH-TOCSY were then analyzed manually to obtain nearly complete side-chain assignments. Chemical shift data were deposited in the Biological Magnetic Resonance Bank (under accession codes BMRB: 18489 [NFU1 NTD], BMRB: 19068 [NFU1 CTD], and BMRB: 26801 [full-length NFU1]).
NMR Structure Determinations
NMR structure calculations followed standard protocols (Liu et al., 2005). Helical secondary structures were initially identified from chemical shift data using TALOS+ (Shen et al., 2009). Initial NOESY peak lists of expected intraresidue, sequential, and α-helical medium-range nuclear Overhauser effect (NOE) peaks were then generated from the resonance assignments. Subsequent manual peak picking was then used to identify remaining, primarily long-range NOESY cross-peaks. Backbone dihedral angle restraints were derived from the chemical shifts by TALOS+. The CYANA program (Güntert, 2004) was then used to automatically assign NOEs and to calculate structures. The automatic NOESY analysis program AUTOSTRUCTURE (Huang et al., 2006) was used in parallel to guide iterative cycles of noise/artifact peak removal, peak picking, and NOE assignments. The 20 conformers with the lowest target function value were then refined in explicit water (Linge et al., 2003) using the CNS program (Brünger et al., 1998). Structures were validated by Verify3D and PSVS (ver. 1.4) (Bhattacharya et al., 2007), ProsaII (Sippl, 1993), PROCHECK (Laskowski et al., 1993), and MolProbity, and the RPF program (Huang et al., 2012) was used to determine the global goodness-of-fit of the final structure ensembles with the NOESY peak lists. Coordinates and restraints were deposited in the PDB (under accession codes PDB: 2LTM for NTD and PDB: 2M5O for CTD).
NMR Spectroscopy for Functional Studies
The HND buffer used for NMR samples contained 8% D2O for the frequency lock. All NMR spectra were collected on 600 or 750 MHz (1H) Bruker BioSpin (Billerica) NMR spectrometers equipped with a z-gradient cryogenic probe. All sample temperatures were regulated at 25° C. NMRPipe software was used to process the raw NMR data, and NMRFAM-SPARKY software (Lee et al., 2014) was used to visualize and analyze the processed NMR data.
[U-15N]-apo-NFU1 samples (0.3 mM) in HND buffer were prepared in the anaerobic chamber (O2 < 1 ppm) to prevent cysteine oxidation. The samples were then transferred to anaerobic NMR tubes (Wilmad-Labglass) equipped with robust seals, and 1H-15N TROSY-HSQC spectra were collected.
[U-15N]-holo-NFU1 used for NMR spectroscopy was prepared as follows. The SEC fraction containing [U-15N]-holo-NFU1 from the cluster assembly reaction was dialyzed in the anaerobic chamber (O2 < 1 ppm) against HND buffer to remove extra iron and sulfide. The sample was then concentrated by using an Amicon centrifugal filter (Millipore), transferred to an anaerobic NMR tube and a 1H-15N TROSY-HSQC spectrum was collected. The protein sample remained under anaerobic environment throughout the whole process.
To study Fe-S cluster transfer from [U-15N]-holo-NFU1 to apo-AcnA. A 1:1 mix of apo-AcnA and [U-15N]-holo-NFU1 (prepared as above) was incubated in the anaerobic chamber for 2 hr and placed in an anaerobic NMR tube prior to collecting the 1H-15N TROSY-HSQC spectrum.
SAXS Data Acquisition and Analysis
All samples containing proteins or protein complexes to be used for SAXS were purified by SEC and dialyzed extensively against HND buffer with three buffer changes. The dimeric apo-NFU1 sample was prepared in air, whereas monomeric apo-NFU1, holo-NFU1, and NFU1-ISCU were prepared in the anaerobic chamber. Protein samples were clarified by passage through a 0.2 mm filter. SAXS data for each protein or protein complex were collected at three concentrations ranging from 2 to 8 mg/mL. No significant interparticle interactions were observed for any of the concentrations used in our SAXS studies. SAXS experiments were carried out on a Bruker Nanostar benchtop SAXS system (Bruker AXS) at the National Magnetic Resonance Facility at Madison (NMRFAM) equipped with a rotating anode (Cu) Turbo X-ray Source and a Vantec-2000 (2048 × 2048 pixel) detector. The sample-to-detector distance was set at ~1 m, allowing for the detection range: 0.012 > q > 0.300 A−1. Forty microliters of protein and buffer samples were loaded separately into a capillary cell with 1 mm diameter, and scattering data were collected for 3 hr with frames recorded every hour. Each frame was compared to check for radiation damage, and none was detected over the course of the experiments. The SAXS datasets were then averaged and converted to 1D scattering profiles for further analysis.
The ATSAS software suite (Petoukhov et al., 2012) was used to process the SAXS data. The Rg for each protein or protein complex was determined by using the Guinier approximation in the q range (qmax·Rg) < 1.3. Pairwise distance distribution functions (Pr) were obtained using the GNOM software (Svergun, 1992). The output from GNOM was then used in conjunction with DAMMIF (Franke and Svergun, 2009) to generate 20 independent ab initio dummy atom models to assess the molecular shape of each sample. Most of the models exhibited excellent agreement with experimental data and had a normalized spatial discrepancy (NSD) < 1. We used the SASREF software (Petoukhov and Svergun, 2005) to carry out rigid body modeling simulations, and CRYSOL software (Svergun et al., 1995) to compare the models resulting from the rigid body modeling simulations with experimental data.
Aconitase Activity Assay
Porcine mitochondrial AcnA (98% sequence identity to human mitochondrial AcnA) was purchased from Sigma-Aldrich and further purified by SEC using a Superdex 200 10/300 GL SEC column (GE Healthcare). The apo-form of AcnA was obtained by incubating holo-AcnA with EDTA and potassium ferricyanide (Unciuleac et al., 2007). Extra EDTA and potassium ferricyanide were removed by extensive dialysis against HND buffer, and apo-AcnA was further purified on a Superdex 200 10/300 GL SEC column (GE Healthcare). The AcnA activation mixtures contained 5 μM apo-AcnA and between 0 and 15 μM [4Fe-4S]-NFU1 to make the final volume 1 mL. [4Fe-4S]-NFU1 was isolated as described above, and its concentration was determined spectrophotometrically by assuming a molar extinction coefficient of ε400 = 15,000 M−1 cm−1 per [4Fe-4S]2+ cluster (Bandyopadhyay et al., 2008). The AcnA activation mixtures were incubated in the anaerobic chamber (O2 < 1 ppm) at room temperature, and the activation reaction was initiated by adding 100 μM sodium citrate to make the final volume 1 mL. The reaction was then carried out at 25° C in a 10 mm pathlength quartz cuvette sealed with a rubber septum. AcnA activity was monitored at 240 nm (following the formation of cis-aconitate from citrate) at 25° C on a UV-1700 UV-visible spectrophotometer (Shimadzu) equipped with a temperature-control utility. UVProbe 2.21 software (Shimadzu) was used to collect and analyze the data.
Mass Spectrometry
Mass spectra were obtained on a 4800 MALDI-TOF/TOF Analyzer (Applied Biosystems) at the University of Wisconsin-Madison Biotechnology Center.
Electron Paramagnetic Resonance
The EPR spectrum of holo-NFU1 sample reduced with one equivalent of dithionite was recorded at the Department of Chemistry, University of Wisconsin-Madison, on a Bruker Elexsys E500 spectrometer equipped with an ER 4122SHQ cavity and a continuous-flow liquid-helium cryostat (ESR900, Oxford Instruments) for temperature control. Acquisition parameters were as follows: sample temperature, 10 K; microwave frequency, 9.40 GHz; microwave power, 5 mW; modulation frequency, 100 kHz; modulation amplitude, 2.0 G; acquisition time constant, 163.84 ms; number of points 1,024; number of scans 8; and magnetic field range, 2,300–4,300 G.
Analytical Ultracentrifugation
The sedimentation equilibrium study was carried out in the Beckman Coulter XL-A analytical ultracentrifuge located in the Biophysics Instrumentation Facility in the Biochemistry Department, University of Wisconsin-Madison. Three different concentrations of holo-NFU1 were used: 4 mg/mL, 2 mg/mL (mid), and 1 mg/mL. The three solutions contained 20 mM HEPES buffer, (pH 7.6), 150 mM NaCl, and 3 mM Tris(2-carboxyethyl)phosphine (TCEP) as the reducing agent. The three samples were placed in each of three sectors in the ultracentrifuge. The reference sectors contained buffer. Equilibrium data were collected at 25° C at 4,800 rpm, followed by depletion at 32,000 rpm to evaluate non-sedimenting absorbance. Optical spectra of the sectors acquired during the ultracentrifugation run showed evidence of severe cluster loss from the most concentrated sample, probably as the result of its lower ratio of TCEP/holo-NFU1. Results from this sector were not used in estimating the molecular weight of holo-NFU1.
Supplementary Material
Highlights.
We determined NMR structures of the isolated N- and C-terminal domains of human NFU1
The domain structures combined with SAXS data yielded a model for full-length apo-NFU1
A holo-NFU1 preparation was shown to be functional in activating apo-aconitase
A structural model for holo-NFU1 was derived from combined NMR and SAXS data
In Brief.
Cai et al. used NMR spectroscopy and small-angle X-ray scattering data to determine the 3D structure of human mitochondrial NFU1 in its apo- and iron-sulfur cluster-containing holo-form. Apo-NFU1 is monomeric, whereas holo-NFU1 consists of a trimer of three [4Fe-4S] cluster-linked dimers.
ACKNOWLEDGMENTS
We thank other members of the Center of Eukaryotic Structure Genomics, the Mitochondrial Proteome Project, and the Northeast Structural Genomics Consortium for their support. We also thank Prof. James Prestegard, University of Georgia, for providing 15N-1H residual dipolar coupling data on NFU1 NTD and CTD samples, Dr. Marco Tonelli for assistance in data collection at the National Magnetic Resonance Facility at Madison (NMRFAM), and Dr. Darrell R. McCaslin for carrying out the sedimentation equilibrium study. Work carried out in at the University of Wisconsin-Madison was supported by US NIH grant U01GM94622 (to J.L.M.). NMR and SAXS data were collected at NMRFAM, which is supported by US NIH grant P41GM103399 (to J.L.M.). Work carried out at Rutgers University was supported by US NIH grants U54GM094597 and 1S10OD018207 (to G.T.M.).
Footnotes
ACCESSION NUMBERS
The accession numbers for the data reported in this paper are PDB: 2LTM, 2M5O; BMRB: 18489, 19068.
SUPPLEMENTAL INFORMATION
Supplemental Information includes six figures and can be found with this article online at http://dx.doi.org/10.1016/j.str.2016.08.020.
AUTHOR CONTRIBUTIONS
Conceptualization, J.L.M., G.T.M., and K.C.; Methodology, K.C., J.L.M., and G.T.M.; Investigation, K.C., G.L., R.O.F., and R.X.; Writing – Original Draft, K.C., J.L.M., and G.T.M.; Writing – Review & Editing, K.C., R.O.F., J.L.M., and G.T.M.; Funding Acquisition, J.L.M. and G.T.M.; Resources, J.L.M. and G.T.M.; Supervision, J.L.M. and G.T.M.
REFERENCES
- Acton TB, Xiao R, Anderson S, Aramini J, Buchwald WA, Ciccosanti C, Conover K, Everett J, Hamilton K, Huang YJ, et al. Preparation of protein samples for NMR structure, function, and small-molecule screening studies. Methods Enzymol. 2011;493:21–60. doi: 10.1016/B978-0-12-381274-2.00002-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreini C, Banci L, Rosato A. Exploiting bacterial operons to illuminate human iron-sulfur proteins. J. Proteome Res. 2016;15:1308–1322. doi: 10.1021/acs.jproteome.6b00045. [DOI] [PubMed] [Google Scholar]
- Aramini JM, Ma LC, Zhou L, Schauder CM, Hamilton K, Amer BR, Mack TR, Lee HW, Ciccosanti CT, Zhao L, et al. Dimer interface of the effector domain of non-structural protein 1 from influenza A virus: an interface with multiple functions. J. Biol. Chem. 2011;286:26050–26060. doi: 10.1074/jbc.M111.248765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bandyopadhyay S, Naik SG, O'Carroll IP, Huynh BH, Dean DR, Johnson MK, Dos Santos PC. A proposed role for the Azotobacter vinelandii NfuA protein as an intermediate iron-sulfur cluster carrier. J. Biol. Chem. 2008;283:14092–14099. doi: 10.1074/jbc.M709161200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartels C, Xia TH, Billeter M, Güntert P, Wüthrich K. The program XEASY for computer-supported NMR spectral-analysis of biological macromolecules. J. Biomol. NMR. 1995;5:1–10. doi: 10.1007/BF00417486. [DOI] [PubMed] [Google Scholar]
- Beinert H. Iron-sulfur proteins: ancient structures, still full of surprises. J. Biol. Inorg. Chem. 2000;5:2–15. doi: 10.1007/s007750050002. [DOI] [PubMed] [Google Scholar]
- Beinert H, Kennedy MC, Stout CD. Aconitase as iron-sulfur protein, enzyme, and iron-regulatory protein. Chem. Rev. 1996;96:2335–2373. doi: 10.1021/cr950040z. [DOI] [PubMed] [Google Scholar]
- Bhattacharya A, Tejero R, Montelione GT. Evaluating protein structures determined by structural genomics consortia. Proteins. 2007;66:778–795. doi: 10.1002/prot.21165. [DOI] [PubMed] [Google Scholar]
- Brünger AT, Adams PD, Clore GM, Delano WL, Gros P, Grossekunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, et al. Crystallography and NMR system – a new software suite for macromolecular structure determination. Acta Crystallogr. D Biol. Crystallogr. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- Cameron JM, Janer A, Levandovskiy V, Mackay N, Rouault TA, Tong WH, Ogilvie I, Shoubridge EA, Robinson BH. Mutations in iron-sulfur cluster scaffold genes NFU1 and BOLA3 cause a fatal deficiency of multiple respiratory chain and 2-oxoacid dehydrogenase enzymes. Am. J. Hum. Genet. 2011;89:486–495. doi: 10.1016/j.ajhg.2011.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen VB, Arendall WB, 3rd, Headd JJ, Keedy DA, Immormino RM, Kapral GJ, Murray LW, Richardson JS, Richardson DC. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr. 2010;66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crack JC, Green J, Thomson AJ, Le Brun NE. Iron-sulfur cluster sensor-regulators. Curr. Opin. Chem. Biol. 2012;16:35–44. doi: 10.1016/j.cbpa.2012.02.009. [DOI] [PubMed] [Google Scholar]
- Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. NMRPIPE – a multidimensional spectral processing system based on UNIX Pipes. J. Biomol. NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- Eletsky A, Jeong MY, Kim H, Lee HW, Xiao R, Pagliarini DJ, Prestegard JH, Winge DR, Montelione GT, Szyperski T. Solution NMR structure of yeast succinate dehydrogenase flavinylation factor Sdh5 reveals a putative Sdh1 binding site. Biochemistry. 2012;51:8475–8477. doi: 10.1021/bi301171u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franke D, Svergun DI. DAMMIF, a program for rapid ab-initio shape determination in small-angle scattering. J. Appl. Crystallogr. 2009;42:342–346. doi: 10.1107/S0021889809000338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fushman D, Weisemann R, Thuring H, Rüterjans H. Backbone dynamics of ribonuclease T1 and its complex with 2’-GMP studied by two-dimensional heteronuclear NMR spectroscopy. J. Biomol. NMR. 1994;4:61–78. doi: 10.1007/BF00178336. [DOI] [PubMed] [Google Scholar]
- Gao H, Subramanian S, Couturier J, Naik SG, Kim SK, Leustek T, Knaff DB, Wu HC, Vignols F, Huynh BH, et al. Arabidopsis thaliana Nfu2 accommodates [2Fe-2S] or [4Fe-4S] clusters and is competent for in vitro maturation of chloroplast [2Fe-2S] and [4Fe-4S] cluster-containing proteins. Biochemistry. 2013;52:6633–6645. doi: 10.1021/bi4007622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Güntert P. Automated NMR structure calculation with CYANA. Methods Mol. Biol. 2004;278:353–378. doi: 10.1385/1-59259-809-9:353. [DOI] [PubMed] [Google Scholar]
- He L, Niemeyer B. A novel correlation for protein diffusion coefficients based on molecular weight and radius of gyration. Biotechnol. Prog. 2003;19:544–548. doi: 10.1021/bp0256059. [DOI] [PubMed] [Google Scholar]
- Huang YJ, Powers R, Montelione GT. Protein NMR recall, precision, and F-measure scores (RPF scores): structure quality assessment measures based on information retrieval statistics. J. Am. Chem. Soc. 2005;127:1665–1674. doi: 10.1021/ja047109h. [DOI] [PubMed] [Google Scholar]
- Huang YJ, Tejero R, Powers R, Montelione GT. A topology-constrained distance network algorithm for protein structure determination from NOESY data. Proteins. 2006;62:587–603. doi: 10.1002/prot.20820. [DOI] [PubMed] [Google Scholar]
- Huang YJ, Rosato A, Singh G, Montelione GT. RPF: a quality assessment tool for protein NMR structures. Nucleic Acids Res. 2012;40:W542–W546. doi: 10.1093/nar/gks373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansson M, Li YC, Jendeberg L, Anderson S, Montelione GT, Nilsson B. High-level production of uniformly 15N- and 13C-enriched fusion proteins in Escherichia coli. J. Biomol. NMR. 1996;7:131–141. doi: 10.1007/BF00203823. [DOI] [PubMed] [Google Scholar]
- Johnson DC, Dean DR, Smith AD, Johnson MK. Structure, function, and formation of biological iron-sulfur clusters. Annu. Rev. Biochem. 2005;74:247–281. doi: 10.1146/annurev.biochem.74.082803.133518. [DOI] [PubMed] [Google Scholar]
- Kay LE, Torchia DA, Bax A. Backbone dynamics of proteins as studied by 15N inverse detected heteronuclear NMR spectroscopy: application to staphylococcal nuclease. Biochemistry. 1989;28:8972–8979. doi: 10.1021/bi00449a003. [DOI] [PubMed] [Google Scholar]
- Kim JH, Tonelli M, Markley JL. Disordered form of the scaffold protein IscU is the substrate for iron-sulfur cluster assembly on cysteine desulfurase. Proc. Natl. Acad. Sci. USA. 2012;109:454–459. doi: 10.1073/pnas.1114372109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koradi R, Billeter M, Wüthrich K. MOLMOL – a program for display and analysis of macromolecular structures. J. Mol. Graph. 1996;14:51–55. doi: 10.1016/0263-7855(96)00009-4. [DOI] [PubMed] [Google Scholar]
- Laskowski RA, MacArthur MW, Moss DS, Thornton JM. PROCHECK: a program to check the stereochemical quality of protein structures. J. Appl. Crystallogr. 1993;26:283–291. [Google Scholar]
- Lee W, Tonelli M, Markley JL. NMRFAM-SPARKY: enhanced software for biomolecular NMR spectroscopy. Bioinformatics. 2014;31:1325–1327. doi: 10.1093/bioinformatics/btu830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Romero P, Rani M, Dunker AK, Obradovic Z. Predicting protein disorder for N-, C-, and internal regions. Genome Inform. Ser. Workshop Genome Inform. 1999;10:30–40. [PubMed] [Google Scholar]
- Lill R. Function and biogenesis of iron-sulphur proteins. Nature. 2009;460:831–838. doi: 10.1038/nature08301. [DOI] [PubMed] [Google Scholar]
- Lill R, Mühlenhoff U. Iron-sulfur-protein biogenesis in eukaryotes. Trends Biochem. Sci. 2005;30:133–141. doi: 10.1016/j.tibs.2005.01.006. [DOI] [PubMed] [Google Scholar]
- Lill R, Mühlenhoff U. Iron-sulfur protein biogenesis in eukaryotes: components and mechanisms. Annu. Rev. Cell Dev. Biol. 2006;22:457–486. doi: 10.1146/annurev.cellbio.22.010305.104538. [DOI] [PubMed] [Google Scholar]
- Linge JP, Williams MA, Spronk CA, Bonvin AM, Nilges M. Refinement of protein structures in explicit solvent. Proteins. 2003;50:496–506. doi: 10.1002/prot.10299. [DOI] [PubMed] [Google Scholar]
- Liu G, Shen Y, Atreya HS, Parish D, Shao Y, Sukumaran DK, Xiao R, Yee A, Lemak A, Bhattacharya A, et al. NMR data collection and analysis protocol for high-throughput protein structure determination. Proc. Natl. Acad. Sci. USA. 2005;102:10487–10492. doi: 10.1073/pnas.0504338102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maio N, Rouault TA. Iron-sulfur cluster biogenesis in mammalian cells: new insights into the molecular mechanisms of cluster delivery. Biochim. Biophys. Acta. 2015;1853:1493–1512. doi: 10.1016/j.bbamcr.2014.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maio N, Singh A, Uhrigshardt H, Saxena N, Tong WH, Rouault TA. Cochaperone binding to LYR motifs confers specificity of iron sulfur cluster delivery. Cell Metab. 2014;19:445–457. doi: 10.1016/j.cmet.2014.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Markley JL, Aceti DJ, Bingman CA, Fox BG, Frederick RO, Makino S, Nichols KW, Phillips GN, Jr., Primm JG, Sahu SC, et al. The center for eukaryotic structural genomics. J. Struct. Funct. Genomics. 2009;10:165–179. doi: 10.1007/s10969-008-9057-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mashruwala AA, Pang YY, Rosario-Cruz Z, Chahal HK, Benson MA, Mike LA, Skaar EP, Torres VJ, Nauseef WM, Boyd JM. Nfu facilitates the maturation of iron-sulfur proteins and participates in virulence in Staphylococcus aureus. Mol. Microbiol. 2015;95:383–409. doi: 10.1111/mmi.12860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moseley HN, Monleon D, Montelione GT. Automatic determination of protein backbone resonance assignments from triple resonance nuclear magnetic resonance data. Methods Enzymol. 2001;339:91–108. doi: 10.1016/s0076-6879(01)39311-4. [DOI] [PubMed] [Google Scholar]
- Navarro-Sastre A, Tort F, Stehling O, Uzarska MA, Arranz JA, Del Toro M, Labayru MT, Landa J, Font A, Garcia-Villoria J, et al. A fatal mitochondrial disease is associated with defective NFU1 function in the maturation of a subset of mitochondrial Fe-S proteins. Am. J. Hum. Genet. 2011;89:656–667. doi: 10.1016/j.ajhg.2011.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neri D, Szyperski T, Otting G, Senn H, Wüthrich K. Stereospecific nuclear magnetic resonance assignments of the methyl groups of valine and leucine in the DNA-binding domain of the 434 repressor by biosynthetically directed fractional 13C labeling. Biochemistry. 1989;28:7510–7516. doi: 10.1021/bi00445a003. [DOI] [PubMed] [Google Scholar]
- Petoukhov MV, Svergun DI. Global rigid body modeling of macromolecular complexes against small-angle scattering data. Biophys. J. 2005;89:1237–1250. doi: 10.1529/biophysj.105.064154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petoukhov MV, Franke D, Shkumatov AV, Tria G, Kikhney AG, Gajda M, Gorba C, Mertens HD, Konarev PV, Svergun DI. New developments in the program package for small-angle scattering data analysis. J. Appl. Crystallogr. 2012;45:342–350. doi: 10.1107/S0021889812007662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rambo RP, Tainer JA. Accurate assessment of mass, models and resolution by small-angle scattering. Nature. 2013;496:477–481. doi: 10.1038/nature12070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rouault TA. Mammalian iron-sulphur proteins: novel insights into biogenesis and function. Nat. Rev. Mol. Cell Biol. 2015;16:45–55. doi: 10.1038/nrm3909. [DOI] [PubMed] [Google Scholar]
- Sheftel AD, Wilbrecht C, Stehling O, Niggemeyer B, Elsasser HP, Mühlenhoff U, Lill R. The human mitochondrial ISCA1, ISCA2, and IBA57 proteins are required for [4Fe-4S] protein maturation. Mol. Biol. Cell. 2012;23:1157–1166. doi: 10.1091/mbc.E11-09-0772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Y, Atreya HS, Liu G, Szyperski T. G-matrix Fourier transform NOESY-based protocol for high-quality protein structure determination. J. Am. Chem. Soc. 2005;127:9085–9099. doi: 10.1021/ja0501870. [DOI] [PubMed] [Google Scholar]
- Shen Y, Delaglio F, Cornilescu G, Bax A. TALOS+: a hybrid method for predicting protein backbone torsion angles from NMR chemical shifts. J. Biomol. NMR. 2009;44:213–223. doi: 10.1007/s10858-009-9333-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sippl MJ. Recognition of errors in three-dimensional structures of proteins. Proteins. 1993;17:355–362. doi: 10.1002/prot.340170404. [DOI] [PubMed] [Google Scholar]
- Stehling O, Wilbrecht C, Lill R. Mitochondrial iron-sulfur protein biogenesis and human disease. Biochimie. 2014;100:61–77. doi: 10.1016/j.biochi.2014.01.010. [DOI] [PubMed] [Google Scholar]
- Studier FW. Protein production by auto-induction in high-density shaking cultures. Protein Expr. Purif. 2005;41:207–234. doi: 10.1016/j.pep.2005.01.016. [DOI] [PubMed] [Google Scholar]
- Svergun DI. Determination of the regularization parameter in indirect-transform methods using perceptual criteria. J. Appl. Crystallogr. 1992;25:495–503. [Google Scholar]
- Svergun D, Barberato C, Koch MHJ. CRYSOL – a program to evaluate x-ray solution scattering of biological macromolecules from atomic coordinates. J. Appl. Crystallogr. 1995;28:768–773. [Google Scholar]
- Tejero R, Snyder D, Mao B, Aramini JM, Montelione GT. PDBStat: a universal restraint converter and restraint analysis software package for protein NMR. J. Biomol. NMR. 2013;56:337–351. doi: 10.1007/s10858-013-9753-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tjandra N, Grzesiek S, Bax A. Magnetic field dependence of nitrogen-proton J splittings in 15N-enriched human ubiquitin resulting from relaxation interference and residual dipolar coupling. J. Am. Chem. Soc. 1996;118:6264–6272. [Google Scholar]
- Tong WH, Jameson GNL, Huynh BH, Rouault TA. Subcellular compartmentalization of human Nfu, an iron-sulfur cluster scaffold protein, and its ability to assemble a [4Fe-4S] cluster. Proc. Natl. Acad. Sci. USA. 2003;100:9762–9767. doi: 10.1073/pnas.1732541100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unciuleac MC, Chandramouli K, Naik S, Mayer S, Huynh BH, Johnson MK, Dean DR. In vitro activation of apo-aconitase using a [4Fe-4S] cluster-loaded form of the IscU [Fe-S] cluster scaffolding protein. Biochemistry. 2007;46:6812–6821. doi: 10.1021/bi6026665. [DOI] [PubMed] [Google Scholar]
- Yabe T, Yamashita E, Kikuchi A, Morimoto K, Nakagawa A, Tsukihara T, Nakai M. Structural analysis of Arabidopsis CnfU protein: an iron-sulfur cluster biosynthetic scaffold in chloroplasts. J. Mol. Biol. 2008;381:160–173. doi: 10.1016/j.jmb.2008.05.072. [DOI] [PubMed] [Google Scholar]
- Yang J, Yan R, Roy A, Xu D, Poisson J, Zhang Y. The I-TASSER suite: protein structure and function prediction. Nat. Methods. 2015;12:7–8. doi: 10.1038/nmeth.3213. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.