Abstract
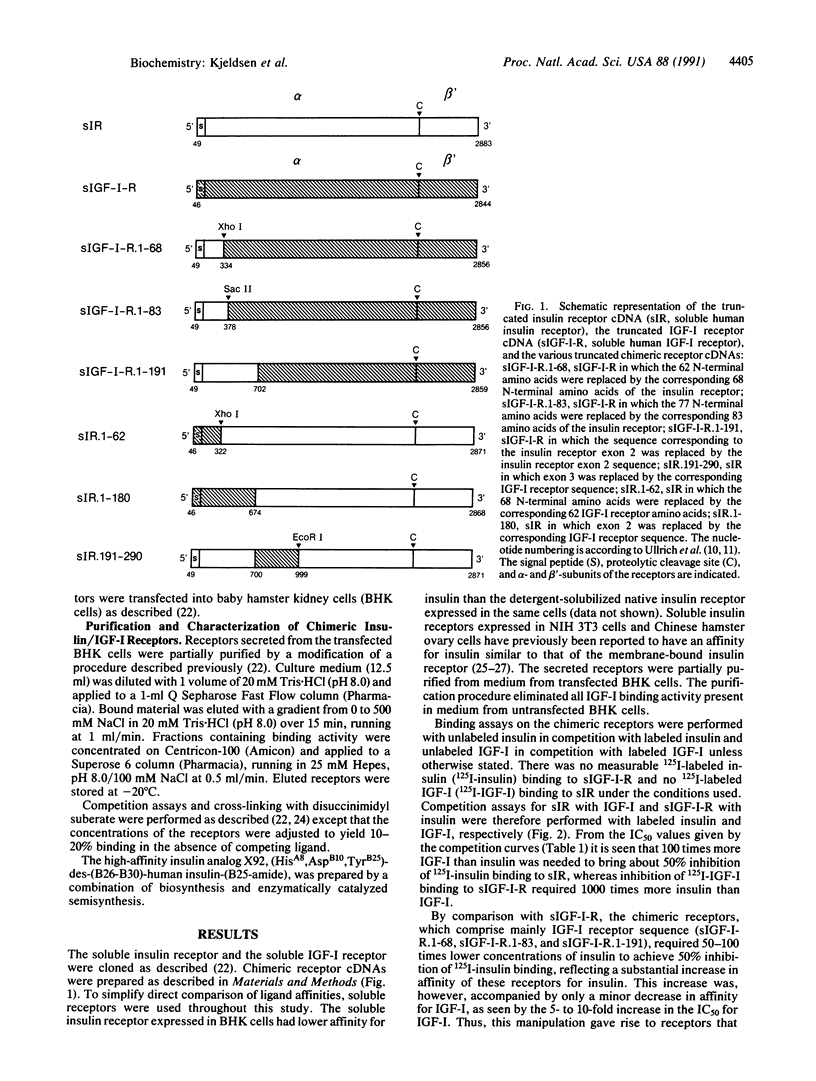
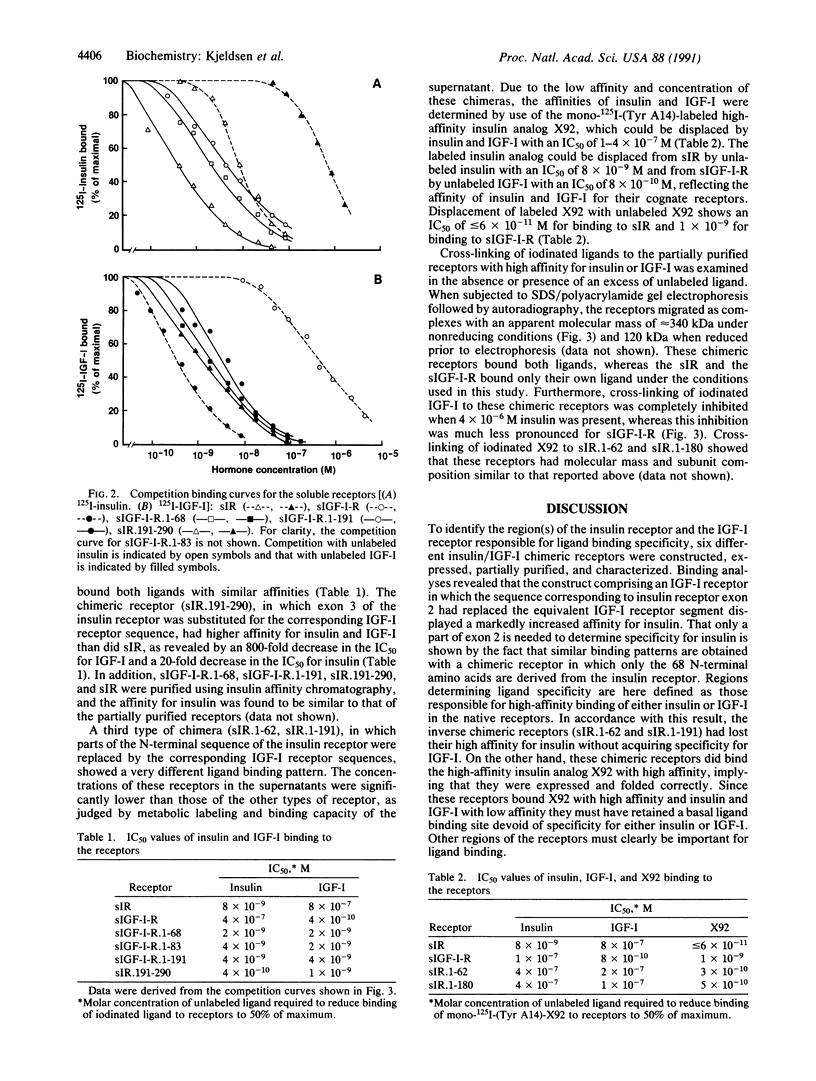
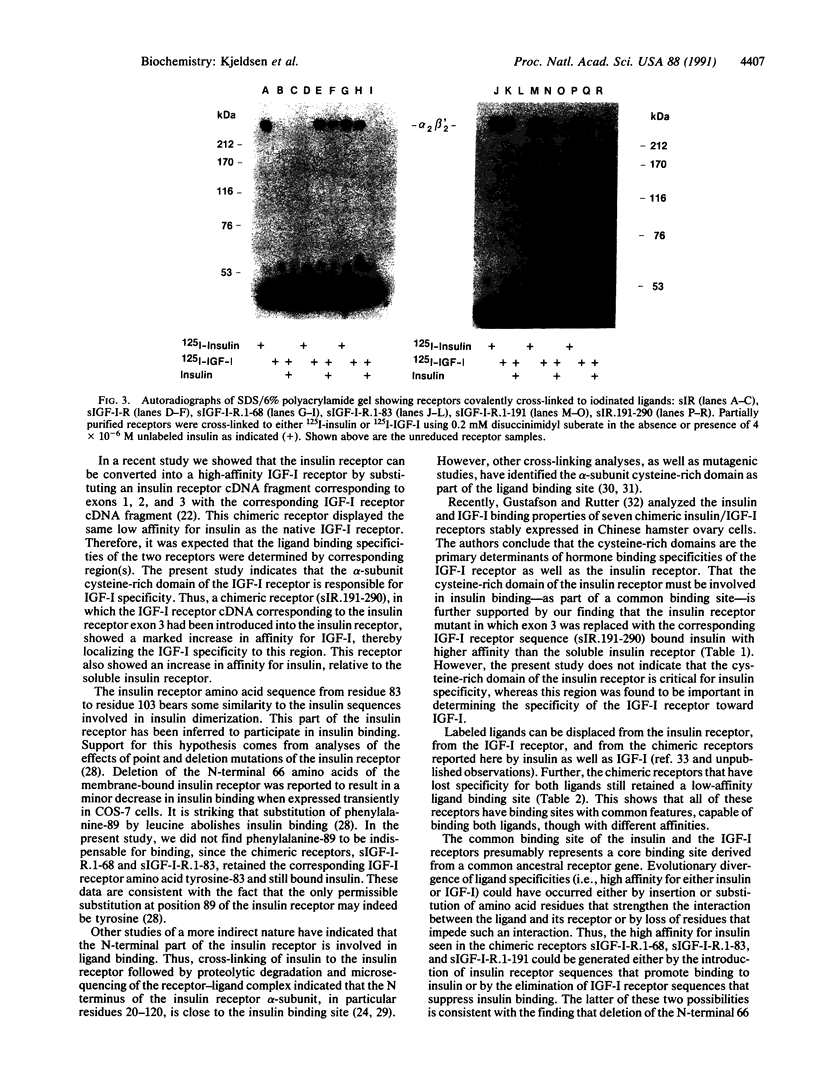
To identify the region(s) of the insulin receptor and the insulin-like growth factor I (IGF-I) receptor responsible for ligand specificity (high-affinity binding), expression vectors encoding soluble chimeric insulin/IGF-I receptors were prepared. The chimeric receptors were expressed in mammalian cells and partially purified. Binding studies revealed that a construct comprising an IGF-I receptor in which the 68 N-terminal amino acids of the insulin receptor alpha-subunit had replaced the equivalent IGF-I receptor segment displayed a markedly increased affinity for insulin. In contrast, the corresponding IGF-I receptor sequence is not critical for high-affinity IGF-I binding. It is shown that part of the cysteine-rich domain determines IGF-I specificity. We have previously shown that exchanging exons 1, 2, and 3 of the insulin receptor with the corresponding IGF-I receptor sequence results in loss of high affinity for insulin and gain of high affinity for IGF-I. Consequently, it is suggested that the ligand specificities of the two receptors (i.e., the sequences that discriminate between insulin and IGF-I) reside in different regions of a binding site with common features present in both receptors.
Full text
PDF

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Andersen A. S., Kjeldsen T., Wiberg F. C., Christensen P. M., Rasmussen J. S., Norris K., Møller K. B., Møller N. P. Changing the insulin receptor to possess insulin-like growth factor I ligand specificity. Biochemistry. 1990 Aug 14;29(32):7363–7366. doi: 10.1021/bi00484a002. [DOI] [PubMed] [Google Scholar]
- Cuatrecasas P. Interaction of concanavalin A and wheat germ agglutinin with the insulin receptor of fat cells and liver. J Biol Chem. 1973 May 25;248(10):3528–3534. [PubMed] [Google Scholar]
- Cuatrecasas P. Isolation of the insulin receptor of liver and fat-cell membranes (detergent-solubilized-( 125 I)insulin-polyethylene glycol precipitation-sephadex). Proc Natl Acad Sci U S A. 1972 Feb;69(2):318–322. doi: 10.1073/pnas.69.2.318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Czech M. P. Molecular basis of insulin action. Annu Rev Biochem. 1977;46:359–384. doi: 10.1146/annurev.bi.46.070177.002043. [DOI] [PubMed] [Google Scholar]
- Czech M. P. Signal transmission by the insulin-like growth factors. Cell. 1989 Oct 20;59(2):235–238. doi: 10.1016/0092-8674(89)90281-x. [DOI] [PubMed] [Google Scholar]
- De Meyts P., Gu J. L., Shymko R. M., Kaplan B. E., Bell G. I., Whittaker J. Identification of a ligand-binding region of the human insulin receptor encoded by the second exon of the gene. Mol Endocrinol. 1990 Mar;4(3):409–416. doi: 10.1210/mend-4-3-409. [DOI] [PubMed] [Google Scholar]
- Ebina Y., Ellis L., Jarnagin K., Edery M., Graf L., Clauser E., Ou J. H., Masiarz F., Kan Y. W., Goldfine I. D. The human insulin receptor cDNA: the structural basis for hormone-activated transmembrane signalling. Cell. 1985 Apr;40(4):747–758. doi: 10.1016/0092-8674(85)90334-4. [DOI] [PubMed] [Google Scholar]
- Ellis L., Sissom J., Levitan A. Truncation of the ectodomain of the human insulin receptor results in secretion of a soluble insulin binding protein from transfected CHO cells. J Mol Recognit. 1988 Feb;1(1):25–31. doi: 10.1002/jmr.300010106. [DOI] [PubMed] [Google Scholar]
- Goldfine I. D. The insulin receptor: molecular biology and transmembrane signaling. Endocr Rev. 1987 Aug;8(3):235–255. doi: 10.1210/edrv-8-3-235. [DOI] [PubMed] [Google Scholar]
- Gustafson T. A., Rutter W. J. The cysteine-rich domains of the insulin and insulin-like growth factor I receptors are primary determinants of hormone binding specificity. Evidence from receptor chimeras. J Biol Chem. 1990 Oct 25;265(30):18663–18667. [PubMed] [Google Scholar]
- Horton R. M., Hunt H. D., Ho S. N., Pullen J. K., Pease L. R. Engineering hybrid genes without the use of restriction enzymes: gene splicing by overlap extension. Gene. 1989 Apr 15;77(1):61–68. doi: 10.1016/0378-1119(89)90359-4. [DOI] [PubMed] [Google Scholar]
- Johnson J. D., Wong M. L., Rutter W. J. Properties of the insulin receptor ectodomain. Proc Natl Acad Sci U S A. 1988 Oct;85(20):7516–7520. doi: 10.1073/pnas.85.20.7516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kahn C. R. The molecular mechanism of insulin action. Annu Rev Med. 1985;36:429–451. doi: 10.1146/annurev.me.36.020185.002241. [DOI] [PubMed] [Google Scholar]
- Kasuga M., Karlsson F. A., Kahn C. R. Insulin stimulates the phosphorylation of the 95,000-dalton subunit of its own receptor. Science. 1982 Jan 8;215(4529):185–187. doi: 10.1126/science.7031900. [DOI] [PubMed] [Google Scholar]
- Kasuga M., Zick Y., Blith D. L., Karlsson F. A., Häring H. U., Kahn C. R. Insulin stimulation of phosphorylation of the beta subunit of the insulin receptor. Formation of both phosphoserine and phosphotyrosine. J Biol Chem. 1982 Sep 10;257(17):9891–9894. [PubMed] [Google Scholar]
- Kobilka B. K., Kobilka T. S., Daniel K., Regan J. W., Caron M. G., Lefkowitz R. J. Chimeric alpha 2-,beta 2-adrenergic receptors: delineation of domains involved in effector coupling and ligand binding specificity. Science. 1988 Jun 3;240(4857):1310–1316. doi: 10.1126/science.2836950. [DOI] [PubMed] [Google Scholar]
- Lammers R., Gray A., Schlessinger J., Ullrich A. Differential signalling potential of insulin- and IGF-1-receptor cytoplasmic domains. EMBO J. 1989 May;8(5):1369–1375. doi: 10.1002/j.1460-2075.1989.tb03517.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lax I., Bellot F., Howk R., Ullrich A., Givol D., Schlessinger J. Functional analysis of the ligand binding site of EGF-receptor utilizing chimeric chicken/human receptor molecules. EMBO J. 1989 Feb;8(2):421–427. doi: 10.1002/j.1460-2075.1989.tb03393.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massague J., Pilch P. F., Czech M. P. Electrophoretic resolution of three major insulin receptor structures with unique subunit stoichiometries. Proc Natl Acad Sci U S A. 1980 Dec;77(12):7137–7141. doi: 10.1073/pnas.77.12.7137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massagué J., Czech M. P. The subunit structures of two distinct receptors for insulin-like growth factors I and II and their relationship to the insulin receptor. J Biol Chem. 1982 May 10;257(9):5038–5045. [PubMed] [Google Scholar]
- Pilch P. F., Czech M. P. Interaction of cross-linking agents with the insulin effector system of isolated fat cells. Covalent linkage of 125I-insulin to a plasma membrane receptor protein of 140,000 daltons. J Biol Chem. 1979 May 10;254(9):3375–3381. [PubMed] [Google Scholar]
- Rafaeloff R., Patel R., Yip C., Goldfine I. D., Hawley D. M. Mutation of the high cysteine region of the human insulin receptor alpha-subunit increases insulin receptor binding affinity and transmembrane signaling. J Biol Chem. 1989 Sep 25;264(27):15900–15904. [PubMed] [Google Scholar]
- Rosen O. M. After insulin binds. Science. 1987 Sep 18;237(4821):1452–1458. doi: 10.1126/science.2442814. [DOI] [PubMed] [Google Scholar]
- Ullrich A., Bell J. R., Chen E. Y., Herrera R., Petruzzelli L. M., Dull T. J., Gray A., Coussens L., Liao Y. C., Tsubokawa M. Human insulin receptor and its relationship to the tyrosine kinase family of oncogenes. 1985 Feb 28-Mar 6Nature. 313(6005):756–761. doi: 10.1038/313756a0. [DOI] [PubMed] [Google Scholar]
- Ullrich A., Gray A., Tam A. W., Yang-Feng T., Tsubokawa M., Collins C., Henzel W., Le Bon T., Kathuria S., Chen E. Insulin-like growth factor I receptor primary structure: comparison with insulin receptor suggests structural determinants that define functional specificity. EMBO J. 1986 Oct;5(10):2503–2512. doi: 10.1002/j.1460-2075.1986.tb04528.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ullrich A., Schlessinger J. Signal transduction by receptors with tyrosine kinase activity. Cell. 1990 Apr 20;61(2):203–212. doi: 10.1016/0092-8674(90)90801-k. [DOI] [PubMed] [Google Scholar]
- Waugh S. M., DiBella E. E., Pilch P. F. Isolation of a proteolytically derived domain of the insulin receptor containing the major site of cross-linking/binding. Biochemistry. 1989 Apr 18;28(8):3448–3455. doi: 10.1021/bi00434a045. [DOI] [PubMed] [Google Scholar]
- Wedekind F., Baer-Pontzen K., Bala-Mohan S., Choli D., Zahn H., Brandenburg D. Hormone binding site of the insulin receptor: analysis using photoaffinity-mediated avidin complexing. Biol Chem Hoppe Seyler. 1989 Mar;370(3):251–258. doi: 10.1515/bchm3.1989.370.1.251. [DOI] [PubMed] [Google Scholar]
- Whittaker J., Okamoto A. Secretion of soluble functional insulin receptors by transfected NIH3T3 cells. J Biol Chem. 1988 Mar 5;263(7):3063–3066. [PubMed] [Google Scholar]
- Yarden Y., Ullrich A. Molecular analysis of signal transduction by growth factors. Biochemistry. 1988 May 3;27(9):3113–3119. doi: 10.1021/bi00409a001. [DOI] [PubMed] [Google Scholar]
- Yip C. C., Hsu H., Patel R. G., Hawley D. M., Maddux B. A., Goldfine I. D. Localization of the insulin-binding site to the cysteine-rich region of the insulin receptor alpha-subunit. Biochem Biophys Res Commun. 1988 Nov 30;157(1):321–329. doi: 10.1016/s0006-291x(88)80050-0. [DOI] [PubMed] [Google Scholar]
- Yip C. C., Yeung C. W., Moule M. L. Photoaffinity labeling of insulin receptor of rat adiopocyte plasma membrane. J Biol Chem. 1978 Mar 25;253(6):1743–1745. [PubMed] [Google Scholar]




