Abstract
Diatraea lineolata and Diatraea saccharalis (Lepidoptera: Crambidae) are moths with stemboring larvae that feed and develop on economically important grasses. This study investigated whether these moths have diverged from a native host plant, corn, onto introduced crop plants including sorghum, sugarcane, and rice. Diatraea larvae were collected from these four host plants throughout the year in El Salvador and were reared on artificial diet until moths or parasitoids emerged. Adult moths were subsequently identified to species. Amplified fragment length polymorphisms (AFLPs) and mitochondrial DNA cytochrome oxidase I (COI) were used to examine whether or not there was genetic divergence of D. lineolata or D. saccharalis populations on the four host plants. Percent parasitism was also determined for each moth on its host plants. D. lineolata was collected from corn in the rainy season and sorghum in the dry season. D. saccharalis was most abundant on sugarcane in the rainy season and sorghum in the dry season. The AFLP analysis found two genetically divergent populations of both D. lineolata and D. saccharalis. Both moths had high levels of parasitism on their dominant host plant in the rainy season, yet had low levels of parasitism on sorghum in the dry season. The presence of two genotypes of both Diatraea spp. on sorghum suggest that host‐associated differentiation is occurring on this novel introduced crop plant.
Keywords: adaptation, biological control, biotypes, host plant phenology, host plant volatiles, host‐associated differentiation, parasitism
1. Introduction
Insects that colonize novel host plants can undergo divergent selection possibly leading to host‐associated differentiation (Abrahamson et al., 2003; Craig et al., 1993; Dres & Mallet, 2002; Medina, 2012; Via, 1999; Vialatte et al., 2005). Populations in distinct habitats, such as different host plants, may diverge genetically even if they remain in contact, a process called ecological speciation (Nosil, 2012; Schluter, 2001). Along with oviposition preference and performance characters, life‐history parameters such as generation time (e.g., voltinism), parthenogenesis, and variation in diapause contribute to the divergence of host plant‐associated populations of insects (Craig et al., 1993; Dickey & Medina, 2010; Diehl & Bush, 1984; Wood et al., 1999). Studies have demonstrated host‐associated differentiation (HAD) for insects on native plants (Abrahamson et al., 2003; Dickey & Medina, 2010; Scheffer & Hawthorne, 2007; Stireman, Nason, & Heard, 2005) as well as for those on novel host plants (Feder, Hunt, & Bush, 1993; Forbes et al., 2009). Natural enemies of herbivores such as parasitoids may undergo cascading or sequential genetic divergence as they follow herbivores onto novel host plants (Stireman et al., 2005; Forbes et al., 2009), or herbivores may escape parasitism on their newly colonized host plants. Formation of host plant‐associated insect populations has been suggested to be more likely with longer associations between insects and novel host plants (Siemann, Rogers, & Dewalt, 2006). However, rapid adaptation and formation of host‐associated insect populations have occurred on introduced cultivated crop plants (Dres & Mallet, 2002; Vialatte et al., 2005; Feder & Forbes, 2010). Additional examples of HAD on novel plants are likely to be uncovered given the large number of plants and insects, which have been introduced outside of their native range.
Host plant‐associated differentiation of insect populations has been demonstrated in a number of systems. Native insects colonizing introduced crop plants have diverged in the course of two hundred years or so (Feder et al., 1993; Vialatte et al., 2005; Feder & Forbes, 2010). Rhagoletis pomonella (Walsh) (Diptera: Tephritidae) has shifted from the native plant hawthorne onto apple, an introduced crop transplanted from the Old World into the United States. Differences in phenologies between these two host plants contribute to isolating the host races of this insect (Feder & Forbes, 2010). The aphid Sitobion avenae Fabricius (Homoptera: Aphididae) has divergent genotypes on cultivated and uncultivated hosts (Vialatte et al., 2005), formed presumably in the last 100 years since its introduction into the United States. In Europe, corn was introduced in the last 500 years. Currently, there are two host plant races of Ostrinia nubilalis Hübner (Lepidoptera: Crambidae), the European corn borer, which have unique pheromone blends and oviposition preference for their natal hosts (Bethenod et al., 2005). Similarly, the fall armyworm Spodoptera frugiperda J.E. Smith (Lepidoptera: Noctuidae) has been found to have two host plant strains, one feeding on corn and one on the introduced host rice (Pashley, Hardy, Hammond, & Mihm, 1990). The moths mate at different times of night and have unique pheromone blends, which contribute to ecological isolation (Groot, Marr, Heckel, & Schofl, 2010).
The sugarcane borer moth, Diatraea saccharalis (Fabricius) (Lepidoptera: Crambidae), is native in the Western Hemisphere and broadly distributed especially in regions associated with sugarcane (Bleszynski, 1969; Box, 1931; CAB 1989; Dyar & Heinrich, 1927). Larvae are endophagous, developing in host plant stems until they pupate and emerge as adults. This moth is considered an introduced species in the southern United States and can be a pest of cultivated plants including corn (Zea mays L.), sorghum (Sorghum bicolor L.), and rice (Oryza sativa L.) in various parts of its range (Cherry & Nuessly, 1993; Fuchs, Huffman, & Smith, 1979; Gifford & Mann, 1967; Vargas, Lastra, & Solis, 2013; White et al., 2001). Few population genetic studies of this insect have been conducted (Joyce et al., 2014; Lange, Scott, Graham, Sallam, & Allsopp, 2004; Pashley et al., 1990). Pashley et al. (1990) found that populations of D. saccharalis from the southern United States and Mexico were divergent from those of Brazil. D. saccharalis has been considered to be a single species with a wide distribution, but a recent study suggests that at least three or more species may exist (Joyce et al., 2014). In the southern United States, at least two putative species were found to occur, one in Texas and Louisiana, and a divergent genotype in Florida (Joyce et al., 2014). Exploring the genetic variability of D. saccharalis in a potential region of origin such as Central America would permit investigation of whether divergence has occurred between populations occurring on native crops such as corn or introduced crops. In Central America, another Diatraea species, D. lineolata (Walker) (Lepidoptera: Crambidae) also feeds on corn (Box, 1951; Quezada, 1978; Solis, 2004; Solis & Metz, 2015), a crop plant considered native in Mexico and Central America and domesticated in the last 5,000–10,000 years (Matsuoka et al., 2002). Because both D. saccharalis and D. lineolata are associated with corn and also feed on introduced crops, they may be subject to disruptive selection from crop plants introduced in the last few hundred years.
El Salvador has a tropical climate with a pronounced dry season and is classified as tropical savannah (Peel, Finlayson, & McMahon, 2007). During the rainy season from May until October, about 95% of annual rainfall occurs, with the months of April and November serving as transition months to and from the dry season. A widespread cropping rotation pattern in El Salvador is corn cultivation in the rainy season, followed by sorghum (an introduced crop) in the dry season (Quezada, 1978; Serrano‐Cervantes et al., 1986). Rice and sugarcane, both nonnative cultivars, are also planted in the rainy season, while plantings of sugarcane persist throughout the year during all 12 months, essentially being cultivated as a perennial crop plant. As the tropical climate transitions from the rainy to the dry season and plants dry out, developing larvae of both moth species become quiescent and shift to a dormant form where they no longer feed and larvae lose their pigmented colored spots (pinacula) on the cuticle (Dyar & Heinrich, 1927; Kevan, 1944; Quezada, 1978). The climatic extremes between the rainy and dry season along with novel host plant availability could exert strong selective pressure on insect populations, contributing to insect populations adapting to novel host plants.
The objective was to determine whether either of two moth species, D. saccharalis and D. lineolata, has genetically divergent host plant‐associated strains feeding on a native host, corn, and the introduced Old World crop plants, sugarcane, rice or sorghum. To test this, D. lineolata and D. saccharalis larvae were collected from available host plants in the rainy season and the dry season in El Salvador, Central America. If host‐associated differentiation had occurred on novel host plants, insects from two host plants (such as corn and sorghum) in the same location should be more genetically divergent than insects collected from a single host plant species such as corn from distant geographic locations.
2. Materials and Methods
2.1. Geographic location of study and field collection of larvae from host plants
The study was conducted in El Salvador, Central America, from August 2011 to May 2012 with an additional collection in rice in October 2013. We only used adult moths reared from field‐collected, host plant‐associated larvae. At each field site, host plants were searched for evidence of larval feeding in stems, indicated by insect frass exuding from a hole in the plant stem. Host plant stems with larvae were cut, and larvae were removed and placed individually in 60‐ml plastic cups on artificial diet (Southland Products, Lake Village Arkansas) in order to rear larvae to adult moths. Larvae were transported to the laboratory for rearing at the University of El Salvador and observed at least twice a week. Any adult moths or parasitoid wasps or flies that emerged from larvae were preserved by freezing or by storage in 80% ethanol. We used the emerged parasitoids to determine the parasitism rate for each Diatraea species on each plant type by dividing the number of parasitoids emerged by the sum of moths and parasitoids emerged.
2.2. Identification of adult moths by morphology
Reared adult Diatraea were identified to species by examining the adult male and female genitalia and comparing them to the key by Dyar and Heinrich (1927). Abdomens of adult moths were prepared for study by soaking the abdomen in cold 10% potassium hydroxide (KOH) overnight to be able to study the sclerotized structures for identification of the genitalia (Robinson, 1976). They were then placed in polyethylene genitalia vials with glycerin for future study and slide mounting. Voucher specimens are at the National Museum of Natural History, Smithsonian Institution, Washington, DC.
2.3. DNA extraction
Following the identification of adult moths, the six legs of each moth were used for DNA extractions using the Qiagen DNeasy Blood and Tissue kit (Venlo, Netherlands) following the protocols for animal tissue with an incubation time of 2 h at 65°C (Qiagen 2006). Final products were eluted in 100 μl of AE buffer. The DNA quantity was measured using the Qubit® dsDNA HS Assay kit (Life Technologies). The quantity of DNA in samples averaged 2–5 ng of DNA per μl. Both male and female adults were used for molecular work.
2.4. Population genetics: amplified fragment length polymorphisms (AFLPS)
Amplified fragment length polymorphisms were produced as described by Vos et al. (1995) and Joyce et al. (2010). Two primer combinations were used, (1) M‐CAT and E‐ACT, and (2) M‐CAA and E‐ACT. Individuals from the four host plant populations (corn, sugarcane, sorghum, and rice) were randomized on two 96‐well plates for AFLP reactions.
Each restriction ⁄ ligation reaction (well) consisted of the following: 0.05 μl each of EcoRI and MseI, 1.1 μl of T4 DNA ligase buffer, 1.1 μl of 0.5 mol/L NaCl, 0.55 μl of diluted BSA (bovine serum albumin), 0.03 μl of T4 DNA ligase, 1.0 μl each of EcoRI and MseI adaptor pairs (Life Technologies‐Thermo Fisher Scientific, Waltham, MA, USA), and 0.61 μl of sterile distilled water. The plate with restriction⁄ ligation reactions was held at room temperature overnight (ca. 12 h at 25°C) to ensure complete digestion (Saunders, Mischke, & Hemeida, 2001). The amplified product was diluted 20‐fold using 15 mmol/L Tris‐HCl buffer (pH 8.0) containing 0.1 mmol/L EDTA. Pre‐selective PCR amplification was performed on a ThermoFisher Arktik thermal cycler. Each reaction contained 15 μl of AFLP preselective mix (all Life Technologies/Thermo Fisher), 1 μl of each amplification primer (Life Technologies), along with 4 μl of the diluted restriction⁄ ligation mixture. The PCR program for pre‐selective amplification consisted of an initial warm‐up of 95°C for 1 min followed by 20 cycles at 95°C for 20 s, 56°C for 30 s, and 72°C for 90 s with a final hold at 75°C for 5 min. The amplified product was diluted 20‐fold using 15 mmol/L Tris‐HCl buffer (pH 8.0) containing 0.1 mmol/L EDTA. Selective amplification was conducted using two primer combinations. For each selective amplification, a reaction consisted of 15 μl of AFLP platinum supreme mix, 1.0 μl of EcoRI + 3 selective primer, and 1.0 μl of MseI + 3 selective primer (all Life Technologies). The PCR program for selective amplification consisted of an initial warm‐up of 95°C for 1 min, 12 cycles of 95°C for 20 s, 65°C for 40 s with a lowering of 0.7°C per cycle, 72°C for 90 s, followed by 35 cycles of 95°C for 20 s, 56°C for 40 s, 72°C for 90 s, and finally a hold of 72°C for 7 min before storing the samples at 4°C. Prior to capillary electrophoresis, 0.4 μl of the GeneScan LIZ 500 size standard and 0.9 μl of HiDi formamide (all Life Technologies) were added to 1 μl of the final product of each sample. Sample fragments were separated using automated capillary electrophoresis by the ABI 3730 XL automated capillary DNA sequencer. GeneMapper version 5.0 (Life Technologies) was used to determine presence or absence of fragments. The peak detection threshold was set for each primer combination and was typically 100 luminescent units. Each AFLP marker was considered a locus and assumed to have two possible alleles (0 = absent, 1 = present). Bands not present in more than one individual were eliminated (i.e., private alleles) prior to further analyses, as they were not considered informative. Structure 2.3.4 software (Pritchard, Wen, & Falush, 2007) was used to group individuals with similar genotypes within each species. Structure uses a Bayesian algorithm to cluster individuals into K, which is defined as the number of genetically distinct populations in a data set. Parameters used for the analyses include the following: no a priori assignment of individuals to a known population, analysis for diploid insects, a burn‐in of 10,000 iterations, an admixture model, and independent loci.
If collection locations were fewer than 5 miles apart, they were considered one location for the population genetic analysis of AFLPs with Structure software (Figure 1). The following two sites were combined and considered one collection site for genetic analysis with Structure; Cooperative El Nilo, and Santa Cruz Porrillo (D. saccharalis sites 1, 5). Five collecting locations were used for Structure genetic analysis for both D. lineolata and D. saccharalis (Figure 1). The number of potential populations for K was estimated as the number of geographic sampling locations (5) plus 4 (K = 9) for both species as suggested by Pritchard, Stephens, and Donnelly (2000), and each iteration was run 20 times. At the completion of Structure runs, K was calculated for each species using Structure Harvester (Earl & VonHoldt, 2012; Evanno, Regnaut, & Goudet, 2005), to determine the most likely number of population clusters (K) for each species.
Figure 1.
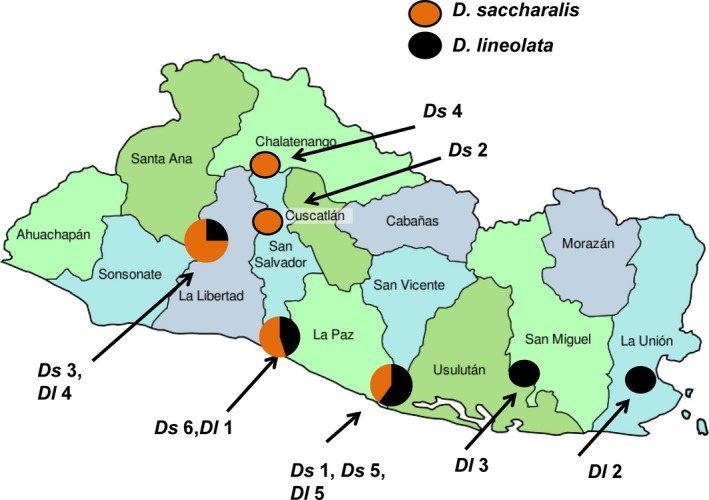
Collection site codes correspond to the collection locations on Tables 1 and 2 for each Diatraea species in El Salvador. See Tables 1 and 2 for collection details including date, location name, longitude, latitude, and host plant type. The proportion of Diatraea saccharalis and Diatraea lineolata collected at each site are shown in brown (Ds) or black (Dl)
Analyses of molecular variance (AMOVA) tests were run using the AFLP data using GenAlEx 6.0 (Peakall & Smouse, 2006). For D. lineolata, all individuals collected were included in a comparison of host plant‐associated individuals collected from corn and sorghum (Table 1). The individuals collected from corn were obtained in the rainy season, and those from sorghum were collected in the dry season; the AMOVA of season consists of the same individuals and produced identical results as that obtained when comparing host plant populations. A separate AMOVA was run to examine genetic variation among four host plant–site populations. We did not include sites with less than five individuals in AMOVA; therefore, the one adult D. lineolata from site 3 in San Miguel was not included. The four host plant–site populations compared were (1) corn at UES‐EE, (2) corn at La Union, (3) sorghum at La Union, and (4) sorghum at Santa Cruz Porillo. The host plant–site analysis was chosen as there was an unbalanced design with different numbers of host plants at each site (Sword, Joern, & Senior, 2005). Pairwise comparisons were made of the genetic distance (F ST) and significance among the four host plant–site populations. All AMOVAs were run with 999 permutations. For D. saccharalis, AMOVA was run to compare three host plant‐associated populations, rice, sugarcane, and sorghum (corn was not included as there were only 3 Ds adults). Subsequently, four host plant–site populations were compared; (1) rice at El Nilo, (2) sugarcane at Ingenio La Cabana El Paisnal, (3) sorghum at San Andres Centa, and (4) sorghum at Santo Tomas. Similarly, (F ST) values were determined for pairs of populations and their significance was tested. An AMOVA was also run to compare variation of D. saccharalis populations between the rainy season and the dry season. Finally, for each Diatraea species, a Mantel test was run for the four host plant–site populations to determine whether genetic distance was significantly correlated with geographic distance.
Table 1.
Collection sites for Diatraea lineolata larvae in El Salvador by host plant
| Site number | Site code | Collection date 2011–2012 | Identification by morphology | Host plant | Collection site | Municipio, departmento | Latitude/Longitude |
|---|---|---|---|---|---|---|---|
| 1 | UES‐corn | Sept. 8, 2011 | D. lineolata | Corn | UES, Estacion Experimental | San Luis Talpa, La Paz | N13°28′, W89°06′ |
| Oct. 7, 2011 | D. lineolata | Corn | UES, Estacion Experimental | San Luis Talpa, La Paz | N13°28′, W89°06′ | ||
| 2 | LaU‐corn | Oct. 12, 2011 | D. lineolata | Corn | Canton Sirama | Sirama, La Union | N13°21′, W87°54′3″ |
| Jan. 11, 2012 | D. lineolata | Sorghum | Canton Sirama | Sirama, La Union | N13°21′, W87°54′3″ | ||
| 3 | SM‐sorg | Sept. 29, 2011 | D. lineolata | Sorghum | Lotte Amatillo | San Miguel | N13°20′29.554, W88°16′4.957 |
| 4 | SaC‐sorg | Dec. 19, 2011 | D. lineolata | Sorghum | CENTA Field 1, Lot 11 | San Andes La Libertad | N13°48.365′, W89°23.71 |
| 5 | SCP‐sorg | Feb. 7, 2012 | D. lineolata | Sorghum | CENTA Research Station | Santa Cruz Porrillo, San Vicente | N13°26′12.2, W88°48.10.7 |
| March 7, 2012 | D. lineolata | Sorghum | CENTA Research Station | Santa Cruz Porrillo, San Vicente | N13°26′12.2, W88°48.10.7 |
UES, University of El Salvador; CENTA, Centro Nacional de Tecnologίa Agropecuaria y Forestal.
2.5. Mitochondrial DNA‐COI
A 658‐base pair region (the “bar code”) of the mitochondrial COI gene region was sequenced from 26 D. lineolata and 23 D. saccharalis including a few individuals from each collection site (Tables 1 and 2, Figure 1). The DNA used for sequencing COI was extracted as described above. The bar‐code region of the COI gene was amplified using primers for the mitochondrial DNA “bar code” of Lepidoptera (Hajibabaei, Janzen, Burns, Hallwachs, & Hebert, 2006). The forward primer LepF was 5_‐ATTCAACCAATCATAAAGATATTGG‐3, and the reverse primer sequence of LepR was 5_‐ TAAACTTCTGGATGTCCAAAAAATCA‐3 (Hajibabaei et al., 2006). The touchdown PCR program consisted of an initial 2 min at 95°C, then 12 cycles of 95°C for 10 s, 58–46°C for 10 s with a lowering of 1°C temperature each cycle, and 72°C for 60 s. Following PCR and confirmation of amplification on an agarose gel, samples were cleaned up using a USB Exo‐sapit pcr cleanup kit (Affymetrix, Inc., Santa Clara, Cal.). Sequencing was carried out on an ABI 3730XL Genetic Analyzer. DNA sequences were edited using Geneious 7.0 (Biomatters, Aukland, New Zealand) (Kearse et al., 2012). The forward and reverse sequences for each individual were trimmed and assembled into a consensus sequence. Mitochondrial DNA was compared with the COI sequences available for D. saccharalis available in GenBank and the Bar Code of Life Data System (BOLD) (Milton, Pierossi, & Ratnasignham, 2013). We did not find any existing D. lineolata mitochondrial DNA sequences in GenBank or BOLD available for comparison. We aligned our 23 consensus sequences from D. saccharalis with others previously obtained from D. saccharalis from the southern USA (Joyce et al., 2014) and with 34 other D. saccharalis COI sequences available in GenBank and BOLD. Alignments were made in Geneious 7.0 using the Clustal W alignment function and used to make an unrooted neighbor‐joining tree. Bootstrap support values were obtained by 500 pseudoreplicates of the aligned dataset.
Table 2.
Collection sites for Diatraea saccharalis larvae by host plant
| Site Number | Site code | Collection date 2011–2012, 2013 | Identification by morphology | Host plant | Collection site | Municipio, department | Latitude/Longitude |
|---|---|---|---|---|---|---|---|
| 1 | Nil‐rice | Oct. 7, 2011 | D. saccharalis | Rice | El Nilo 1 | Zacatecaluca, La Paz | N13°23.700, W 88°52.860 |
| Oct. 18, 2013 | D. saccharalis | Rice | El Nilo 1 | Zacatecaluca, La Paz | N13°23.700, W88°52.860 | ||
| Dec. 12, 2011 | D. saccharalis | Cane | El Nilo 1 | Zacatecaluca, La Paz | N13°23.700′, W88°52.860 | ||
| Feb. 12, 2012 | D. saccharalis | Sorghum | El Nilo 1 | Zacatecaluca, La Paz | N13°23.700′, W88°52.860 | ||
| 2 | InC‐cane | Nov. 22, 2011 | D. saccharalis | Cane | Ingenio La Cabaña | Paisnal, San Salvador | N13°59′56.8″, W 89°11′09.5″ |
| Dec. 6, 2011 | D. saccharalis | Cane | Ingenio La Cabaña | Paisnal, San Salvador | N13°59′56.8″ W89°11′09.5″ | ||
| 3 | SaC‐sorg | Dec. 19, 2011 | D. saccharalis | Sorghum | CENTA | San Andres, La Libertad | N13°48.365′, W89°23.717′ |
| 4 | CJC‐sorg | Jan. 26, 2012 | D. saccharalis | Sorghum | Cooperative Juan Chacon | Chilamate, Chaletanango | N14°5.310′, W89°12.582 |
| 5 | SCP‐sorg | Feb. 7, 2012 | D. saccharalis | Sorghum | CENTA | Santa Cruz Porrillo, San Vicente | N13°26′12.2, W88°48.10.7 |
| March 7, 2012 | D. saccharalis | Sorghum | CENTA | SCP, SV | N13°26′12.2, W88°48.10.7 | ||
| 6 | SaT | March 28, 2012 | D. saccharalis | Sorghum | Santo Tomas | Santo Tomas, La Paz | N13°28′35.23″ W89°6′33.32″ |
| April 19, 2012 | D. saccharalis | Corn | Santo Tomas | St. Tm, La Paz | N13°28′35.23″, W89°6′33.32″ |
CENTA, Centro Nacional de Tecnologίa Agropecuaria y Forestal.
3. Results
Diatraea lineolata and D. saccharalis larvae were collected at sites throughout El Salvador (Tables 1 and 2, Figure 1). We attempted to visit collection sites multiple times so that we could collect from host plants cultivated throughout the year in the rainy season and the dry season. Diatraea lineolata larvae collected from the rainy season had black pinacula (spots), while larvae of D. saccharalis from the rainy season had brown pinacula. Larvae of both species collected in the dry season typically had pale cream colored pinacula that blended in color with the integument. All larvae were reared as previously described, and subsequent identification of adults (described below) found that larvae with black pinacula were D. lineolata, while larvae with brown pinacula were D. saccharalis.
Adult identification was confirmed using genitalia and the key by Dyar and Heinrich (1927) (Figure 2a,b). Diatraea lineolata was collected from corn in the rainy season, and sorghum in the dry season, but was not found in any collections from sugarcane or rice. Diatraea saccharalis was collected from sugarcane and rice in the rainy season, and sorghum and corn in the dry season. Although D. saccharalis was collected in rice, another stemborer Rupela albinela (Cramer) (Lepidoptera: Pyralidae) was much more abundant in rice.
Figure 2.
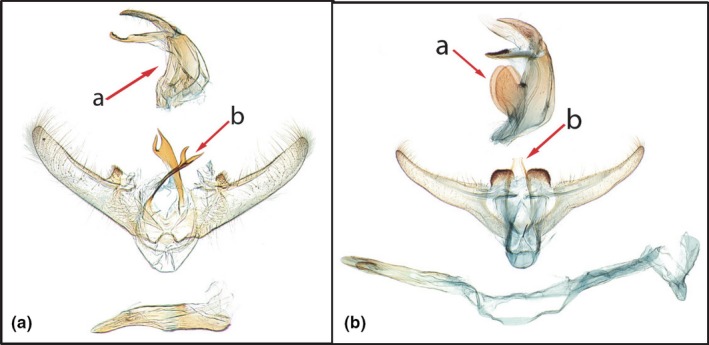
(a) Diatraea lineolata male genitalia, USNM #113649, (a.a) lateral lobe of tegumen absent; (a.b) apex of juxta arms with two distinct points, apical teeth subequal in size, and clawlike (b) Diatraea saccharalis male genitalia, USNM #114649, (b.a) lateral lobe of tegumen present, round, and as long as wide; (b.b) apex of juxta arms blunt, not bidentate
3.1. Parasitism of Diatraea lineolata and Diatraea saccharalis
Parasitoid wasps and parasitoid flies were reared from the collected Diatraea larvae. D. lineolata were collected from corn and sorghum, yet 37% parasitism of D. lineolata occurred on corn, with only 7% parasitism on sorghum (Table 3). Two parasitoid flies, Billea (Paratheresia spp.) and Palpozenillia (Diptera: Tachinidae), were the dominant parasitoids of D. lineolata on corn. Diatraea saccharalis was parasitized most heavily on sugarcane with 67% parasitism, with most parasitism by Billea spp. (Table 3). Only 5% parasitism of D. saccharalis occurred on sorghum and 10% on rice (Table 3).
Table 3.
Number of adult Diatraea lineolata and Diatraea saccharalis moths or parasitoids reared from larvae on the four host plants
| Moth species | Parasitoid | Host plant | |||
|---|---|---|---|---|---|
| Corn | Sugarcane | Sorghum | Rice | ||
| D. lineolata | 22 | – | 43 | ||
| Billea spp. | 5 | – | 3 | – | |
| Palpozenillia spp. | 7 | – | 0 | – | |
| Hyperparasitoid | 1 | – | 0 | – | |
| Total percent parasitism | (Par/Moth + Par) | 37% | – | 7% | – |
| D. saccharalis | 3 | 20 | 19 | 10 | |
| Billea spp. | 0 | 32 | 0 | 0 | |
| Apanteles spp. | 0 | 1 | 0 | 0 | |
| Iphaulaux spp | 0 | 1 | 0 | 0 | |
| Hyperparasitoid | 0 | 7 | 0 | 0 | |
| Tachinidae | 0 | 0 | 1 | 0 | |
| Ceraphronidae | 0 | 0 | 0 | 1 | |
| Total percent parasitism | (Par/Moth + Par) | 0% | 67% | 5% | 10% |
Billea, Palpozenillia = (Diptera: Tachinidae), Others = Hymenoptera. No D. lineolata were found in collections from sugarcane or rice.
3.2. Population comparisons using AFLPs
We produced 125 unique AFLP markers using two primer combinations for 65 D. lineolata moths. The 65 individuals consisted of 22 adults reared from corn and 43 adults reared from sorghum. Individuals from corn were reared from two sites (Table 1), while individuals from sorghum were obtained from three sites (Table 1). Structure Harvester found that K = 2, indicating two genetically distinct groups of D. lineolata (Figure 3). Collections from corn were assigned primarily to one cluster (green, Figure 3), while collections from sorghum had two genetically distinct clusters (red and green, Figure 3). To visualize geographic variation in genotypes, the individual bars from structure, which represent individual moths, were mapped to their collection sites (Figure 4). Individuals reared from corn came from the University of El Salvador Experimental Station in San Luis Talpa and also from Canton Sirama in La Union. D. lineolata collected from corn from geographically distant sites are almost entirely assigned to the same genetic cluster illustrated in green. At Canton Sirama in La Union, individuals collected from corn and sorghum had more genetic differentiation (with no geographic separation) than those collected from corn at two sites at a substantial distance from each other (Figure 4, see AMOVA results below).
Figure 3.
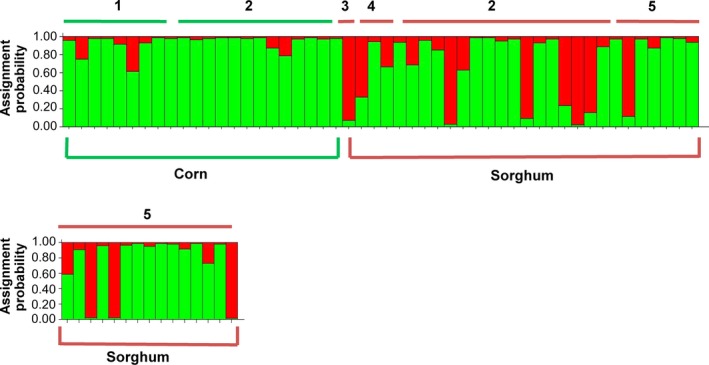
Structure analysis of AFLPs from Diatraea lineolata collected in El Salvador from corn and sorghum from September 2011 to March 2012. Structure 2.3.4 was run using the following parameters: diploid individuals, 10,000 iterations, admixed data, and independent loci. Structure Harvester found that K = 2. The number above the bars represents the collection site for larval collections (see Table 1, Figure 1). The host plant that each larva was collected from is below the bars
Figure 4.
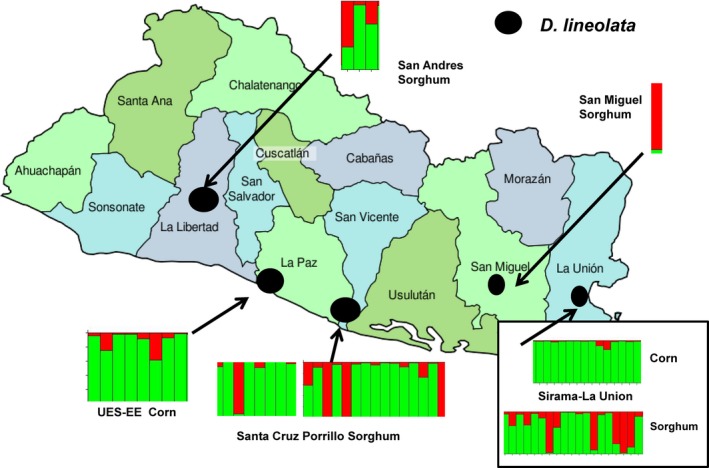
Map of El Salvador with results from Structure analysis for individual Diatraea lineolata moths collected on corn or sorghum, mapped to their respective collection sites
For the second moth species, D. saccharalis, we produced 112 AFLP markers for 52 adults reared from larvae collected on rice, sugarcane, sorghum, and corn. For the Structure software analysis, we used 10 adults reared from larvae collected on rice, 20 adults from sugarcane, 19 adults from sorghum, and three from corn. In the rainy season, sugarcane was the dominant host plant of D. saccharalis, and D. saccharalis was not found on corn in the rainy season as we had expected. Collections from corn were dominated by the other Diatraea species, D. lineolata. Structure Harvester found for D. saccharalis that K = 2, indicating there were two genetically distinct groups (Figure 5). Samples from rice from two sites were all assigned to the green cluster, as were those from corn. Most individuals reared from sugarcane in the rainy season were assigned to the green cluster; collections from sorghum had a mix of two genotypes (Figure 5).
Figure 5.
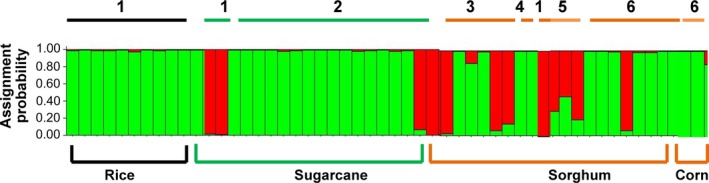
Structure analysis for Diatraea saccharalis moths raised from larvae collected from host plants throughout the year. Each vertical bar represents an individual. Collection site numbers are above the bars, while host plants are below the bars. Details on collections are listed in Table 2. Structure Harvester found that K = 2
3.3. Analysis of molecular variation (AMOVA)
AMOVA of host plant‐associated populations of D. lineolata found that host plants had a significant effect on genetic variation (Table 4); host plant populations accounted for 2% of variation, while host plant × site populations contributed 6% of variation. Pairwise F ST values between the four groups found that genetic distance between the two corn populations (UES, La Union, 0.024) was not significant, but genetic distances between corn and sorghum populations were significant (0.039–0.082), with the F ST genetic distance of 0.068 between the corn and sorghum populations at the Canton Sirama (Table 5). A Mantel test found no significant relationship between the genetic distance and geographic distance for D. lineolata populations from corn and sorghum (r = .201, p = .501).
Table 4.
Analysis of molecular variation (AMOVA) for Diatraea lineolata and Diatraea saccharalis
| Population | df | SS | Variation (%) | p | |
|---|---|---|---|---|---|
| Diatraea lineolata | (a) Among host plant × site | 3 | 77.468 | 6 | .001 |
| Individuals within groups | 57 | 786.630 | 94 | ||
| (b) Among host plants | 1 | 23.78 | 2 | .007 | |
| Individuals within groups | 59 | 840.810 | 98 | ||
| Diatraea saccharalis | (a) Among host plant × site | 3 | 80.794 | 8 | .001 |
| Individuals within groups | 40 | 543.933 | 92 | ||
| (b) Among host plants | 2 | 44.946 | 4 | .001 | |
| Individuals within groups | 46 | 638.850 | 96 | ||
| (c) Among seasons | 1 | 29.557 | 4 | .003 | |
| Individuals within seasons | 47 | 654.239 | 96 |
Table 5.
Pairwise comparisons of genetic divergence estimates (F ST) between Diatraea host plant–site populations
| Host plant × Site | 1 | 2 | 3 | 4 | |
|---|---|---|---|---|---|
| D. lineolata | 1. UESEE–Corn | 0 | |||
| 2. La Union–Corn | 0.024 NS | 0 | |||
| 3. La Union–Sorghum | 0.082a | 0.068a | 0 | ||
| 4. Santa Cruz Porrillo–Sorghum | 0.039a | 0.082a | 0.035a | 0 | |
| D. saccharalis | 1. Nilo–Rice | 0 | |||
| 2. Paisnal–Sugarcane | 0.081a | 0 | |||
| 3. San Andres–Sorghum | 0.072a | 0.104a | 0 | ||
| 4. Santo Tomas–Sorghum | 0.025NS | 0.133a | 0.095a | 0 |
p < .05. Comparison between populations is significant.
For D. saccharalis, the AMOVA test found the three host plant‐associated populations on rice, sugarcane and sorghum accounted for 4% of the variation, and season of collection (rainy or dry season) similarly accounted for 4% of variation. The host plant × site populations accounted for 8% of variation (Table 4). All AMOVA were significant (p < .01) (Table 4). The pairwise comparisons of F ST values between the three host plant‐associated populations found that sugarcane population was significantly divergent from sorghum and marginally divergent from rice (sugarcane vs. sorghum F ST 0.044, p = .01; rice vs. sugarcane F ST 0.046, p = .05; rice vs. sorghum F ST 0.018, p = .16). Pairwise comparisons of F ST values for the four host plant–site populations found D. saccharalis on sugarcane was significantly divergent from the rice populations (F ST = 0.081) and the two sorghum populations (F ST 0.104, F ST 0.133) (Table 5). Genetic divergence was significant between rice and sorghum from San Andres, but not significant between the rice population and sorghum from Santo Tomas; finally, there was significant divergence between sorghum from San Andres and sorghum from Santo Tomas (F ST 0.095). A Mantel test found there was no significant correlation of genetic and geographic distance (r = .505, p = .105).
3.4. Mitochondrial DNA‐COI sequences
We obtained mitochondrial COI sequences from 26 individuals of D. lineolata including a few from each collection site. There was little genetic variation between pairs of sequences of mitochondrial DNA. Most individuals of D. lineolata were 99% or more similar, with 1–2 base pairs differing among some pairs. No previously sequenced COI sequences were available in GenBank for D. lineolata for comparison with our results.
For D. saccharalis, 23 individuals were sequenced including at least one individual from each collection site (Figure 6). Phylogenetic analysis of D. saccharalis sequences, including 34 sequences from GenBank, uncovered substantial geographic structure across the species (Figure 6). The unrooted neighbor‐joining consensus tree had three clades. The first and largest group of D. saccharalis consisted of individuals from El Salvador clustered with those from Mexico and the southern United States. Individuals from El Salvador in the larger clade were collected from all sites in the study and were collected on rice, sugarcane, and sorghum. Collections from El Salvador had a small subgroup of several individuals with 4–5 base pairs differing between them. These divergent individuals came from two interior collections (sites 2, 3) from sugarcane and sorghum. The 4‐to 5‐base pair difference in the sequences from the smaller clade represents almost a 1% divergence from other individuals collected in El Salvador (Table S1). An additional small group of four individuals from East and South Texas were also 1% divergent from other D. saccharalis in that clade. The second large grouping in the phylogenetic tree consists of individuals from South America, including Brazil, Argentina, and Bolivia. This group of moths was 2–3% divergent from the other two clades (Table S1); similarly, the third group of D. saccharalis from Florida was 2–3% divergent from the other two clusters.
Figure 6.
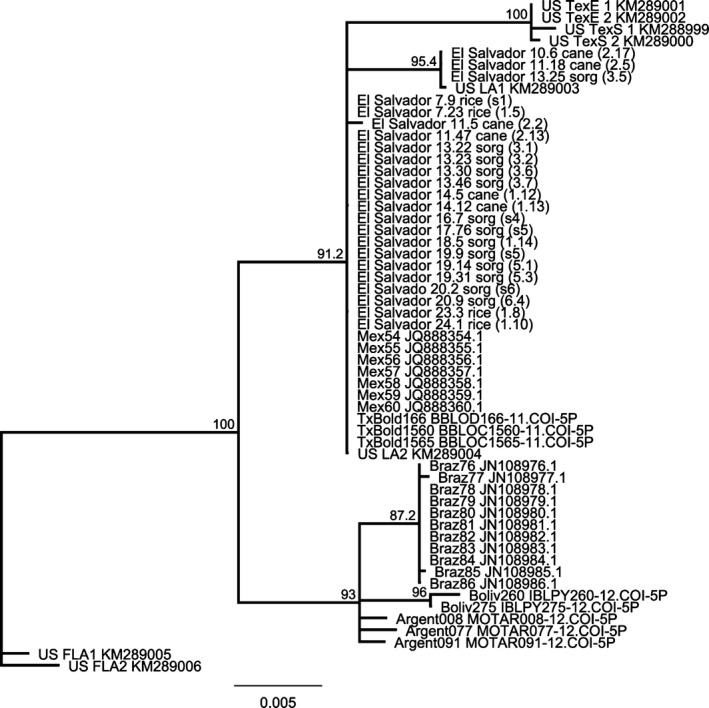
Diatraea saccharalis collected in El Salvador combined with individuals previously sequenced from GenBank and Bold. Bootstrap support values are based on 500 pseudoreplicates, and those above 80% are shown above supported nodes. Previously sequenced individuals have GenBank accession numbers or Barcode of Life identification numbers. El Salvador individuals are followed by an original collection number, followed by host plant of collection, and then collection site. Collection sites correspond to the six sites listed in Table 2. Collection sites followed by a number (i.e., rice 1.5) correspond to the individual bars in the AFLP structure analysis in Figure 5
4. Discussion
Diatraea larvae were collected from host plants and reared to adults to provide definitive evidence of their host plant association. D. lineolata was the predominant Diatraea species feeding on corn. Diatraea saccharalis was common on sugarcane in the rainy season and occasionally found on rice. Both D. saccharalis and D. lineolata were abundant on sorghum in the dry season, with nearly twice as many larvae of D. lineolata collected from sorghum as on were collected on corn. Diatraea lineolata is native to the Western Hemisphere and has had a much longer association with corn than with sorghum, which was introduced from Africa in the mid‐1800s (Dillon et al., 2007). The association of Diatraea spp. and sorghum in Central America is relatively recent on an evolutionary time scale, and little genetic variability would be expected in the mitochondrial DNA between individuals of D. lineolata collected on corn or sorghum. Most mitochondrial DNA sequences obtained from the D. lineolata reared in this study were nearly identical. In contrast, results from the AFLP markers for D. lineolata from corn and sorghum found the presence of two genetically distinct groups (Figure 3), which was not related to geographic distance among the populations. Individuals collected on corn represented one genetic group, while those collected from sorghum had two genetic groups present. The AFLP data suggest the populations of D. lineolata may be experiencing some degree of host plant‐associated differentiation.
Two populations of D. lineolata from La Union from corn and sorghum were collected several months apart in the same field, yet the sorghum field had an additional genotype present which was not collected in corn fields (Figure 3). The corn and sorghum populations from La Union had an F ST value of 0.082, higher than the genetic distance between other host‐associated populations of D. lineolata. D. lineolata populations on corn are synchronized with the host plant, which is typically grown in May through November. As plants senesce, D. lineolata pupae enter diapause in plant stems or roots, and adult moths emerge when the rainy season begins. Some D. lineolata larvae have asynchronous development and develop slower or faster than the rest of the population; asynchrony would normally be disadvantageous and have negative fitness consequences (van Asch & Visser, 2007; Mopper, 2005; Singer & Parmesan, 2010). For example, adult D. lineolata which emerge as corn plants senesce would oviposit on corn but larvae would not survive. The availability of a novel host plant such as sorghum when corn was no longer available would provide a potential host plant for D. lineolata oviposition, larval feeding and reproduction. The corn and sorghum populations of D. lineolata from La Union are separated in time as the two host plants grow in the rainy and dry season. Availability of a novel host plant such as sorghum at the start of the dry season could impose strong selection on moths to adapt to available host plant resources. Host plant shifts can occur in sympatry. For example, the pea aphid, Acyrthosiphon pisum Harris which was introduced into the USA about 100 years ago, shows host plant preferences for alfalfa and red clover when there is no spatial separation of the two host plant species (Via, 1999).
The mechanisms that may contribute to host‐associated differentiation and maintenance of the isolation of insects on two or more host plant have been explored in other systems. Insects feeding on a novel host plants can develop host fidelity with oviposition preference for the natal plant (Diehl & Bush 1984; Wood et al., 1999), and assortative mating could occur between individuals from the same host plant. Host plant fidelity could be tested for D. lineolata populations from corn and sorghum, to determine whether adult moths prefer to oviposit on or prefer the odor of their respective host plants. Rhagoletis pomonella host plant‐associated populations prefer the odors of their natal host plant and avoid odors of non‐natal host plants (Forbes, Fisher, & Feder, 2005). Hybrids of two Rhagoletis host plant‐associated populations did not prefer the host plant of either parent species; the authors suggest this could lead to inability to find suitable oviposition sites and failure to reproduce, contributing to further isolation of host plant‐associated populations (Linn et al., 2004). McBride and Singer (2010) similarly found that hybrids of host plant‐associated populations of Euphydryas editha had intermediate behavioral preferences, leading to reduced performance and reproductive isolation of parental populations. Other behavioral factors that contribute to the isolation of populations are whether or not they have distinct pheromone blends which contribute to assortative mating (Landolt & Phillips, 1997). For example, Ostrinia nubilalis has two host plant races in Europe with different pheromone blends, and there is a host plant preference for oviposition on the natal host as well as assortative mating of the two moth types (Bethenod et al., 2005). Pheromones were not investigated for the host plant‐associated populations of Diatraea lineolata or D. saccharalis in this study, but could be similarly investigated in future studies.
The second moth species, D. saccharalis, was found to have two genetically divergent groups through the use of both mitochondrial DNA and AFLP markers. Sugarcane, the primary host plant of D. saccharalis, is a perennial and grown year round through the wet and dry seasons. Sugarcane harvest occurs in the beginning of the dry season, but some plant portions along with roots remain in the soil where insects can diapause. Perennials have been suggested to favor insect specialization (Via, 1999). More genetic variability was found in the mitochondrial DNA sequences from D. saccharalis individuals than among those of D. lineolata. A recent study found evidence for several possible D. saccharalis species, but host plant‐associated strains have not been previously investigated (Joyce et al., 2014). D. saccharalis mitochondrial DNA sequences from El Salvador had two groups (a large and small clade) that were 1% divergent from each other (Figure 6, Table S1). Most of the sequences from El Salvador (20 of 23 individuals) comprised one large clade, which grouped with individuals previously sequenced from Mexico and the southern United States. The two clades with individuals from El Salvador are not specific to one particular host plant. The El Salvador D. saccharalis in the large clade were collected from sites (1–6) from rice, sugarcane, and sorghum. The smaller clade with individuals from El Salvador contained two individuals from sugarcane site 2, and one moth from site 3 on sorghum; no individuals in the small clade were associated with rice. Both clades with D. saccharalis individuals from El Salvador contained individuals of the two AFLP genotypes (Figures 5 and 6).
The AFLPs used in the Structure analysis found that all D. saccharalis from rice were one genotype, most larvae from sugarcane were one genotype, and collections from sorghum consisted of two genotypes (Figure 5). The AMOVA of the four host plant × site D. saccharalis populations found the sugarcane, and sorghum populations were significantly distant from each other, but this was not attributable to isolation by distance. The highest F ST values were between sugarcane and the two sorghum populations at San Andres and Santo Tomas (F ST 0.133, F ST 0.104) (Table 5), followed by an F ST of 0.095 between the two sorghum populations (site 3, 4). The AMOVA found that season accounted for 4% of genetic variation among populations (Table 4). The genetic divergence among populations of D. saccharalis from sugarcane and sorghum suggests some level of host‐associated differentiation. Host plant phenology may contribute to host‐associated differentiation of D. saccharalis populations on sugarcane and sorghum as was suggested for D. lineolata on corn and sorghum. The availability of sorghum as a novel host plant at the time of sugarcane harvest provides a host plant resource for D. saccharalis oviposition and development, similar to that discussed for D. lineolata.
Both Diatraea species had lower parasitism rates on sorghum than on the other dominant host plant for each moth (D. lineolata corn, D. saccharalis sugarcane). Sorghum could provide these Diatraea spp. a refuge from parasitism and therefore a fitness advantage for offspring. A previous study of Diatraea larvae on corn in El Salvador suggested that parasitoids may be synchronized with their host insects and similarly become dormant in the dry season (Quezada, 1978). Parasitoids in other systems are attracted to the odors associated with their host insects or odors associated with their host herbivores feeding on plants (Vet & Dicke, 1992). It is possible that the parasitoid flies (Tachinidae) attacking Diatraea spp. in El Salvador do not yet recognize volatiles emitted from sorghum because it is a novel plant, as this is the cases for some predators that have shifted host plants (Raffa, Powell, & Townsend, 2013).
Both D. lineolata and D. saccharalis have colonized novel crop plants, which have been introduced for cultivation. D. lineolata and D. saccharalis both appear to be experiencing some degree of host‐associated differentiation on sorghum, perhaps due to the differences in host plant phenology associated with the corn–sorghum and sugarcane–sorghum cropping cycles between the rainy and dry seasons. The low percent parasitism of each moth species on sorghum suggests that natural enemies have not yet followed their host herbivores onto this relatively new host plant resource. Further experiments could explore the preference and performance of host plant‐associated populations of these two Diatraea species, to uncover potential mechanisms, which might contribute to genetic divergence of host plant‐associated populations.
Conflict of Interest
None declared.
Data Accessibility
All mitochondrial DNA‐COI sequences have been submitted to GenBank to request accession numbers.
Genbank numbers of D. saccharalis in this study include the following: El_Salvador_7.9 KX976522, El_Salvador_7.23 KX976523, El_Salvador_10.6 KX976524, El_Salvador_11.5 KX976525, El_Salvador_11.18 KX976526, El_Salvador_11.47 KX976527, El_Salvador_13.22 KX976528, El_Salvador_13.23 KX976529, El_Salvador_13.25 KX976530, El_Salvador_13.30 KX976531, El_Salvador_13.46 KX976532, El_Salvador_14.5 KX976533, El_Salvador_14.12 KX976534, El_Salvador_16.7 KX976535, El_Salvador_17.76 KX976536, El_Salvador_18.5 KX976537, El_Salvador_19.9 KX976538, El_Salvador_19.14 KX976539, El_Salvador_19.31 KX976540, El_Salvador_20.2 KX976541, El_Salvador_20.9 KX976542, El_Salvador_23.3 KX976543, El_Salvador_24.1 KX976544
Genbank numbers of D. lineolata in this study include the following: DL_1.5 KX976545, DL_5.5 KX976546, DL_6.14 KX976547, DL_9.21 KX976548, DL_9.8 KX976549, DL_9.12 KX976550, DL_13.8 KX976551, DL_15.24 KX976552, DL_15.36 KX976553, DL_15.79 KX976554, DL_15.111 KX976555, DL_15.12 KX976556, DL_15.80 KX976557, DL_17.4 KX976558, DL_17.10 KX976559, DL_17.12 KX976560, DL_17.27 KX976561, DL_17.31 KX976562, DL_17.70 KX976563, DL_17.24 KX976564, DL_17.26 KX976565, DL_17.58 KX976566, DL_19.9 KX976567, DL_19.21 KX976568, DL_19.36 KX976569
Supporting information
Acknowledgments
The Fulbright Fellowship provided financial support to conduct field work in El Salvador. The University of El Salvador (UES) Agronomy faculty provided technical and financial support as well as laboratory and office space. The Centro Nacional de Tecnologίa Agropecuaria y Forestal (CENTA) in El Salvador provided technical support, especially Reina Flor Gúzman and Mario Parada Jaco. Many UES faculty collaborators helped find field sites and lead collecting efforts, including Dagoberto Pérez, Gustavo Henrίque Martίnez, and Rafael Menjίvar. Daniel Benίtez of Compañίa Azucarera Salvadoreña (CASSA) and Ever Quiñonez of Ingenio La Cabaña El Paisnal El Salvador provided technical assistance. Students at the University of El Salvador assisted with field work and rearing insects, including Saúl Ovidio González, Cindy Marίn Martίnez, Altagracia Zepeda Aguilar, Lissette Hernandez, Rosa Marίa Estrada, Rubén Lόpez Sorto, Georgina Herrera, Melany Murillo Torres, and Rolando Iraheta. Matthew Lewis, USDA, ARS, Systematic Entomology Laboratory, Beltsville, MD, helped with mitochondrial DNA, and University of California Merced undergraduate research assistants Etienne Melese and Ashley Valley Arevalo assisted with DNA extraction. D.M. (Monty) Wood of the Canadian National Collection of Insects confirmed the identity of Billea and Palpozenillia parasitoids. William White of USDA Sugarcane in Houma Louisiana read an earlier version of the manuscript.
Joyce, A. L. , Sermeno Chicas, M. , Serrano Cervantes, L. , Paniagua, M. , Scheffer, S. J. and Solis, M. A . (2016), Host‐plant associated genetic divergence of two Diatraea spp. (Lepidoptera: Crambidae) stemborers on novel crop plants. Ecology and Evolution, 6: 8632–8644. doi: 10.1002/ece3.2541
References
- Abrahamson, W. G. , Blair, C. P. , Eubanks, M. D. , & Morehead, S. A . (2003). Sequential radiation of unrelated organisms: The gall fly Eurosta solidaginis and the tumbling flower beetle Mordellistena convicta . Journal of Evolutionary Biology, 16, 781–789. [DOI] [PubMed] [Google Scholar]
- van Asch, M. , & Visser, M. E. (2007). Phenology of forest caterpillars and their host trees: The importance of synchrony. Annual Review of Entomology, 52, 37–55. [DOI] [PubMed] [Google Scholar]
- Bethenod, M. T. , Thomas, Y. , Rousset, F. , Frerot, B. , Pelozuelo, L. , Genestier, G. , & Bourguet, D . (2005). Genetic isolation between two sympatric host plant races of the European corn borer, Ostrinia nubilalis Hubner. II: Assortative mating and host‐plant preferences for oviposition. Heredity, 94, 264–270. [DOI] [PubMed] [Google Scholar]
- Bleszynski, S. (1969). The taxonomy of crambinae moth borers of sugarcane In Williams J. R., Metcalf J. R., Mungomery R. W., & Mathes R. (Eds.), Pests of sugarcane (pp. 11–59). New York, NY: Elsevier. [Google Scholar]
- Box, H. E. (1931). The crambine genera Diatraea and Xanthopherne (Lep. Pyralidae). Bulletin of Entomological Research, 22, 1–50. [Google Scholar]
- Box, H. E. (1951). New species and records of Diatraea Guild from northern Venezuela (Lepid:Pyral.). Bulletin of Entomological Research, 42, 379–398. [Google Scholar]
- CAB International (1989). Distribution maps of plant pests, Diatraea saccharalis. Series A Agricultural Map 5 (revised). London: CABI. [Google Scholar]
- Cherry, R. H. , & Nuessly, G. S. (1993). Insect management in sugarcane. Gainesville, FL: Univ Florida IFAS Extension. [Google Scholar]
- Craig, T. P. , Itami, J. K. , Abrahamson, W. G. , & Horner, J. D . (1993). Behavioral evidence for host‐race formation in Eurosta solidaginis . Evolution, 47, 1696–1710. [DOI] [PubMed] [Google Scholar]
- Dickey, A. M. , & Medina, R. F. (2010). Testing host‐associated differentiation in a quasi‐endophage and a parthenogen on native trees. Journal of Evolutionary Biology, 23, 945–956. [DOI] [PubMed] [Google Scholar]
- Diehl, S. R. , & Bush, G. L. (1984). An evolutionary and applied perspective to insect biotypes. Annual Review of Entomology, 29, 471–504. [Google Scholar]
- Dillon, S. , Shapter, F. M. , Henry, R. J. , Cordeiro, G. , Izquierdo, L. , & Lee, L. S. (2007). Domestication to crop improvement: Genetic resources for Sorghum and Saccharum (Andropogoneae). Annals of Botany, 100, 975–989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dres, M. , & Mallet, J. (2002). Host races in plant‐feeding insects and their importance in sympatric speciation. Philosophical transactions of the Royal Society of London. Series B, Biological Sciences, 357, 471–492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dyar, H. G. , & Heinrich, C. (1927). The American moths of the genus Diatraea and allies. Proceedings United States National Museum, 71, 1–48. [Google Scholar]
- Earl, D. A. , & VonHoldt, B. M. (2012). STRUCTURE HARVESTER: A website and program for visualizing STRUCTURE output and implementing the Evanno method. Conservation Genetics Resources, 4, 359–361. [Google Scholar]
- Evanno, G. , Regnaut, S. , & Goudet, J. (2005). Detecting the number of clusters of individuals using the software STRUCTURE: A simulation study. Molecular Ecology, 14, 2611–2620. [DOI] [PubMed] [Google Scholar]
- Feder, J. L. , & Forbes, A. A. (2010). Sequential speciation and the diversity of parasitic insects. Ecological Entomology, 35, 67–76. [Google Scholar]
- Feder, J. L. , Hunt, T. A. , & Bush, G. L. (1993). The effect of climate, host plant phenology, and host fidelity on the genetics of apple and hawthorn‐infesting races of Rhagoletis pomonella . Entomologia Experimentalis Et Applicata, 69, 117–135. [Google Scholar]
- Forbes, A. A. , Fisher, J. , & Feder, J. L. (2005). Habitat avoidance: Overlooking an important aspect of host specific mating and sympatric speciation? Evolution, 59, 1552–1559. [DOI] [PubMed] [Google Scholar]
- Forbes, A. A. , Powell, T. H. Q. , Stelinski, L. L. , Smith, J. J. , & Feder, J. L . (2009). Sequential sympatric speciation across trophic levels. Science, 323, 776–779. [DOI] [PubMed] [Google Scholar]
- Fuchs, T. W. , Huffman, F. R. , & Smith, J. W. (1979). Introduction and establishment of Apanteles flavipes (Hymenoptera: Braconidae) on Diatraea saccharalis (Lep: Pyralidae) in Texas. Entomophaga, 24, 109–114. [Google Scholar]
- Gifford, J. R. , & Mann, G. A. (1967). Biology, rearing and a trial release of Apanteles flavipes in the Florida everglades to control the sugarcane borer. Journal of Economic Entomology, 60, 44–47. [Google Scholar]
- Groot, A. , Marr, M. , Heckel, D. G. , & Schofl, G. (2010). The roles and interactions of reproductive isolation mechanisms in fall armyworm (Lepidoptera: Noctuidae) host strains. Ecological Entomology, 35, 105–118. [Google Scholar]
- Hajibabaei, M. , Janzen, D. H. , Burns, J. M. , Hallwachs, W. , & Hebert, P. D. N. (2006). DNA barcodes distinguish species of tropical Lepidoptera. Proceedings of the National Academy of Sciences of the United States of America, 103, 968–971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joyce, A. L. , Bernal, J. S. , Vinson, S. B. , Hunt, R. E. , Schulthess, F. , & Medina, R. F . (2010). Geographic variation in male courtship acoustics and reproductive isolation of populations of Cotesia sesamiae (Hymenoptera: Braconidae) and Cotesia flavipes . Entomologia Experimentalis Et Applicata, 137, 153–164. [Google Scholar]
- Joyce, A. L. , White, W. H. , Nuessly, G. S. , Solis, M. A. , Scheffer, S. J. , Lewis, M. L. , & Medina, R. F . (2014). Geographic population structure of the sugarcane borer, Diatraea saccharalis (F.) (Lepidoptera: Crambidae), in the Southern United States. PLoS ONE, 9(10), e110036. doi:10.1371/journal.pone.0110036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kearse, M. , Moir, R. , Wilson, A. , Stones‐Havas, S. , Cheung, M. , Sturrock, S. , … Drummond, A . (2012). Geneious Basic: An integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics, 28, 1647–1649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kevan, D. K. Mc. E. (1944). The Bionomics of the neotropical cornstalk borer, Diatraea lineolata, WLK. (Lepidoptera: Pyralidae) in Trinidad B.W.I. Bulletin of Entomological Research, 35, 23–30. [Google Scholar]
- Landolt, P. , & Phillips, T. (1997). Host plant influences on sex pheromone behavior of phytophagous insects. Annual Review of Entomology, 42, 371–391. [DOI] [PubMed] [Google Scholar]
- Lange, C. L. , Scott, K. D. , Graham, G. C. , Sallam, M. N. , & Allsopp, P. G. (2004). Sugarcane moth borrers (Lepidoptera: Noctuidae and Pyraloidea): Phylogenetics constructed using COII and 16S mitochondrial partial gene sequences. Bulletin of Entomological Research, 94, 457–464. [DOI] [PubMed] [Google Scholar]
- Linn, C. E. , Dambroski, H. R. , Feder, J. L. , Berlocher, S. , Nojima, S. , & Roelofs, W. L. (2004). Postzygotic isolating factor in sympatric speciation in Rhagoletis flies: Reduced response of hybrids to parental host‐fruit odors. Proceedings of the National Academy of Sciences of the United States of America, 101, 17753–17758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuoka, Y. , Vigouroux, Y. , Goodman, M. M. , Sanchez, G. J. , Buckler, E. , & Doebley, J . (2002). A single domestication for maize shown by multilocus microsatellite genotyping. Proceedings of the National Academy of Sciences of the United States of America, 99, 6080–6084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McBride, C. S. , & Singer, M. C. (2010). Field studies reveal strong postmating isolation between ecologically divergent butterfly populations. PLoS Biology, 8, e1000529. doi:10.1371/journal.pbio.1000529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medina, R. F. (2012). Implications of host‐associated differentiation in the control of pest species In: Barbosa P., Letourneau D. K. & Agrawal A. A. (Eds.), Insect outbreaks revisited (pp. 291–310). Hoboken: Wiley‐Blackwell. [Google Scholar]
- Milton, M. , Pierossi, P. , & Ratnasignham, S. (2013). Bar code of life datasystems handbook v.3.6. Guelph, Ontario, Canada: Bold Systems, Biodiversity Institute of Ontario; Boldsystems.org website. Retrieved from: http://www.boldsystems.org/index.php/resources Accessed 2015 November 10. [Google Scholar]
- Mopper, S. (2005). Phenology: How time creates spatial structure in endophagous insect populations. Annales Zoologici Fennici, 42, 327–333. [Google Scholar]
- Nosil, P. (2012). Ecological speciation. Oxford, UK: Oxford Press. 280 pp. [Google Scholar]
- Pashley, D. P. , Hardy, T. N. , Hammond, A. M. , & Mihm, J. A. (1990). Genetic evidence for sibling species within the sugarcane borer (Lepidoptera: Pyralidae). Annals of the Entomological Society of America, 83, 1048–1053. [Google Scholar]
- Peakall, R. , & Smouse, P. E. (2006). GENALEX 6: Genetic analysis in Excel. Population genetic software for teaching and research. Molecular Ecology Notes, 6, 288–295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peel, M. C. , Finlayson, B. L. , & McMahon, T. A. (2007). Updated world map of the Köppen‐Geiger climate classification. Hydrology and Earth System Sciences, 11, 1633–1644. [Google Scholar]
- Pritchard, J. K. , Stephens, M. , & Donnelly, P. (2000). Inference of population structure using multilocus genotype data. Genetics, 155, 945–950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pritchard, J. K. , Wen, X. , & Falush, D. (2007). Documentation for structure software: Version 2.2. Chicago, Ill: Univ. of Chicago. [Google Scholar]
- Qiagen (2006). DNeasy tissue handbook. Valencia, CA: Qiagen. [Google Scholar]
- Quezada, J. R. (1978). Poblaciones remanentes de Barrenadores en canas de maíz. XXIV Congreso, PCCMCA Programa Cooperativa Centro Americana para el mejoramiento de cultivos y animales, San Salvador, El Salvador. 10‐14 Julio, M23.1‐23.9.
- Raffa, K. F. , Powell, E. N. , & Townsend, P. A. (2013). Temperature‐driven range expansion of an irruptive insect heightened by weakly coevolved plant defenses. Proceedings of the National Academy of Sciences of the United States of America, 110, 2193–2198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson, G. S. (1976). The preparation of slides of Lepidoptera genitalia with special reference to the microlepidoptera. Entomologists Gazette, 27, 127–133. [Google Scholar]
- Saunders, J. , Mischke, S. , & Hemeida, A. A. (2001). The use of AFLP techniques for DNA fingerprinting in plants. A‐1910A. Beckman Coulter Application Notes. Fullerton, CA: Beckman Coulter; pp. 1–9. [Google Scholar]
- Scheffer, S. S. , & Hawthorne, D. J. (2007). Molecular evidence of host‐associated genetic divergence in the holly leafminer Phytomyza glabricola (Diptera: Agromyzidae): Apparent discordance among marker systems. Molecular Ecology, 16, 2627–2637. [DOI] [PubMed] [Google Scholar]
- Schluter, D. (2001). Ecological causes of speciation. Trends in Ecology & Evolution, 16, 372–380. [DOI] [PubMed] [Google Scholar]
- Serrano‐Cervantes, L. , Martinez, G. H. , Najera‐Montes, J. A. , Reyes, R. , & Sequeira, R. A . (1986). Determinacion de la ocurrencia de barrenadores, Diatraea (Lepidoptera: Pyralidae) y del nivel de control biológico nativo en El Salvador. XXXII Congreso, PCCMCA Programa Cooperativa Centro Americana para el mejoramiento de cultivos y animales, San Salvador, El Salvador. S 15.1‐10.
- Siemann, E. , Rogers, W. E. , & Dewalt, S. J. (2006). Rapid adaptation of insect herbivores to an invasive plant. Proceedings of the Royal Society of London B: Biological Sciences, 273, 2763–2769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singer, M. C. , & Parmesan, C. (2010). Phenological asynchrony between herbivorous insects and their hosts: Signal of climate change or pre‐existing adaptive strategy? Philosophical Transactions of the Royal Society B: Biological Science, 365, 3161–3176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solis, M. A. (2004). Systematics of Mexican stalkboring crambine Pyraloidea In: Rodrıguez del Bosque L. A., Vejar Cota G. & Cortez Mondaca E. (Eds.), Taller internacional sobre barrenadores del tallo de cana de azucar, Los Mochis, Sinaloa, Mexico (pp. 6–22). Mexico, D. F.: Sociedad Mexicana de Control Biologico. [Google Scholar]
- Solis, M. A. , & Metz, M. A. (2015). An illustrated guide to the identification of the known species of Diatraea Guilding (Lepidoptera: Crambidae: Crambinae). ZooKeys, 565, 73–121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stireman, J. O. III , Nason, J. D. , & Heard, S. B. (2005). Host‐associated genetic differentiation in phytophagous insects: General phenomenon or isolated exceptions? Evidence from a goldenrod‐insect community. Evolution, 59, 2573–2587. [PubMed] [Google Scholar]
- Sword, G. A. , Joern, A. , & Senior, L. B. (2005). Host plant‐associated genetic differentiation in the snakeweed grasshopper, Hesperotettix viridis (Orthoptera: Acrididae). Molecular Ecology, 14, 2197–2205. [DOI] [PubMed] [Google Scholar]
- Vargas, G. , Lastra, L. A. , & Solis, M. A. (2013). First record of Diatraea tabernella Lepidoptera: Crambidae) in the Cauca river Valley of Colombia. Florida Entomologist, 96, 1198–1201. [Google Scholar]
- Vet, L. E. M. , & Dicke, M. (1992). Ecology of infochemical use by natural enemies in a tritrophic context. Annual Review of Entomology, 37, 141–172. [Google Scholar]
- Via, S. (1999). Reproductive isolation between sympatric host races of pea aphids. I. Gene flow restriction and habitat choice. Evolution, 53, 1446–1457. [DOI] [PubMed] [Google Scholar]
- Vialatte, A. , Dedryver, C. A. , Simon, J. C. , Galman, M. , & Plantegenest, M . (2005). Limited genetic exchange between populations of an insect pest living on uncultivated and related cultivated host plants. Proceedings of the Royal Society of London B: Biological Sciences, 272, 1075–1082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vos, P. , Hogers, R. , Bleeker, M. , Reijans, M. , van de Lee, T. , Hornes, M. , … Zabeau, M . (1995). AFLP: A new technique for DNA fingerprinting. Nucleic Acids Research, 23, 4407–4414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White, W. H. , Miller, J. D. , Milligan, S. B. , Burner, D. M. , & Legendre, B. L . (2001). Inheritance of sugarcane borer resistance in sugar cane derived from two measures of insect damage. Crop Science, 41, 1706–1710. [Google Scholar]
- Wood, T. K. , Tilmon, K. J. , Shantz, A. B. , Harris, C. K. , & Pesek, J . (1999). The role of host‐plant fidelity in initiating insect race formation. Evolutionary Ecology Research, 1, 317–332. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All mitochondrial DNA‐COI sequences have been submitted to GenBank to request accession numbers.
Genbank numbers of D. saccharalis in this study include the following: El_Salvador_7.9 KX976522, El_Salvador_7.23 KX976523, El_Salvador_10.6 KX976524, El_Salvador_11.5 KX976525, El_Salvador_11.18 KX976526, El_Salvador_11.47 KX976527, El_Salvador_13.22 KX976528, El_Salvador_13.23 KX976529, El_Salvador_13.25 KX976530, El_Salvador_13.30 KX976531, El_Salvador_13.46 KX976532, El_Salvador_14.5 KX976533, El_Salvador_14.12 KX976534, El_Salvador_16.7 KX976535, El_Salvador_17.76 KX976536, El_Salvador_18.5 KX976537, El_Salvador_19.9 KX976538, El_Salvador_19.14 KX976539, El_Salvador_19.31 KX976540, El_Salvador_20.2 KX976541, El_Salvador_20.9 KX976542, El_Salvador_23.3 KX976543, El_Salvador_24.1 KX976544
Genbank numbers of D. lineolata in this study include the following: DL_1.5 KX976545, DL_5.5 KX976546, DL_6.14 KX976547, DL_9.21 KX976548, DL_9.8 KX976549, DL_9.12 KX976550, DL_13.8 KX976551, DL_15.24 KX976552, DL_15.36 KX976553, DL_15.79 KX976554, DL_15.111 KX976555, DL_15.12 KX976556, DL_15.80 KX976557, DL_17.4 KX976558, DL_17.10 KX976559, DL_17.12 KX976560, DL_17.27 KX976561, DL_17.31 KX976562, DL_17.70 KX976563, DL_17.24 KX976564, DL_17.26 KX976565, DL_17.58 KX976566, DL_19.9 KX976567, DL_19.21 KX976568, DL_19.36 KX976569


