Abstract
Glycosylation of certain proteins at the mammalian cell surface is an essential event in carcinogenesis. Sialylation, one type of glycosylation, can act on multiple cell-behaviors, such as migration, growth, and malignant invasion. Two polysialyltransferases, ST8Sia II (STX) and ST8Sia IV (PST), are responsible for synthesis of polysialic acid on neural cell adhesion molecule. We showed previously that STX and PST are oppositely expressed in normal murine mammary gland cells undergoing transforming growth factor-β-induced epithelial-mesenchymal transition. The molecular basis for regulation of STX and PST remained unclear. In the present study, we observed that transcription factor Pax3 upregulates STX expression, downregulates PST expression, and modulates upregulated expression of PSA, which attaches primarily to neural cell adhesion molecule to form PSA-NCAM. Overexpression of Pax3 in normal murine mammary gland cells transformed the expression of epithelial-mesenchymal transition markers E-cadherin and N-cadherin, and significantly promoted cell migration, but had no effect on cell proliferation.
Keywords: Pax3, polysialyltransferase, STX, PST, epithelial-mesenchymal transition, PSA-NCAM
Introduction
Glycans, important compositions of animal cell membranes, play pivotal roles in cell–cell interactions, cell–extracellular matrix interactions, and signal transduction. Glycans occur in various forms, and their changes are involved in pivotal pathophysiological events during tumor progression.1 Polysialic acid (PSA) exists in a linear homopolymer of α2,8-linked form and reduces the interaction of neural cell adhesion molecule (NCAM) based on its large negative charge,2 and thus plays crucial roles in neural cell migration, neurite outgrowth,3 and synaptogenesis.4 PSA expression shows temporal specificity, presenting as high expression during embryogenesis, and a barely detectable level during postnatal development.
PSA synthesis is usually catalyzed synergistically by two polysialyltransferases, STX, and PST,5 which are highly homologous (59% amino acid sequence similarity).6 Analysis of genomic structure and promoter activity of mouse STX and PST genes7–9 revealed their proximal promoter regions and putative binding sites for transcriptions factors. Microarray analysis showed that STX is one of the target genes of transcription factor Pax3.10 It was also reported that the transcription factor, AP-2delta, regulates STX expression in chick retina.11 Activation of cAMP-CREB cascade increases STX expression, but has no effect on PST expression.12. In postnatal mouse visual cortex, expression of STX and PST at the mRNA level is positively regulated by PKC-mediated signaling.13
We previously demonstrated significantly higher PSA expression in breast cancer (BC) tissue samples and malignant breast cells in comparison with normal breast tissue samples and nonmalignant cells.14 We found that STX and PST were differentially expressed in epithelial-mesenchymal transition (EMT) of normal murine mammary gland (NMuMG) cells induced by transforming growth factor-β (TGF-β).15
In the current study, we analyzed the promoter region of PST and found a potential binding motif of Pax3, which can also modulate STX expression.10 Our findings demonstrate dual roles of Pax3 in regulating opposite expression of STX and PST in NMuMG cells. Overexpression of Pax3 had a significant promoting effect on cell migration, but no effect on cell proliferation.
Materials and methods
Cells and cell culture
NMuMG cells and mouse mammary tumor 4T1 cells were cultured as described previously.14 For induction of EMT, NMuMG cells (∼30% confluence) were suffered with 2 ng/ml TGF-β1 (R&D Systems) for 48 h.
Antibodies and reagents
Mouse anti-E-cadherin IgG2a mAb and anti-NCAM IgG mAb were obtained from BD Biosciences, goat anti-Pax3 IgG polyclonal antibody (C-20) X and mouse anti-N-cadherin IgG1 mAb were purchased from Santa Cruz Biotechnology, mouse anti-β-tubulin IgG1, anti-vimentin IgG1 mAb, and anti-FN polyclonal antibody mAb were obtained from Sigma-Aldrich, and mouse anti-PSA-NCAM IgM mAb was acquired from Developmental Studies Hybridoma Bank. Secondary antibodies with HRP-conjugated were obtained from Beyotime.
Quantitative real-time PCR analysis
Total RNA was isolated with an RNApure Tissue kit (CoWin Biotech; Beijing, China). An RNA sample (A260/A280>1.8) was reversed transcribed to cDNA using ReverTra Ace-α- ® (Toyobo; Osaka, Japan). Specific primers used for various genes were as follows: forward 5′-GCTCGTGGTCTTCCTCATCT and reverse 5′-GCGGTG AAGAGCCATTTATT for STX; forward 5′-GCGAACTGCCTATCCATCAC and reverse 5′-CATGAGGAGACCTGTGCTAGG for PST; forward 5′-GGC GGATCTAGAAAGGAAGG and reverse 5′-CGGAGCCTTCATCTGACTG for Pax3; forward 5′-CAGTGCGGCAACCAGATAG and reverse 5′-GGAGTGGCACTGTCAACCTC for β-tubulin. Quantitative real-time PCR (qRT-PCR) was performed following the procedure described previously,16 and the data were analyzed by 2–ΔΔCt method.17
Plasmid construction
The Pax3 coding sequence was amplified with the forward primer 5′-CTAGCTAGCATGACCACGCTGGCCG and reverse primer 5′-GGAATT CCGGGCTCCAAGTGGACAGTT, then inserted into the vector pcDNA3.1(+) (Invitrogen; Carlsbad, CA, USA) at the XhoI and EcoRI sites. The 5′-flanking promoter region (−443/−162) of PST was cloned using forward primer 5′-GGGGTACCCTCACAACGACTCTCCGAGC and reverse primer 5′-CGGAATTCAGCTCTCCCGGTTCTCCAG, and constructed into plasmid TOPFlash at the KpnI and EcoRI sites. All plasmids were confirmed by DNA sequencing.
Transfection and RNA interference
NMuMG cells were stably transfected with the constructed plasmid pcDNA3.1-Pax3 and control vector using Lipofectamine 2000 (Invitrogen), selected with G418 (700 µg/ml), confirmed by Western blot analysis, and designated as NMuMG/Pax3 and NMuMG/Mock.
Three short interfering RNAs (siRNAs) for Pax3 and negative control dicer substrate duplex were synthesized by RiboBio Co. (Guangzhou, China). The sequences of validated siRNA for Pax3 were: sense 5′-GCCACAAGAUAGUGGAGAUdTdT, antisense 5′-AUCUCCACUAUCUUGUGGCdTdT.
Western blot analysis
Cells were harvested and lysed in the pre-cooled radioimmunoprecipitation assay (RIPA) buffer containing with 1% protease inhibitor cocktail (Biotool, Houston, TX, USA). Samples (30 µg protein) were separated by SDS-PAGE, transferred onto PVDF membrane, incubated with primary antibody as indicated and blotted with HRP-conjugated secondary antibody. Protein bands were analyzed by ChemiDoc XRS+ system (Bio-Rad).
Luciferase assay
NMuMG cells were transiently co-transfected with specific expression vectors for 48 h, then the cells were washed twice with PBS, and lysed in 100 µl lysis buffer. Luciferase reporter assay system (Promega; Madison, WI, USA) was applied to detect the fluorescence. Luminescence intensity was acquired by Synergy H4 Multi-Mode Microplate Reader (Bio Tek; Winooski, VT, USA). Relative luciferase activities were normalized relative to control. Results were expressed as mean ± SD from triplicate experiments.
Electrophoretic mobility shift assays
Nuclear extracts were separated using nuclear and cytoplasmic extraction kit (CoWin Biotech), and protein concentrations were measured using BCA Protein Quantitation Kit (CoWin Biotech). Electrophoretic mobility shift assays (EMSAs) were performed and analyzed as described previously.18 In this assay, single-stranded probes covering two putative binding sites were synthesized, annealed to double-stranded oligonucleotide probes, labeled with DIG, and designated as P-1: 5′-CCACCTCCAATGCACAAGG TGTCACATTTG. Mutation probes were designated as P-1-m: 5′-CCACCTCCAATGCAACCTTTGTCACATTTGAAAAG.
Immunofluorescence staining
Cell immunofluorescence staining was performed as described previously.19 In brief, cells were fixed with 4% fresh paraformaldehyde on glass coverslips and blocked with 3% BSA, incubated at 4℃ with primary antibody for 16 h, and then stained with secondary antibody. DAPI (Invitrogen; Paisley, UK) was applied to mark the cell nuclei. Laser confocal fluorescence microscopy (model Eclipse Ti-U; Nikon; Tokyo, Japan) was used to detect stained cells.
Wound healing assay
NMuMG/Pax3 and mock cells were seeded into 6-well plates at 5 × 105 cells per well. Pipette tips were used to scratch on the confluent monolayer to form linear wounds. Cells were washed with PBS and the images were captured at 0 h. After 24 h culture in medium without FBS, cells were washed with PBS and linear wounds were captured again.
Proliferation (MTT) assay
Cells (2 × 103/well) were plated on 96-well plates and after culture for different hours, 4 µL MTT solution (Cers, Yantai, China) was added to form formazan, then the reaction was stopped by addition of 100 µL DMSO, and absorbance at 595 nm was assessed immediately as previously described.16
Statistical analysis
Data were determined by one-way analysis of variance using the software program GraphPad Prism 5. Differences between means with P < 0.05 were considered statistically significant.
Results
Pax3 expression was upregulated during EMT
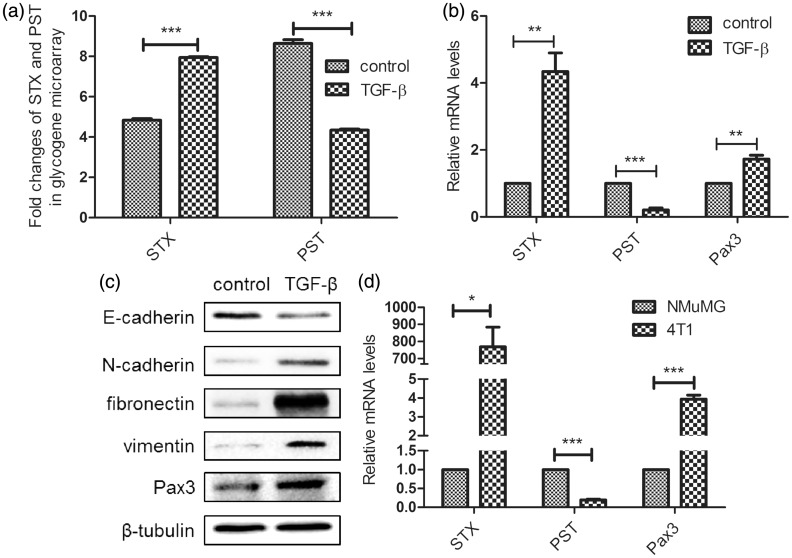
We demonstrated previously that two polysialyltransferases, STX and PST, show significant differential expression during TGF-β-induced EMT process in NMuMG cells, in contrast with the expression in non-treated cells15 (Figure 1(a)). This finding was confirmed by RT-PCR (Figure 1(b)). Bremer et al. showed that transcription factor Pax3, one member of a paired box (PAX), regulates expression of STX by binding to its promoter region.10 We observed significantly increased Pax3 expression in TGF-β-induced EMT in NMuMG cells (Figure 1(b) and (c)). Metastatic 4T1 cells, in comparison with untreated NMuMG cells, showed higher expression of STX and Pax3 and lower expression of PST (Figure 1(d)), similarly to NMuMG cells undergoing EMT.
Figure 1.
Pax3 is upregulated in NMuMG cells undergoing TGF-β-induced EMT. (a) Fold changes of STX and PST in control vs. treated cells were assessed by glycogene chip analysis (GlycoV4 GeneChip). (b) Expression of STX, PST, and Pax3 in control vs. treated cells was analyzed by qRT-PCR. (c) Western blot analysis of expression of EMT markers and Pax3 in cells undergoing EMT. Cells were cultured in 6-well plates, treated (or not) with TGF-β for 48 h, harvested, and lysed in RIPA buffer. Lysates (30 µg protein/well) were subjected to SDS-PAGE and analysis of E-cadherin, N-cadherin, fibronectin, vimentin, and Pax3 expression. (d) Expression of STX, PST, and Pax3 in NMuMG and 4T1 cells was analyzed by qRT-PCR, with β-tubulin as internal control. Data were analyzed by the 2−ΔΔCt method. *P < 0.05; **P < 0.01; ***P < 0.001; NS: not significant
Pax3 was responsible for both upregulation of STX and downregulation of PST in EMT
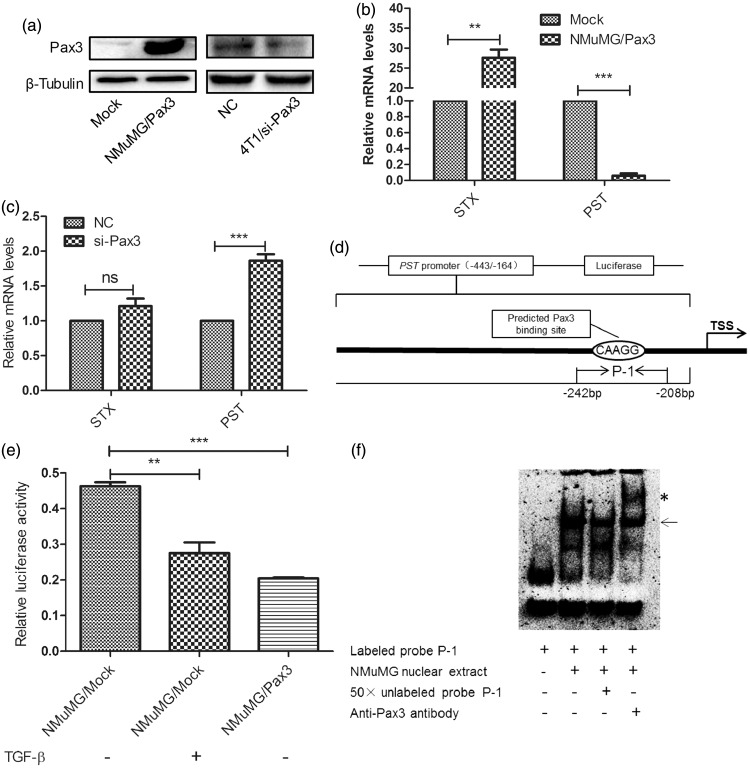
Bremer et al.20 showed that Pax3 caused specific increase of STX. To clarify whether alteration of STX expression was modulated by Pax3 in NMuMG cells undergoing TGF-β-induced EMT, we established stable Pax3-overexpressing cell line NMuMG/Pax3 (Figure 2(a)). Pax3 overexpression significantly increased STX expression, but reduced PST expression (Figure 2(b)). In 4T1 cells, transient silencing of Pax3 enhanced PST expression, but had no effect on STX expression (Figure 2(a) and (c)). Pax3 was previously shown to positively regulate STX expression through binding to a CAAGG sequence complementary to its paired domain sequence GTTCC.21,22 However, the effect of Pax3 on PST expression has not been studied. Our analysis of the PST gene promoter region revealed a CAAGG motif between positions −443 and −162 upstream of the transcriptional start point (TSS) (Figure 2(d)). We cloned this promoter region sequence into a luciferase reporter. Luciferase activity was reduced significantly in both TGF-β-treated and Pax3-overexpressing cells co-transfected with luciferase and Renilla expression vectors (Figure 2(e)). These findings suggest that Pax3 interacts with the PST promoter.
Figure 2.
Pax3 causes upregulation of STX and downregulation of PST. (a) Western blot analysis of Pax3 expression. Mock: pcDNA3.1(+). NC: scrambled sequence control. (b) and (c) Changes in STX and PST expression levels in NMuMG/Pax3 and 4T1/si-Pax3 cells, respectively, were analyzed by qRT-PCR, with β-tubulin as internal control. Data were analyzed by the 2−ΔΔCt method and presented as fold change. **P < 0.01; ***P < 0.001; NS: not significant. (d) Schematic summary of PST promoter sequence analysis, showing two predicted Pax3 binding sites. TSS: translation start site. (e) Luciferase activity analysis of PST promoter (−443/−162) for untreated NMuMG/pcDNA3.1(+) cells (lane 1), TGF-β-treated NMuMG/pcDNA3.1(+) cells (lane 2), and NMuMG/Pax3 cells (lane 3). Firefly luciferase activity was normalized to Renilla luciferase activity, and presented as mean ± SD from three independent experiments. (f) EMSA was performed using DIG-labeled probe P-1 and nuclear extracts from NMuMG cells. Arrow: DNA-protein complexes. Competitive assay was performed with 50-fold unlabeled probe P-1 (lane 3). Supershift analysis was performed by adding anti-Pax3 antibody (lane 4; *)
The binding capacity of Pax3 with PST promoter was evaluated using EMSAs. Treatment with probe P-1 resulted in appearance of a shifted band. Then, 50-fold unlabeled probes were added in competition assay to remove the non-specific binding, resulting in significant reduction of retarded bands. A supershifted band following addition of anti-Pax3 polyclonal antibody indicated specific binding by probe P-1 (Figure 2(f)). The shifted band was still present following addition of labeled mutation probe P-1-m (data not shown).
Results of luciferase assays and EMSAs, taken together, indicate that Pax3 binds to the PST promoter and thereby suppresses PST expression.
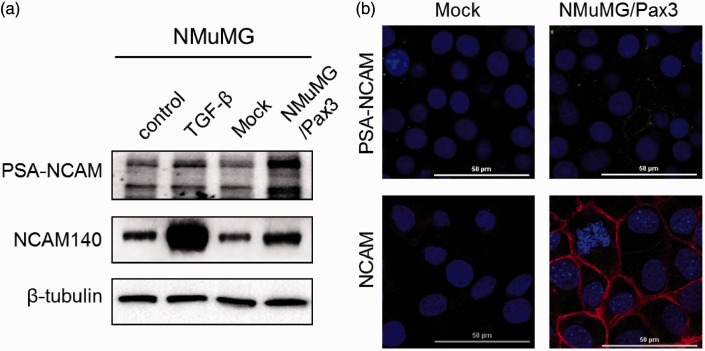
Effects of Pax3 on expression of PSA-NCAM and NCAM
Previously observation showed an enhanced PSA expression levels during TGF-β-induced EMT in both NMuMG and MCF10A cells.14 STX and PST are both involved in addition of PSA to NCAM, the major PSA substrate. We were interested in the effect of Pax3 on expression of PSA-NCAM. In comparison with control cells, PSA-NCAM expression was enhanced in NMuMG/Pax3 cells and in NMuMG cells undergoing EMT (Figure 3(a) and (b)). NCAM was moderately increased in NMuMG/Pax3 cells, and greatly increased in NMuMG cells undergoing EMT. These findings, taken together, suggest that Pax3 enhances NCAM expression and promotes NCAM sialylation.
Figure 3.
Effects of Pax3 on PSA-NCAM and NCAM expression. (a) Western blot analysis of PSA-NCAM and NCAM in NMuMG cells. (b) Immunofluorescence staining of PSA-NCAM and NCAM in NMuMG/Mock and NMuMG/Pax3 cells. Nuclei were visualized by DAPI staining. Bars: 50 µm. (A color version of this figure is available in the online journal.)
Effects of Pax3 on migration and proliferation of NMuMG cells
It has been revealed that Pax3 can contribute to regulation of migratory events in embryogenesis, particularly in malignant melanoma and neuroblastoma. We examined the impacts of Pax3 on migration and proliferation in NMuMG cells. Pax3 overexpression obviously receded expression of epithelial cell marker E-cadherin, meanwhile, enhanced expression of mesenchymal cell marker N-cadherin (Figure 4(a) and (b)), and significantly increased cell migration evaluated by wound healing assay (Figure 4(c) and (d)), but had no effect on cell proliferation (Figure 4(e)).
Figure 4.
Effects of Pax3 on migration and proliferation of NMuMG cells. (a) Western blot analysis of EMT markers. Cells were cultured in 6-well plates, harvested, and lysed in RIPA buffer. Lysates (30 µg protein per lane) were subjected to SDS-PAGE and analysis of E-cadherin, N-cadherin, fibronectin, and vimentin expression. (b) Immunofluorescence staining of E-cadherin and N-cadherin in NMuMG/Mock and NMuMG/Pax3 cells, and visualization of nuclei by DAPI staining. Bars: 50 µm. (c) Representative phase-contrast images showing increased migration of NMuMG/Pax3 cells, with NMuMG/Mock cells as control. After cells reached 100% confluence, scratches were made, regions were photographed (0 h), and the same regions were photographed at 24 h to assess the degree of wound healing. (d) Results of wound healing assay were analyzed using the Scion Image software program, and expressed as mean ± SD percentage area covered by moving cells at 24 h. All experiments were performed in triplicate. ***P < 0.001. (e) Proliferation (MTT) assays. NMuMG/Mock and NMuMG/Pax3 cells were cultured, and MTT assays were performed as described in Materials and Methods. Data are presented as mean ± SD from three independent experiments. (A color version of this figure is available in the online journal.)
Discussion
Synthesis of heavily sialylated glycans is an event very frequently associated with malignant transformation. The change of terminal sialic acid structures, in particular, can be used as a tumor biomarker.23 NCAM is the major substrate of PSA, and PSA-NCAM plays crucial function in neuronal differentiation and maturation.24 There is steadily increasing evidence that aberrantly re-expressed PSA-NCAM modulates cell adhesion, migration, and invasion in many kinds of tumor,25–27 and shows a strong relationship with severe clinical prognosis.28 The polysialyltransferases STX and PST catalyze synthesis of PSA on NCAM, either individually or synergistically. Results of the present study show that PST expression is significantly reduced but PSA-NCAM expression is increased in NMuMG cells undergoing TGF-β-induced EMT, implying that change of STX played a dominant role over PST, and upregulation of STX was primarily accounted for the increase of PSA-NCAM in this model.
Pax3 could be re-detected in some kinds of tumors, including melanoma, neuroblastoma, and rhabdomyosarcoma, and promotes tumor progression via multiple signaling pathways.29–31 Knockdown of Pax3 in neuroblastoma and melanoma cells strongly inhibited proliferation and migration.26 In the present study, Pax3 overexpression strikingly enhanced cell migration (Figure 4(c) and (d)), but had no effect on cell proliferation (Figure 4(e)). In a study by Bremer and his co-wokers,22 Pax3 bound a CAAGG motif on the sense strand of STX and promoted STX expression, in agreement with the promoting effect of Pax3 on STX expression in the present study (Figure 2(b)). The following observations indicate that PST is a downstream target gene of Pax3: (i) the promoter region of PST includes a possible Pax3 binding site; (ii) PST expression was strikingly reduced in Pax3-overexpressing NMuMG cells (Figure 2(b)), but increased when endogenous Pax3 was inhibited (Figure 2(c)); (iii) luciferase assays and EMSAs showed that Pax3 can interact with the PST promoter region and thereby decrease its expression (Figure 2(e) and (f)). Pax3 evidently plays dual roles (activation or suppression of gene expression) that are dependent on the cellular “context". Dual roles of transcription factors have been reported previously.32 For example, to remodel a tumor microenvironment, Snail is acetylated and then transactivates its target gene; however, Snail also can act as a repressor of E-cadherin and induces EMT.33 Pax3 can function as a repressor by binding to a target gene promoter through its paired domain directly, or through interaction with other protein.34,35 The present findings suggest a negative regulatory role of Pax3 in PST expression. Studies are in progress to elucidate the molecular mechanism whereby Pax3 interacts with the PST promoter.
The mRNA levels of STX and PST expressed differently in both tissue- and time-specific patterns. STX is the major polysialyltransferase during embryogenesis, and plays dominated role in PSA synthesis in cancer cells,28 whereas PST is mainly expressed in mouse or human adult brain.36,37 In our current study, mRNA level of STX was obviously higher than that of PST in NMuMG cells undergoing TGF-β-induced EMT, as a result of Pax3 upregulation.
In conclusion, our findings demonstrate that the transcription factor Pax3 enhances expression of PSA on NCAM in NMuMG cells by upregulating STX expression and downregulating PST expression.
Acknowledgments
This study was funded by the National Natural Science Foundation of China (No. 81672537), the Natural Science Foundation of Jiangsu Province, China (No.BK20161132, 20160173) and the Fundamental Research Funds for the Central Universities (No. JUSRP51619B, JUSRP116032). We thank Dr. S. Anderson for the English revision of the manuscript.
Authors’ contributions
FG and XL designed the experiments; DG and JG carried out the experiments and analyzed the data; DG, FG and XL wrote the paper.
Declaration of Conflicting Interests
The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
References
- 1.Li M, Song L, Qin X. Glycan changes: cancer metastasis and anti-cancer vaccines. J Biosci 2010; 35: 665–73. [DOI] [PubMed] [Google Scholar]
- 2.Troy FA. Polysialylation: from bacteria to brains. Glycobiology 1992; 2: 5–23. [DOI] [PubMed] [Google Scholar]
- 3.Doherty P, Cohen J, Walsh FS. Neurite outgrowth in response to transfected N-CAM changes during development and is modulated by polysialic acid. Neuron 1990; 5: 209–19. [DOI] [PubMed] [Google Scholar]
- 4.Muller D, Wang C, Skibo G, Toni N, Cremer H, Calaora V, Rougon G, Kiss JZ. PSA–NCAM is required for activity-induced synaptic plasticity. Neuron 1996; 17: 413–22. [DOI] [PubMed] [Google Scholar]
- 5.Ong E, Nakayama J, Angata K, Reyes L, Katsuyama T, Arai Y, Fukuda M. Developmental regulation of polysialic acid synthesis in mouse directed by two polysialyltransferases, PST and STX. Glycobiology 1998; 8: 415–24. [DOI] [PubMed] [Google Scholar]
- 6.Close BE, Wilkinson JM, Bohrer TJ, Goodwin CP, Broom LJ, Colley KJ. The polysialyltransferase ST8Sia II/STX: posttranslational processing and role of autopolysialylation in the polysialylation of neural cell adhesion molecule. Glycobiology 2001; 11: 997–1008. [DOI] [PubMed] [Google Scholar]
- 7.Yoshida Y, Kurosawa N, Kanematsu T, Kojima N, Tsuji S. Genomic structure and promoter activity of the mouse polysialic acid synthase gene (mST8Sia II). Brain-specific expression from a TATA-less GC-rich sequence. J Biol Chem 1996; 271: 30167–73. [DOI] [PubMed] [Google Scholar]
- 8.Takashima S, Yoshida Y, Kanematsu T, Kojima N, Tsuji S. Genomic structure and promoter activity of the mouse polysialic acid synthase (mST8Sia IV/PST) gene. J Biol Chem 1998; 273: 7675–83. [DOI] [PubMed] [Google Scholar]
- 9.Eckhardt M, Gerardy-Schahn R. Genomic organization of the murine polysialyltransferase gene ST8SiaIV (PST-1). Glycobiology 1998; 8: 1165–72. [DOI] [PubMed] [Google Scholar]
- 10.Mayanil CS, George D, Freilich L, Miljan EJ, Mania-Farnell B, McLone DG, Bremer EG. Microarray analysis detects novel Pax3 downstream target genes. J Biol Chem 2001; 276: 49299–309. [DOI] [PubMed] [Google Scholar]
- 11.Li X, Persad AR, Monckton EA, Godbout R. Transcription factor AP-2delta regulates the expression of polysialyltransferase ST8SIA2 in chick retina. FEBS Lett 2014; 588: 770–5. [DOI] [PubMed] [Google Scholar]
- 12.Nakagawa S, Kim JE, Lee R, Chen J, Fujioka T, Malberg J, Tsuji S, Duman RS. Localization of phosphorylated cAMP response element-binding protein in immature neurons of adult hippocampus. J Neurosci 2002; 22: 9868–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tell F, Bélanger MC, Di Cristo G. Sensory experience differentially modulates the mRNA expression of the polysialyltransferases ST8SiaII and ST8SiaIV in postnatal mouse visual cortex. PloS One 2011; 6: e24874–e24874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang X, Li X, Zeng YN, He F, Yang XM, Guan F. Enhanced expression of polysialic acid correlates with malignant phenotype in breast cancer cell lines and clinical tissue samples. Int J Mol Med 2015; 37: 197–206. [DOI] [PubMed] [Google Scholar]
- 15.Guan F, Schaffer L, Handa K, Hakomori S. Functional role of gangliotetraosylceramide in epithelial-to-mesenchymal transition process induced by hypoxia and by TGF-β. FASEB J 2010; 24: 4889–903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Guan F, Wang X, He F. Promotion of cell migration by neural cell adhesion molecule (NCAM) is enhanced by PSA in a polysialyltransferase-specific manner. PloS One 2015; 10: e0124237–e0124237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-DeltaDeltaC(T)) method. Methods 2001; 25: 402–8. [DOI] [PubMed] [Google Scholar]
- 18.Guo J, Song B, Li X, Hepsilon C, Yang G, Yang X, Guan F. Downregulation of gangliotetraosylceramide and beta1,3-galactosyltransferase-4 gene expression by Smads during transforming growth factor beta-induced epithelial-mesenchymal transition. Mol Med Rep 2015; 11: 2241–7. [DOI] [PubMed] [Google Scholar]
- 19.Yang G, Xu Z, Lu W, Li X, Sun C, Guo J, Xue P, Guan F. Quantitative analysis of differential proteome expression in bladder cancer vs. normal bladder cells using SILAC method. PloS One 2015; 10: e0134727–e0134727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mayanil CS, George D, Mania-Farnell B, Bremer CL, McLone DG, Bremer EG. Overexpression of murine Pax3 increases NCAM polysialylation in a human medulloblastoma cell line. J Biol Chem 2000; 275: 23259–66. [DOI] [PubMed] [Google Scholar]
- 21.Chalepakis G, Gruss P. Identification of DNA recognition sequences for the Pax3 paired domain. Gene 1995; 162: 267–70.. [DOI] [PubMed] [Google Scholar]
- 22.Mayanil CSK, George D, Freilich L, Miljan EJ, Mania-Farnell B, McLone DG, Bremer EG. Microarray analysis detects novel pax3 downstream target genes. J Biol Chem 2001; 276: 49299–309. [DOI] [PubMed] [Google Scholar]
- 23.Amon R, Reuven EM, Leviatan Ben-Arye S, Padler-Karavani V. Glycans in immune recognition and response. Carbohyd Res 2014; 389: 115–22. [DOI] [PubMed] [Google Scholar]
- 24.Bonfanti L, Theodosis DT. Polysialic acid and activity-dependent synapse remodeling. Cell Adhes Migrat 2009; 3: 43–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hromatka BS, Drake PM, Kapidzic M, Stolp H, Goldfien GA, Shih Ie M, Fisher SJ. Polysialic acid enhances the migration and invasion of human cytotrophoblasts. Glycobiology 2013; 23: 593–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tanaka F, Otake Y, Nakagawa T, Kawano Y, Miyahara R, Li M, Yanagihara K, Inui K, Oyanagi H, Yamada T, Nakayama J, Fujimoto I, Ikenaka K, Wada H. Prognostic significance of polysialic acid expression in resected non-small cell lung cancer. Cancer Res 2001; 61: 1666–70. [PubMed] [Google Scholar]
- 27.Fernandez-Briera A, Garcia-Parceiro I, Cuevas E, Gil-Martin E. Effect of human colorectal carcinogenesis on the neural cell adhesion molecule expression and polysialylation. Oncology 2010; 78: 196–204. [DOI] [PubMed] [Google Scholar]
- 28.Falconer RA, Errington RJ, Shnyder SD, Smith PJ, Patterson LH. Polysialyltransferase: a new target in metastatic cancer. Curr Cancer Drug Targ 2012; 12: 925–39. [DOI] [PubMed] [Google Scholar]
- 29.Fang WH, Ahmed M, Wang Q, Li HM, Kumar P, Kumar S. PAX3 promotes tumor progression via CD105 signaling. Microvasc Res 2013; 86: 42–3. [DOI] [PubMed] [Google Scholar]
- 30.Schmitt-Ney M, Camussi G. The PAX3-FOXO1 fusion protein present in rhabdomyosarcoma interferes with normal FOXO activity and the TGF-beta pathway. PloS One 2015; 10: e0121474.–e0121474.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mayanil CS, Pool A, Nakazaki H, Reddy AC, Mania-Farnell B, Yun B, George D, McLone DG, Bremer EG. Regulation of murine TGFbeta2 by Pax3 during early embryonic development. J Biol Chem 2006; 281: 24544–52. [DOI] [PubMed] [Google Scholar]
- 32.Valin A, Gill G. Regulation of the dual-function transcription factor Sp3 by SUMO. Biochem Soc Transac 2007; 35: 1393–6. [DOI] [PubMed] [Google Scholar]
- 33.Hsu DS, Wang HJ, Tai SK, Chou CH, Hsieh CH, Chiu PH, Chen NJ, Yang MH. Acetylation of snail modulates the cytokinome of cancer cells to enhance the recruitment of macrophages. Cancer Cell 2014; 26: 534–48. [DOI] [PubMed] [Google Scholar]
- 34.Liu Y, Zhu H, Liu M, Du J, Qian Y, Wang Y, Ding F, Gu X. Downregulation of Pax3 expression correlates with acquired GFAP expression during NSC differentiation towards astrocytes. FEBS Lett 2011; 585: 1014–20. [DOI] [PubMed] [Google Scholar]
- 35.Hsieh MJ, Yao YL, Lai IL, Yang WM. Transcriptional repression activity of PAX3 is modulated by competition between corepressor KAP1 and heterochromatin protein 1. Biochem Biophys Res Commun 2006; 349: 573–81. [DOI] [PubMed] [Google Scholar]
- 36.Oltmann-Norden I, Galuska SP, Hildebrandt H, Geyer R, Gerardy-Schahn R, Geyer H, Muhlenhoff M. Impact of the polysialyltransferases ST8SiaII and ST8SiaIV on polysialic acid synthesis during postnatal mouse brain development. J Biol Chem 2008; 283: 1463–71. [DOI] [PubMed] [Google Scholar]
- 37.Angata K, Nakayama J, Fredette B, Chong K, Ranscht B, Fukuda M. Human STX polysialyltransferase forms the embryonic form of the neural cell adhesion molecule. Tissue-specific expression, neurite outgrowth, and chromosomal localization in comparison with another polysialyltransferase, PST. J Biol Chem 1997; 272: 7182–90. [DOI] [PubMed] [Google Scholar]